Introduction
Cancer tissue develops through the proliferation,
migration, invasion and angiogenesis of cancer cells (1,2).
However, the interactions between cancer cells and the surrounding
stromal cells (cancer-stromal interaction) have also been
demonstrated to promote the progression of cancer (1,2).
Furthermore, cancer-associated fibroblasts (CAFs), which are the
main components of stromal cells, are attracting attention as novel
targets for anti-cancer treatment (3-5).
CAFs, which are different from normal fibroblasts (NFs), are
activated fibroblasts observed in the cancer stroma and have the
characteristics of myofibroblasts, including expression of α-smooth
muscle actin (α-SMA) (6). CAFs
can be isolated from various types of cancer, such as colorectal
cancer, gastric cancer, pancreatic cancer, prostate cancer, breast
cancer and lung cancer (7-12).
However, to the best of our knowledge, the mechanisms of CAF
development have not been clarified, and several types of cells
have been reported as the origin of CAFs (4). For example, conversion from NFs
(13), cancer cells that have
undergone the epithelial-mesenchymal transition (14,15), endothelial cells that have
undergone the endothelial-mesenchymal transition (16), bone marrow-derived mesenchymal
stem cells (17,18) and adipose tissue-derived stem
cells (19) are considered to be
potential origins of CAFs. However, the specific origins of CAFs
have not yet been clarified.
CAFs have been reported to affect tumor progression,
including proliferation, migration, invasion and angiogenesis
(3,5,7,14,20,21). Some of them have been demonstrated
to be mediated in part by the secretion of cell adhesion molecules,
growth factors and cytokines, such as IL-6 and vascular endothelial
growth factor-A (VEGFA) for angiogenesis (7), basic fibroblast growth factor (bFGF)
for proliferation (12), and
monocyte chemoattractant protein-1 (MCP-1) for migration (20). In addition, some reports have
revealed that CAFs are involved in drug resistance acquisition and
cancer apoptosis (9,22-24). Therefore, although these reports
(3,5,7,9,14,20-24) suggested that CAFs are involved in
tumor progression through multiple mechanisms, the details of these
mechanisms have not yet been elucidated.
Chitinase 3-like 1 (CHI3L1), a 40 kDa secreted
glycoprotein also known as YKL-40 (25,26), was originally considered to be
associated with inflammatory diseases, such as asthma, liver
fibrosis and arthritis (27-29). A number of reports have described
the involvement of CHI3L1 in cancer (11,30-35). For example, serum CHI3L1 levels
are associated with prognosis in patients with colorectal cancer
(30), and upregulation of CHI3L1
expression in tumors promotes tumor angiogenesis, proliferation,
migration, invasion and radiation resistance (31-33). Furthermore, CHI3L1 has been
reported to be associated with tumor-associated macrophages
(11,33,34). However, the mechanisms of CHI3L1
production and the mechanisms through which CHI3L1 induces tumor
progression have not been clarified. Although two reports have
described the association of CHI3L1 and CAFs, the mechanisms were
found to involve immune cells or exosomes rather than cytokine
secretion (11,35).
Therefore, the present study evaluated the
interactions between CAFs and colorectal cancer cells mediated by
CHI3L1 and cytokine secretion. The present findings clarified that
CHI3L1, which was mainly secreted from CAFs, acted on CAFs
themselves to increase the secretion of IL-8, which may promote
tumor angiogenesis in colorectal cancer. To the best of our
knowledge, this is the first report of the roles of CAFs in
angiogenesis promoted by CHI3L1 in the microenvironment of
colorectal cancer.
Materials and methods
Cell lines
The HT-29, HCT116 and DLD-1 human colorectal cancer
cell lines and the EA.hy926 human umbilical vein endothelial cell
line were purchased from American Type Culture Collection. These
cancer cell lines have been authenticated (no. KBN0811) using short
tandem repeat DNA analysis by the Japanese Collection of Research
Bioresources Cell Bank. HT-29, HCT116 and EA.hy926 cells were
cultured in DMEM (Sigma-Aldrich; Merck KGaA) supplemented with 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin solution (Gibco; Thermo Fisher Scientific,
Inc.). DLD-1 cells were cultured in RPMI-1640 medium
(Sigma-Aldrich; Merck KGaA) supplemented with 10% FBS and 1%
penicillin-streptomycin solution. All cells were incubated at 37°C
in an atmosphere containing 5% CO2.
Isolation and culture of human colon
fibroblasts
Human colon fibroblasts used in present study were
established from a specimen resected from a 70-year-old Japanese
man with advanced well-differentiated colon adenocarcinoma at the
Department of Gastroenterological Surgery, Nagoya City University
(Nagoya, Japan) in April 2020. In the selection of the case,
adenocarcinoma (grade of differentiation not considered) was used
as an inclusion criterion, and small tumor for which the stromal
components around the cancer could not be sufficiently sampled and
perforation from which cancer and stool contamination could be
detected were used as exclusion criteria. The technical procedure
was the same as that described in a previous report (7). After obtaining written informed
consent, tissues were retrieved from two separate areas:
Near-cancerous tissue and normal colon tissue (10 cm from the
cancer tissue). To avoid cancer and stool contamination, the
tissues were collected from the serosal side with care to not
penetrate the mucosa and to not cut into the cancer tissue. The
tissue was fragmented with scissors into cubes measuring ~2
mm3 and then incubated at 37°C for 2 h in DMEM
containing 1,000 PU/ml Dispase (Godo Shusei Co., Ltd.). The
fragments were cultured in DMEM containing 5% FBS and 1%
penicillin-streptomycin solution and incubated at 37°C in an
atmosphere containing 5% CO2. These fibroblasts were
used at passages 4-7.
Agents
Recombinant human CHI3L1 (cat. no. 2599-CH),
recombinant human IL-8 (cat. no. 208-IL) and recombinant human
VEGFA (cat. no. 293-VE) were purchased from R&D Systems, Inc.,
and reconstituted with PBS to a concentration of 100 µg/ml
and stored at −20°C. In the experiments, the recombinant CHI3L1,
IL-8 and VEGFA were diluted in DMEM or RPMI-1640 medium as
appropriate for each cell line to a final concentration of 100, 100
and 50 ng/ml, respectively, according to previous reports (11,31-33,36-39). Human CHI3L1 neutralizing antibody
(mAY; mouse monoclonal antibody; cat. no. MABC196) was purchased
from Sigma-Aldrich; Merck KGaA, and IgG control (cat. no. MAB002)
was purchased from R&D Systems, Inc. Both were reconstituted
with PBS to concentrations of 1 and 0.5 mg/ml, respectively, and
stored at -20°C. For experiments, mAY and IgG control were diluted
in medium and used at a concentration of 10 µg/ml. The
duration of action of the assays was based on previous reports
(7,11).
Immunostaining and immunofluorescence
staining
The primary antibodies against vimentin (V9; cat.
no. ab8069; Abcam), cytokeratin (AE1/AE3; cat. no. ab27988; Abcam),
CD90 (5E10; cat. no. 555593; BD Biosciences), α-SMA (1A4; cat. no.
ab7817; Abcam) and CHI3L1 (cat. no. ab77528; Abcam) were used for
immunostaining and immunofluorescence staining.
Immunohistochemistry was performed as previously
described (40). The resected
specimen was fixed with 10% formalin for 2 days at room
temperature. Formalin-fixed, paraffin-embedded and
4-µm-thick sections were deparaffinized with xylene and
hydrated with ethanol at 100% twice, 90, 80 and 70% for 5 min each.
After washing with running water, samples were soaked in 10 mM
citric acid buffer and boiled using a microwave for 10 min at
100°C. Subsequently, the slides were soaked in 100% methanol and
0.3% hydrogen peroxide mixed solution for 30 min to block
endogenous peroxidase activity, and blocked with 4% Block Ace
Powder (cat. no. UKB80; DS Pharma Biomedical Co., Ltd.) for 10 min
in a humidity box at room temperature. Afterwards, slides were
stained with the primary antibodies [anti-vimentin (dilution,
1:1,000), anti-cytokeratin (dilution, 1:100), anti-α-SMA (dilution,
1:500) and anti-CHI3L1 (dilution, 1:500)] overnight at 4°C.
Subsequently, the sections were incubated with undiluted
anti-mouse/rabbit EnVision+/HRP-labeled polymer (cat. no.
K4001/K4003; Dako; Agilent Technologies, Inc.), as the secondary
antibody, for 45 min at room temperature. The tissues were stained
with 3,3′-diaminobendizine substrate (cat. no. K3467; Dako; Agilent
Technologies, Inc.) for 10 min at room temperature and
counterstained with hematoxylin for 30 sec at room temperature. The
slides were observed and analyzed using the BZ-X710 fluorescence
microscope and BZ-X Analyzer software version 1.4.0.1 (both from
Keyence Corporation) at a magnification of ×200.
Immunostaining and immunofluorescence staining of
cultured cells were performed as previously described (7). For immunostaining, cells grown in
chamber slides were fixed with 4% paraformaldehyde buffer for 20
min at room temperature, treated with 0.1% Triton X for 3 min and
blocked with 3% BSA (FUJIFILM Wako Pure Chemical Corporation)/PBS
for 1 h at room temperature. Then, the primary antibodies
[anti-vimentin (dilution, 1:80), anti-cytokeratin (dilution, 1:80),
anti-CD90 (dilution, 1:100)] were applied for 1 h at room
temperature. Subsequently, anti-mouse EnVision+/HRP-labeled polymer
was applied undiluted as the secondary antibody for 1 h at room
temperature. The cells were stained with anti-mouse
3,3′-diaminobendizine substrate (Dako; Agilent Technologies, Inc.)
for 15 min at room temperature and counterstained with hematoxylin
for 30 sec at room temperature. The slides were observed and
analyzed using the BZ-X710 fluorescence microscope and BZ-X
Analyzer software version 1.4.0.1 at a magnification of ×40.
For immunofluorescence staining, cells grown in
chamber slides were fixed with 4% paraformaldehyde buffer for 20
min at room temperature, treated with 0.1% TritonX for 3 min and
blocked with 3% BSA/PBS for 1 h at room temperature. Then,
anti-α-SMA mouse monoclonal antibody (dilution, 1:150) was applied
as the primary antibody for 2 h at room temperature. Subsequently,
goat anti-mouse IgG H&L (Alexa Fluor 488; dilution, 1:200; cat.
no. ab150113; Abcam) was applied as the secondary antibody for 30
min at room temperature. After washing with PBS, samples were
mounted with ProLong Gold Antifade Reagent with
4′,6-diamidino-2-phenylindole (Invitrogen; Thermo Fisher
Scientific, Inc.). The slides were observed and analyzed using the
BZ-X710 fluorescence microscope and BZ-X Analyzer software version
1.4.0.1 at a magnification of ×200.
Cytokine antibody array
Fibroblasts were seeded in 6-well plates with DMEM
containing 2% FBS (5×104 cells/well). After overnight
incubation, the medium was exchanged (3 ml/well). After 24 h, the
culture supernatants were collected and centrifuged at 400 × g for
5 min at 4°C to remove particulates, and 500 µl of the
supernatant was used for each array. A Proteome Profiler Human XL
Cytokine Array kit (cat. no. ARY022B; R&D Systems, Inc.) was
used according to the manufacturer's protocol. Images were captured
using an LAS-3000 instrument (FUJIFILM Corporation) with each
signal normalized to the positive controls.
RNA interference
CHI3L1 small interfering RNA (siRNA) (s3000),
interleukin-13 receptor α2 (IL-13Rα2) siRNA (s7376), VEGFA siRNA
(s462) and non-targeting negative control siRNA (Silencer Select
Negative Control No. 1; cat. no. 4390843) were pre-designed siRNAs
purchased from Invitrogen; Thermo Fisher Scientific, Inc. IL-8
siRNA (cat. no. sc-39631) was also pre-designed siRNA purchased
from Santa Cruz Biotechnology, Inc. CAFs were seeded at
1×105 cells/well in 6-well plates for RT-qPCR,
2.5×104 cells/well in 24-well plates for ELISA and the
angiogenesis assay, and ~80% confluent in 10-cm dishes for western
blotting the day before siRNA transfection. According to the
manufacturer's instructions, siRNAs and Lipofectamine RNAiMAX
(Invitrogen; Thermo Fisher Scientific, Inc.) were mixed with
Opti-MEM (Invitrogen; Thermo Fisher Scientific, Inc.) and incubated
for 5 min at room temperature. The siRNA-lipid complex was diluted
into DMEM to achieve a final siRNA concentration of 10 nM. Cells
were incubated for 24 h in a 5% CO2 incubator at 37°C.
For RT-qPCR and western blotting, cell pellets were collected
immediately after 24-h transfection. For ELISA and the angiogenesis
assay, the medium was exchanged (1 ml/well) after 24 h of
transfection, and after an additional 48 h, culture supernatants
were collected.
Reverse transcription-quantitative PCR
(RT-qPCR)
Fibroblasts and colorectal cancer cell lines (HT-29,
HCT116 and DLD-1) were seeded in 6-well plates with DMEM or
RPMI-1640 medium as appropriate for each cell line containing 5%
FBS (1×105 cells/well). After overnight incubation, the
medium was exchanged and the cells were exposed to the medium with
or without 100 ng/ml CHI3L1. After 24 h, cell pellets were
harvested, and total RNA was extracted from each sample using a
QIAcube and RNeasy Plus Mini Kit (Qiagen GmbH) according to the
manufacturer's protocol. The RNA was reverse transcribed using
SuperScript III First-Strand Synthesis SuperMix (Invitrogen; Thermo
Fisher Scientific, Inc.) and a T100 Thermal Cycler (Bio-Rad
Laboratories, Inc.). The temperature protocol of reverse
transcription was as follows: 25°C for 10 min, 50°C for 30 min and
85°C for 5 min. RT-qPCR was performed using TaqMan Gene Expression
Assays (cat. no. 4331182; Applied Biosystems; Thermo Fisher
Scientific, Inc.) and pre-designed primers for CHI3L1
(Hs01072228_m1), IL-13Rα2 (Hs00152924_m1), IL-8
(Hs00174103_m1), VEGFA (Hs00900055_m1) and MCP-1
(Hs00234140_m1) using the CFX Connect Real-Time System (Bio-Rad
Laboratories, Inc.). The thermocycling conditions were as follows:
Initial denaturation at 95°C for 20 sec, followed by 60 cycles at
95°C for 1 sec and 60°C for 20 sec. GAPDH (Hs99999905_m1)
was used as a loading control to normalize mRNA levels and each
sample was quantified using the standard curve method (41).
ELISA
Fibroblasts and colorectal cancer cell lines (HT-29,
HCT116 and DLD-1) were seeded in 24-well plates with DMEM or
RPMI-1640 medium as appropriate for each cell line containing 5%
FBS (2.5×104 cells/well). After overnight incubation,
the medium was exchanged (1 ml/well), and the cells were exposed to
the medium with or without 100 ng/ml CHI3L1. After 48 h, culture
supernatants were collected and centrifuged at 400 × g for 5 min at
4°C to remove particulates. For siRNA transfection, the medium was
exchanged after 24 h of transfection, and culture supernatants were
collected after an additional 48 h. Assays were performed using the
Human Chitinase 3-like 1 Quantikine ELISA Kit (cat. no. DC3L10;
R&D Systems, Inc.), Human IL-8-CXCL8 Quantikine ELISA Kit (cat.
no. D8000C; R&D Systems, Inc.) and Human VEGF Quantikine ELISA
Kit (cat. no. DVE00; R&D Systems, Inc.), and the concentration
of each protein was measured using a SpectraMax ABS microplate
reader (Molecular Devices, LLC) according to the manufacturer's
protocol.
Western blotting
The primary antibodies used for western blotting
were as follows: Anti-CHI3L1 (dilution, 1:1,000; cat. no. ab77528;
Abcam), anti-IL-13Rα2 (dilution, 1:1,000; EPR22978-163; cat. no.
ab260044; Abcam), and anti-GAPDH (dilution, 1:2,000; cat. no.
SC-47724; Santa Cruz Biotechnology, Inc.). GAPDH was used as a
loading control to normalize protein levels. The secondary
antibodies were polyclonal goat anti-rabbit immunoglobulins
HRP-conjugated (dilution, 1:2,000; cat. no. P0448; Dako; Agilent
Technologies, Inc.) and polyclonal goat anti-mouse immunoglobulins
HRP-conjugated (dilution, 1:2,000; cat. no. P0447; Dako; Agilent
Technologies, Inc.).
Fibroblasts and colorectal cancer cell lines (HT-29,
HCT116 and DLD-1) were seeded at 2×105 cells in 10-cm
dishes with DMEM or RPMI-1640 medium as appropriate for each cell
line containing 5% FBS and incubated in a CO2 incubator
at 37°C until cell growth was semi-confluent. Proteins from cell
pellets were collected using radioimmunoprecipitation lysis buffer
with Protease Inhibitor Single Use Cocktail and Phosphatase
Inhibitor Cocktail (all from Thermo Fisher Scientific, Inc.) as
previously described (40), and
the concentrations of total protein from each sample were measured
using a Pierce BCA protein assay kit (Thermo Fisher Scientific,
Inc.). Proteins were suspended in SDS sample buffer (90 mM
Tris-HCL, 9% glycerol, 3% SDS, 150 mM DTT, 0.003% bromophenol blue,
distilled water) and boiled at 90°C for 5 min for denaturation.
Proteins (20 µg/lane) were then separated using SDS-PAGE
with 10% Mini-PROTEAN TGX Precast Gels (Bio-Rad Laboratories, Inc.)
and transferred onto nitrocellulose membranes (Bio-Rad
Laboratories, Inc.). The protein bands on the membrane were blocked
with iBind Flex Solution (iBind Flex 5X Buffer, 100X Additive and
distilled water; Invitrogen; Thermo Fisher Scientific, Inc.) at
room temperature for 10 min. The primary and secondary antibody
reactions were then performed using the iBind Flex Western Device
(Invitrogen; Thermo Fisher Scientific, Inc.) for 2.5 h at room
temperature according to the manufacturer's protocol.
Protein-antibody complex bands were visible on an Amersham Imager
600 (GE Healthcare) using SuperSignal West ECL/Pico/Femto
chemiluminescent substrate (Thermo Fisher Scientific, Inc.). The
results were semi-quantified by densitometry analysis using ImageJ
software version 1.53 (National Institutes of Health).
Angiogenesis assay
To evaluate the direct effects of CHI3L1, IL-8 and
VEGFA on vascular endothelial cells, serum-free DMEM containing
recombinant CHI3L1, IL-8 or VEGFA was used. The control medium for
the addition of recombinant CHI3L1, IL-8 or VEGFA was serum-free
DMEM. In addition, to confirm whether secreted CHI3L1 protein
produced by CAFs affected angiogenesis, culture supernatants of
each cell (NFs, CAFs and HT-29) were used. The control medium for
the culture supernatant was DMEM containing 5% FBS. Culture
supernatants were collected as follows: NFs, CAFs and HT-29 were
seeded in 24-well plates with medium containing 5% FBS
(2.5×104 cells/well). After overnight incubation, the
medium was exchanged (1 ml/well), and the cells were exposed to the
medium with or without mAY or control IgG. After 48 h, culture
supernatants were collected and centrifuged at 400 × g for 5 min at
4°C to remove particulates. In the case of transfection in CAFs,
after overnight incubation, and 24-h transfection with negative
control, CHI3L1, IL-8 or VEGFA siRNA, the medium was exchanged (1
ml/well). After an additional 48 h, culture supernatants were
collected.
Matrigel Matrix (cat. no. 354230; Corning, Inc.) was
added to 96-well plates (50 µl/well) at 4°C. The plates were
incubated at 37°C for 30 min to solidify the Matrigel. EA.hy926
cells were trypsinized, counted, resuspended in each medium under
the various aforementioned conditions and seeded into
Matrigel-coated plates (1×104 cells/well). After 16 h of
incubation at 37°C, the plates were observed at a magnification of
×40 using the BZ-X710 fluorescence microscope, and the number of
endotubes was counted in four fields for each condition.
Cell proliferation assay
Colorectal cancer cell lines (HT-29, HCT116 and
DLD-1) were seeded in 96-well plates with DMEM or RPMI-1640 medium
as appropriate for each cell line containing 5% FBS
(1×104 cells/well). After overnight incubation, the
medium was exchanged and the cells were exposed to the medium with
various concentrations (0-1,000 ng/ml) of CHI3L1 and incubated at
37°C. After 0, 24 and 48 h, medium was exchanged with fresh medium
containing premix
4-[3-(4-Iodophenyl)-2-(4-nitro-phenyl)-2H-5-tetrazolio]-1,3-benzene
sulfonate (WST-1) using a Premix WST-1 Cell Proliferation Assay
System (Takara Bio, Inc.) and incubated for 2 h at 37°C in the
shade. The absorbance was measured at 450 nm using a SpectraMax ABS
microplate reader (Molecular Devices, LLC) according to the
manufacturer's protocol. The absorbance at each timepoint was
compared with the absorbance at 0 h as a standard value for each
cancer cell line. Cells without CHI3L1 treatment were used as
controls.
Wound healing assay
Colorectal cancer cell lines (HT-29, HCT116 and
DLD-1) were cultured in 24-well plates with DMEM or RPMI-1640
medium as appropriate for each cell line containing 10% FBS until
confluency. After the wounds were generated by carefully scratching
the cells with 200-µl pipette tips, the medium was exchanged
with fresh medium containing 2% FBS and the cells were exposed to
the medium with or without 100 ng/ml CHI3L1 at 37°C. After 0 and 24
h, the plates were observed at a magnification of ×40 using the
BZ-X710 fluorescence microscope. The widths of wounds were measured
using the BZ-X Analyzer software version 1.4.0.1 and compared with
that at 0 h as a standard value for each cancer cell line. Cells
without CHI3L1 treatment were used as controls.
Statistical analysis
Statistical analysis was performed using EZR
software (Easy R) version 1.41 (Jichi Medical University Saitama
Medical Center, Saitama, Japan). Data are presented as the mean and
standard error. Statistical significance was evaluated with
unpaired Student's t-tests or one-way ANOVA followed by Tukey's
test. All experiments were repeated at least three times. P<0.05
was considered to indicate a statistically significant
difference.
Results
Characterization of isolated and cultured
CAFs and NFs
Immunohistochemical analysis of colon adenocarcinoma
tissue demonstrated that cancer cells were negative for vimentin,
which is a mesenchymal cell marker, and positive for cytokeratin,
which is an epithelial cell marker (5,17,18,42). On the other hand, stromal cells
were positive for vimentin and negative for cytokeratin (Fig. 1A). Furthermore, to confirm that
the isolated and cultured cells were fibroblasts, immunostaining
for vimentin, CD90 and cytokeratin was performed (5,17,18,42,43). Both cells from near-cancerous
colon tissues (CAFs) and those from normal colon tissues (NFs) were
positive for vimentin and CD90 and negative for cytokeratin,
indicating that all of these cells were fibroblasts (Fig. 1B). In addition, to confirm the
myofibroblast characteristics of CAFs, which express α-SMA,
immunohistochemical and cell immunofluorescence staining of α-SMA
was also performed (6). In the
immunohistochemical analysis, stromal cells were positive for α-SMA
and cancer cells were negative for α-SMA (Fig. 1C). Immunofluorescence staining
indicated that all fibroblasts were positive for α-SMA; however,
α-SMA expression was stronger in fibroblasts from near-cancerous
tissues (CAFs) and weaker in fibroblasts from normal tissues (NFs)
(Fig. 1D). Therefore, fibroblasts
isolated from near-cancerous tissues were referred to as CAFs, and
fibroblasts isolated from normal tissues were referred to as
NFs.
 | Figure 1Localization of fibroblasts in cancer
tissues and isolated fibroblasts. (A) In cancer tissue from a
patient, immunohistochemical staining of vimentin was positive in
the stroma (indicated by S) and negative in cancer cells (indicated
by C). On the other hand, immunohistochemical staining of
cytokeratin was negative in the stroma and positive in cancer cells
(magnification, ×200). (B) All cultivated cells from the same
patient were positive for vimentin and CD90 and negative for
cytokeratin (magnification, ×40). (C) In cancer tissue from the
same patient, the stroma (indicated by S) was positive and cancer
cells (indicated by C) were negative for α-SMA immunohistochemical
staining (magnification, ×200). (D) Fibroblasts from near-cancerous
tissues of the same patient (CAFs) were positive for α-SMA and
fibroblasts from normal tissues of the same patient (NFs) were
weakly positive for α-SMA (magnification, ×200). α-SMA, α-smooth
muscle actin; CAFs, cancer-associated fibroblasts; NFs, normal
fibroblasts. |
Secretion of CHI3L1 from CAFs, NFs and
cancer cell lines
First, analysis of cytokines secreted from each cell
line using a cytokine array of cell culture supernatants
demonstrated that CAFs secreted more CHI3L1 and IL-8 than NFs
(Fig. 2A). Subsequently, to
investigate the origin of CHI3L1 in the cancer microenvironment,
immunohistochemistry, RT-qPCR, western blotting and ELISA were
performed to evaluate CHI3L1 expression in, and secretion from,
fibroblasts and cancer cells or three colorectal cancer cell lines.
Immunohistochemistry demonstrated that CHI3L1 was weakly expressed
in cancer cells and strongly expressed in portions of the stroma
(Fig. 2B). RT-qPCR revealed that
CHI3L1 mRNA expression was >4 times higher in CAFs than
in NFs but was less than half of that in CAFs in DLD-1 cells; HT-29
and HCT116 cells rarely expressed CHI3L1 (Fig. 2C). Furthermore, western blotting
of CHI3L1 demonstrated that CAFs expressed higher levels of CHI3L1
compared with NFs and colorectal cancer cell lines (Fig. 2D). ELISA using the culture
supernatants yielded similar results to those of RT-qPCR and
western blotting. CAFs secreted higher levels of CHI3L1, whereas
NFs and DLD-1 cells secreted lower levels of CHI3L1, and HT-29 and
HCT116 cells secreted little CHI3L1 (Fig. 2E). These results indicated that
CAFs were the main origin of CHI3L1 secretion in the colorectal
cancer microenvironment.
CAFs express the CHI3L1 receptor
IL-13Rα2
A number of CHI3L1 receptors have not yet been
identified; however, IL-13Rα2 has previously been reported as a
CHI3L1 receptor (44-47). Therefore, RT-qPCR and western
blotting were performed to evaluate IL-13Rα2 expression in
fibroblasts and colorectal cancer cell lines. RT-qPCR demonstrated
that both CAFs and NFs expressed IL-13Rα2. In particular,
expression in CAFs was 10 times higher than that in NFs, whereas
expression was rarely observed in the three cancer cell lines
(Fig. 3A). The results of western
blotting were similar to those of RT-qPCR (Fig. 3B).
Effects of CHI3L1 on IL-8 and VEGFA
levels in CAFs
IL-8 and VEGFA are angiogenic factors and it is
possible that CHI3L1 is involved in their expression and secretion
(31,33,38,39). Therefore, RT-qPCR and ELISA were
conducted to evaluate the effect of CHI3L1 on IL-8 and VEGFA
levels. First, the effect of various concentrations of CHI3L1
(1-1,000 ng/ml) on IL-8/VEGFA expression was assessed by
RT-qPCR using CAFs without CHI3L1 treatment as a control (Fig. S1). IL-8 mRNA expression in
CAFs was markedly enhanced after treatment with CHI3L1 at
concentrations ≥10 ng/ml, while VEGFA mRNA expression in
CAFs was not significantly enhanced except at concentrations of
1,000 ng/ml. Based on these results and existing reports (11,32,33,38,39), CHI3L1 at a concentration of 100
ng/ml was used in subsequent experiments. Subsequently, after 100
ng/ml CHI3L1 treatment, the IL-8 and VEGFA mRNA
levels in fibroblasts and colorectal cancer cell lines were
measured by RT-qPCR at 24 h and the secreted protein levels in the
cell culture supernatants were measured by ELISA at 48 h.
RT-qPCR revealed that IL-8 expression in CAFs
was ~16 times higher than that in NFs, and was also markedly higher
than that in the three cancer cell lines (Fig. 4A). Furthermore, treatment with
CHI3L1 increased IL-8 expression in CAFs. ELISA also
demonstrated that CAFs secreted more IL-8 than NFs and cancer cell
lines, and the addition of CHI3L1 to CAFs further increased IL-8
secretion from CAFs (Fig.
4B).
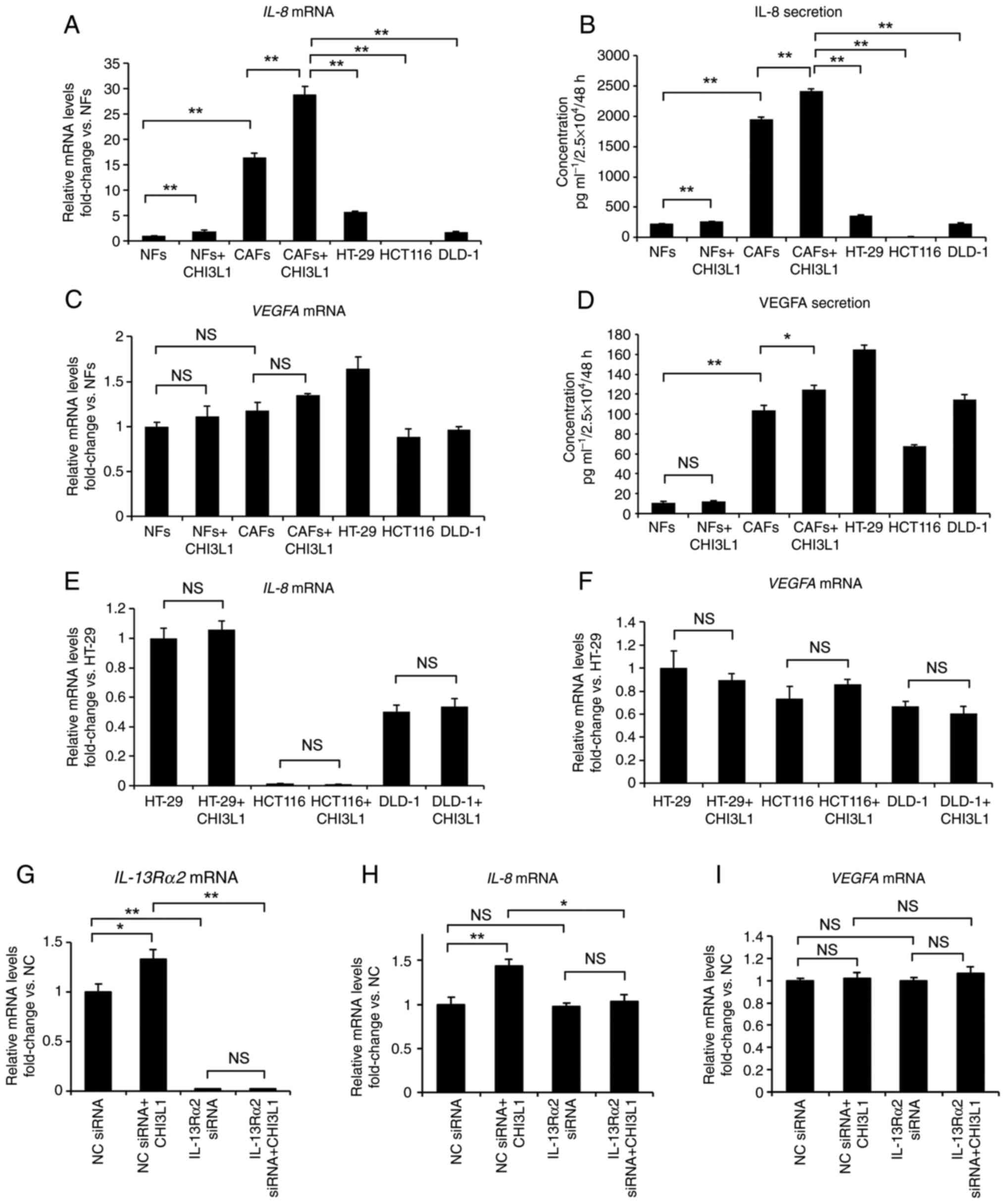 | Figure 4Changes in IL-8 and VEGFA following
the addition of 100 ng/ml CHI3L1. (A) RT-qPCR indicated that IL-8
mRNA expression was higher in CAFs than in NFs and cancer cells and
that 24-h treatment with 100 ng/ml CHI3L1 increased IL-8 expression
in CAFs. (B) ELISA of the cell culture supernatants revealed that
CAFs secreted more CHI3L1 than NFs and cancer cells, and that 48-h
treatment with 100 ng/ml CHI3L1 further increased IL-8 secretion
from CAFs. (C) RT-qPCR demonstrated that VEGFA mRNA
expression was slightly stronger in CAFs than in NFs, although the
difference was not significant. Treatment of CAFs with CHI3L1 did
not affect VEGFA expression. (D) In contrast to RT-qPCR, ELISA of
the cell culture supernatants demonstrated significant differences
in VEGFA secretion between untreated CAFs and CAFs treated with 100
ng/ml CHI3L1; however, VEGFA secretion from CAFs was not
particularly high compared with that from cancer cell lines. (E)
RT-qPCR revealed no significant changes in IL-8 mRNA
expression in the three cancer cell lines, between groups treated
with and without 100 ng/ml CHI3L1. (F) RT-qPCR revealed no
significant changes in VEGFA mRNA expression in the three
cancer cells, between groups treated with and without 100 ng/ml
CHI3L1. (G) RT-qPCR demonstrated that IL-13Rα2 expression in
CAFs was suppressed after transfection with IL-13Rα2 siRNA. (H)
RT-qPCR indicated that IL-13Rα2 knockdown suppressed the enhanced
IL-8 mRNA expression induced by CHI3L1 treatment in CAFs.
(I) RT-qPCR revealed that IL-13Rα2 knockdown and the addition of
CHI3L1 did not change VEGFA mRNA expression in CAFs. Data
are presented as the mean ± SEM. *P<0.05,
**P<0.01. The relative mRNA expression levels of
IL-8, VEGFA and IL-13Rα2 were normalized to
GAPDH expression in each sample. CAFs, cancer-associated
fibroblasts; CHI3L1, chitinase 3-like 1; IL-13Rα2, interleukin-13
receptor α2; NC, negative control; NFs, normal fibroblasts; NS, not
significant; RT-qPCR, reverse transcription-quantitative PCR;
si/siRNA, small interfering RNA; VEGFA, vascular endothelial growth
factor-A. |
RT-qPCR demonstrated that VEGFA expression
was higher in CAFs than in NFs, although this difference was not
significant. Although VEGFA expression in the three cancer
cell lines differed depending on the cell line, there were no clear
differences compared with in CAFs. Furthermore, the addition of
CHI3L1 to CAFs did not cause significant changes (Fig. 4C). ELISA demonstrated significant
differences between CAFs and NFs, and between untreated CAFs and
CHI3L1-treated CAFs. However, VEGFA secretion from CAFs was not
particularly high compared with that from the cancer cell lines
(Fig. 4D). In the three cancer
cell lines, RT-qPCR revealed no significant changes in the
expression levels of IL-8 or VEGFA between groups treated with and
without CHI3L1 in all cell lines (Fig. 4E and F).
To confirm the relationship between CHI3L1 and
IL-13Rα2, a receptor for CHI3L1, the present study also evaluated
IL-8 and VEGFA expression after CHI3L1 treatment and
knockdown of IL-13Rα2 in CAFs. Compared with the negative control
siRNA, transfection with IL-13Rα2 siRNA significantly downregulated
IL-13Rα2 expression in CAFs, as demonstrated by RT-qPCR
(Fig. 4G). IL-13Rα2 siRNA also
suppressed the enhanced IL-8 expression induced by the addition of
CHI3L1 to CAFs (Fig. 4H). On the
other hand, IL-13Rα2 siRNA did not significantly affect
VEGFA expression in CAFs (Fig.
4I).
According to a previous report (33), MCP-1 is related to CHI3L1. The
present study revealed that it is secreted from CAFs and NFs using
a cytokine array; however, RT-qPCR revealed a decrease in
MCP-1 expression after the addition of 100 ng/ml CHI3L1 in
CAFs (Fig. S2). Furthermore,
although some reports have demonstrated that CHI3L1 has effects
other than angiogenesis (11,32,33,38), in the present study, there was no
CHI3L1 dose-dependent effect on proliferation and no significant
effect on migration in colorectal cancer cell lines (Fig. S3).
Changes in IL-8 and VEGFA expression
after CHI3L1 knockdown in CAFs
RT-qPCR, western blotting and ELISA were performed
to evaluate changes in the expression and secretion of IL-8 and
VEGFA in CAFs transfected with CHI3L1 siRNA. Compared with the
negative control siRNA, transfection with CHI3L1 siRNA
significantly downregulated CHI3L1 expression and secretion in CAFs
(Fig. 5A-C). Furthermore, RT-qPCR
and ELISA demonstrated that transfection with CHI3L1 siRNA also
significantly suppressed IL-8 expression and secretion in CAFs
(Fig. 5D and E). Furthermore,
RT-qPCR and ELISA showed that transfection also significantly
suppressed VEGFA expression and secretion in CAFs (Fig. 5F and G).
Effects of CHI3L1 on tube formation by
vascular endothelial cells
Matrigel angiogenesis assays were subsequently
performed to evaluate the effects of CHI3L1 on tube formation in
EA.hy926 vascular endothelial cells using either recombinant
addition or cell culture supernatants.
First, to evaluate the direct effects of CHI3L1,
IL-8 and VEGFA on vascular endothelial cells, tube formation was
compared between control medium (serum-free DMEM) and medium
containing recombinant CHI3L1, IL-8 or VEGFA (Fig. 6A and B). IL-8 and VEGFA, which are
angiogenic factors (48),
significantly increased tube formation of EA.hy926 cells compared
with the control when applied at concentrations of 100 and 50
ng/ml, respectively. In addition, CHI3L1 tended to increase tube
formation of EA.hy926 cells compared with the control when applied
at a concentration of 100 ng/ml, although the difference was not
significant.
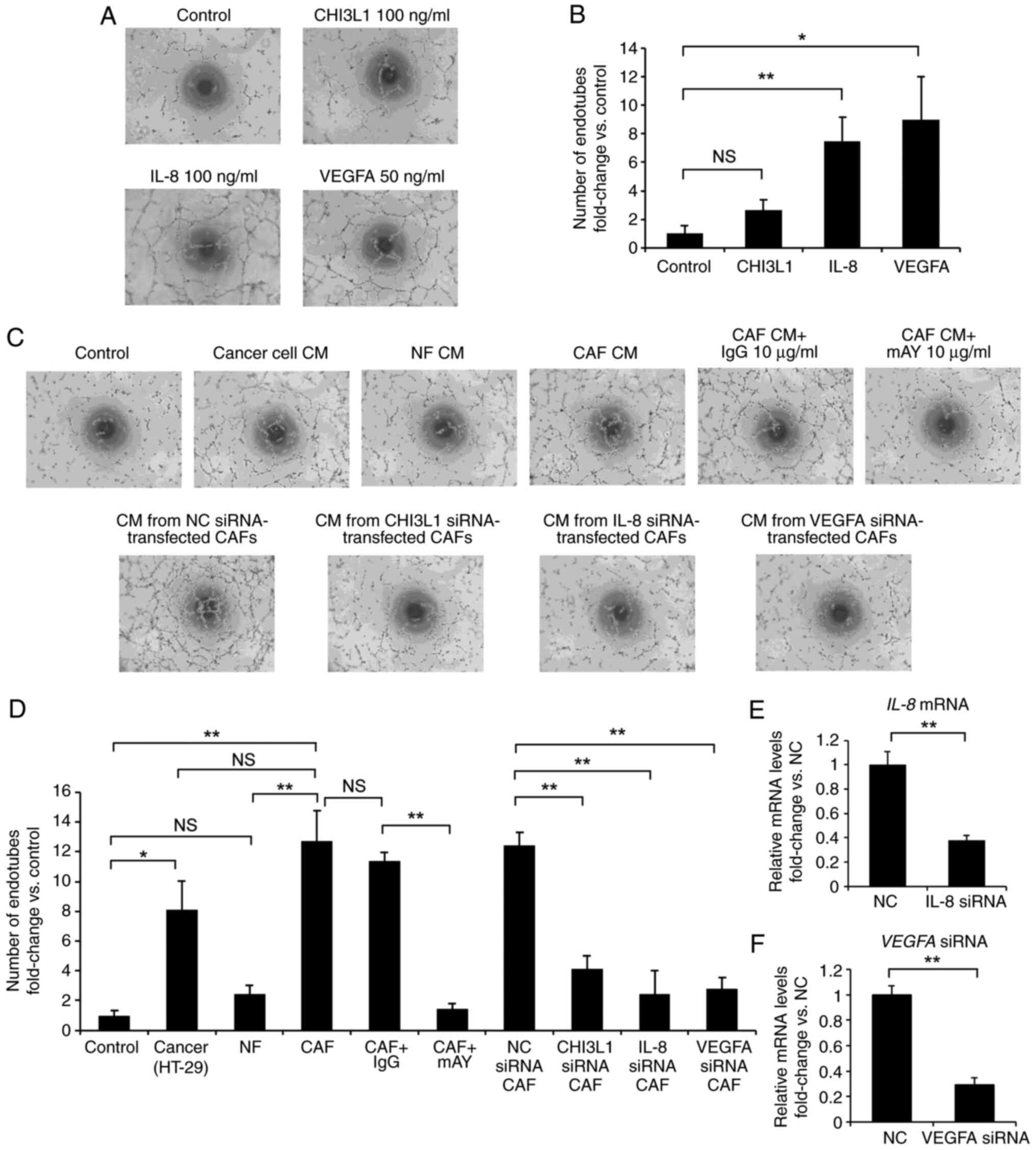 | Figure 6Effects of CHI3L1 on tube formation
in vascular endothelial cells. (A) EA.hy926 cells were cultured in
serum-free DMEM, serum-free DMEM with 100 ng/ml CHI3L1, serum-free
DMEM with 100 ng/ml IL-8 and serum-free DMEM with 50 ng/ml VEGFA.
The cells incubated with serum-free DMEM alone were used as a
control (magnification, ×40). (B) IL-8 and VEGFA increased tube
formation by EA.hy926 cells, and CHI3L1 tended to increase tube
formation by EA.hy926 cells, although no significant difference was
observed. (C) EA.hy926 cells were cultured in DMEM containing 5%
FBS, conditioned medium from cancer cells (HT-29), NF or CAF
cultures, conditioned medium from CAFs treated with control IgG or
mAY at 10 µg/ml, and conditioned medium from CAFs
transfected with negative control, CHI3L1, IL-8 or VEGFA siRNA. The
cells incubated only with DMEM containing 5% FBS were used as
controls (magnification, ×40). (D) CM from CAFs increased tube
formation by EA.hy926 cells compared with control CM and CM from
NFs. The addition of mAY or transfection of CAFs with CHI3L1, IL-8
or VEGFA siRNA suppressed the effects of CAF CM on tube formation.
(E) RT-qPCR demonstrated that IL-8 mRNA expression in CAFs
was suppressed by transfection with IL-8 siRNA. (F) RT-qPCR
demonstrated that VEGFA mRNA expression in CAFs was
suppressed by transfection with VEGFA siRNA. Data are presented as
the mean ± SEM. *P<0.05, **P<0.01. The
relative mRNA expression levels of IL-8 and VEGFA
were normalized to GAPDH expression in each sample. CAFs,
cancer-associated fibroblasts; CHI3L1, chitinase 3-like 1; CM,
conditioned medium; mAY, human CHI3L1 neutralizing antibody; NC,
negative control; NFs, normal fibroblasts; NS, not significant;
RT-qPCR, reverse transcription-quantitative PCR; si/siRNA, small
interfering RNA; VEGFA, vascular endothelial growth factor-A. |
Subsequently, to confirm whether secreted CHI3L1
protein produced by CAFs affected angiogenesis, tube formation was
compared after treatment with control medium (DMEM containing 5%
FBS), conditioned medium from cancer cells (HT-29), NF or CAF
cultures, conditioned medium from CAFs treated with control IgG or
mAY at 10 µg/ml, and conditioned medium from CAFs
transfected with negative control, CHI3L1, IL-8 or VEGFA siRNA
(Fig. 6C and D). CAF supernatants
significantly increased tube formation of EA.hy926 cells compared
with control and NF supernatants. Furthermore, CAF supernatants
tended to increase tube formation compared with HT-29 supernatants,
although the difference was not significant. The addition of
control IgG had no inhibitory effects on tube formation by CAF
supernatants, but the addition of mAY significantly inhibited tube
formation by CAF supernatants compared with control IgG.
Furthermore, tube formation was significantly suppressed by
conditioned medium from CAFs transfected with CHI3L1, IL-8 or VEGFA
siRNA compared with that from CAFs transfected with negative
control siRNA. For reference, compared with the negative control
siRNA, transfection with IL-8 or VEGFA siRNA significantly
suppressed IL-8 or VEGFA expression, respectively, in CAFs
(Fig. 6E and F).
Discussion
CAFs have been demonstrated to have important roles
in the cancer microenvironment, and the involvement of cytokines
and tumor growth factors secreted by CAFs is being actively
investigated (3-5,7).
The present study demonstrated that CHI3L1 was mainly secreted from
CAFs in the cancer microenvironment and was involved in tumor
angiogenesis via IL-8 secretion. Previous reports have suggested
that CHI3L1 is expressed by and secreted from cancer cells
(31-33). Furthermore, CHI3L1 has been
reported to serve a role in cancer progression by CAFs; however,
this mechanism has only been investigated in the context of immune
cells or exosomes, not cytokines (11,35). Accordingly, the present study
focused on cytokines secreted from CAFs and demonstrated that CAFs,
not cancer cells, mainly secreted CHI3L1 and IL-8. However, CAFs
are not the only origin of CHI3L1 and angiogenic factors. As shown
in previous reports (32,33) and the present study, some
colorectal cancer cell lines secrete CHI3L1, IL-8 and VEGFA,
suggesting that both CAFs and cancer cells may be potential origins
of CHI3L1 and angiogenic factors. Furthermore, because CAFs may be
derived from several cell types, the secretion of CHI3L1 and
angiogenic factors from CAFs of different individuals may also
differ (4). However, the present
results suggested that CHI3L1 and angiogenic factors secreted from
CAFs may have roles in the cancer microenvironment.
In the present study, to clarify the mechanisms of
action of CHI3L1 secreted from CAFs in the cancer microenvironment,
CHI3L1 receptor expression in various cell types was also
evaluated. Although the details of CHI3L1 receptors have not been
fully elucidated, a number of previous studies have reported that
IL-13Rα2 is a CHI3L1 receptor (44-47). IL-13Rα2 expression is upregulated
in melanoma, head and neck cancer, and glioma, as well as other
types of malignant tumors (49-51). In colorectal cancer, high IL-13Rα2
expression in cancer cells is associated with a poor prognosis
(52), although individual
differences are also observed. In the present study, expression was
observed in fibroblasts, particularly CAFs, but weak expression was
observed in colon cancer cell lines. In addition, IL-8 expression
and secretion were affected by CHI3L1 treatment in CAFs, but not in
cancer cells. Furthermore, IL-13Rα2 knockdown suppressed the
enhanced IL-8 expression induced by CHI3L1 treatment in CAFs,
suggesting that this receptor may be involved in CHI3L1-dependent
IL-8 production by CAFs. CHI3L1 has also been reported to act
directly on cancer cells and to be involved in cancer progression,
including proliferation and migration (33,38,53). Therefore, CHI3L1 receptors other
than IL-13Rα2 may be involved in mediating the effects of CHI3L1 in
the cancer microenvironment. Indeed, CD44v3 and transmembrane
protein 219 have been reported to act as CHI3L1 receptors (26,45,53). Overall, the results of the present
study indicated that CHI3L1 affected cancer progression by acting
on CAFs, rather than through direct effects on cancer cells.
CHI3L1 affects angiogenesis (31-33), an important mechanism that
supports cancer progression by supplying nutrients and oxygen.
Additionally, anti-angiogenic drugs have been demonstrated to serve
important roles in colorectal cancer treatment (1,54).
VEGFA, IL-8, monocyte chemotactic protein-1, bFGF, platelet-derived
growth factor, hepatocyte growth factor and epidermal growth factor
are angiogenic factors, and our previous studies revealed that
IL-6-dependent VEGFA secretion from CAFs promoted cancer
angiogenesis (7,55). Furthermore, CHI3L1 may be related
to the angiogenic functions of IL-8, VEGFA and MCP-1 (31,33,38,39). The present study focused mainly on
IL-8 signaling by performing cytokine array analysis. Changes in
IL-8 secretion were observed after CHI3L1 addition or CHI3L1
knockdown in CAFs, suggesting that IL-8 secretion from CAFs may be
regulated by CHI3L1 and that CHI3L1 may be related to IL-8-mediated
angiogenesis. The involvement of VEGFA was not clarified in the
present study because not all of the results showed significant
differences. A previous report has suggested that VEGFA is involved
in CHI3L1 signaling (31),
whereas another report has demonstrated that CHI3L1 is involved in
angiogenesis via other mechanisms because CHI3L1 could not be
antagonized by anti-VEGFA antibodies (32). The involvement of MCP-1, which has
been reported previously (33),
was also not clarified in CAFs in the present study. Furthermore,
CHI3L1 itself has been reported to be an angiogenic factor
(32,33). The results of the present
angiogenesis assays also supported this finding, although the
difference was not significant. Therefore, despite the present
findings demonstrating that CHI3L1 was mainly involved in IL-8
secretion from CAFs, further studies on the angiogenic effects of
CHI3L1 are required. To the best of our knowledge, no reports have
demonstrated the roles of fibroblasts in mediating CHI3L1-related
cancer angiogenesis.
In summary, the results of the present study
demonstrated that in the colorectal cancer microenvironment, CHI3L1
is mainly secreted from CAFs and acts on CAFs themselves (Fig. 7). Furthermore, the action of
CHI3L1 on CAFs promoted the secretion of IL-8, which induced cancer
angiogenesis. CAFs have been reported to affect tumor growth,
including cancer cell proliferation, migration, invasion and
angiogenesis through the expression or secretion of various
proteins (3,5,7,14,20,21), and some of these mechanisms may be
related to CHI3L1. Further studies are required to elucidate other
functions of CAFs and CHI3L1 in cancer progression and their
mechanisms, as well as animal experiments to confirm their effects
in vivo. Furthermore, the relationship between the
expression levels of CHI3L1 in CAFs and clinicopathological
factors, such as venous invasion and metastasis, needs to be
clarified in further studies. Treatments targeting the cancer
stroma have been attracting increasing attention (56,57). The present study demonstrated that
CAFs and CHI3L1 served important roles in cancer-stromal
interactions and that knockdown of CHI3L1 or addition of
anti-CHI3L1 antibodies suppressed angiogenesis. Therefore,
targeting the cancer stroma and CHI3L1 could help to establish
novel cancer treatments that differ from conventional drugs.
Supplementary Data
Availability of data and materials
All data generated or analyzed during this study
are included in this published article.
Authors' contributions
KW, KS, YuM, TH, RO, MH, HT, YoM, AMi, MK and ST
designed the study. KW, AMa, SH, TY, TS and HU performed the
experiments and acquired the data. KS, YuM, TH, RO, MH, HT, YoM,
AMi and MK provided technical support in performing the
experiments. KW, AMi, MK and YoM confirmed the authenticity of all
data. KW and KS planned all experiments, analyzed and interpreted
the data, and drafted the manuscript. All authors are equally
responsible for all aspects of the study, including the integrity
and accuracy of the data. All authors have read and approved the
final manuscript.
Ethics approval and consent to
participate
Ethical approval for the use of human tissue was
granted by the Nagoya City University Graduate School of Medical
Sciences and Nagoya City University Hospital Institutional Review
Board (Nagoya, Japan). Written informed consent was obtained from
the patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by a Grant-in-Aid for Scientific
Research (Japan Society for the Promotion of Science; grant no.
21K08716).
Abbreviations:
|
α-SMA
|
α-smooth muscle actin
|
|
bFGF
|
basic fibroblast growth factor
|
|
CAFs
|
cancer-associated fibroblasts
|
|
CHI3L1
|
chitinase 3-like 1
|
|
IL-13Rα2
|
interleukin-13 receptor α2
|
|
mAY
|
human CHI3L1 neutralizing
antibody
|
|
MCP-1
|
monocyte chemoattractant
protein-1
|
|
NFs
|
normal fibroblasts
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
si/siRNA
|
small interfering RNA
|
|
VEGF
|
vascular endothelial growth
factor
|
References
|
1
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liotta LA and Kohn E: The microenvironment
of the tumour-host interface. Nature. 411:375–379. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nakagawa H, Liyanarachchi S, Davuluri RV,
Auer H, Martin EW Jr, de la Chapelle A and Frankel WL: Role of
cancer-associated stromal fibroblasts in metastatic colon cancer to
the liver and their expression profiles. Oncogene. 23:7366–7377.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shiga K, Hara M, Nagasaki T, Sato T,
Takahashi H and Takeyama H: Cancer-associated fibroblasts: Their
characteristics and their roles in tumor growth. Cancers (Basel).
7:2443–2458. 2015. View Article : Google Scholar
|
|
5
|
Koliaraki V, Pallangyo CK, Greten FR and
Kollias G: Mesenchymal cells in colon cancer. Gastroenterology.
152:964–979. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Orimo A and Weinberg RA: Heterogeneity of
stromal fibroblasts in tumors. Cancer Biol Ther. 6:618–619. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nagasaki T, Hara M, Nakanishi H, Takahashi
H, Sato M and Takeyama H: Interleukin-6 released by colon
cancer-associated fibroblasts is critical for tumour angiogenesis:
Anti-interleukin-6 receptor antibody suppressed angiogenesis and
inhibited tumourstroma interaction. Br J Cancer. 110:469–478. 2014.
View Article : Google Scholar
|
|
8
|
Shen J, Zhai J, You Q, Zhang G, He M, Yao
X and Shen L: Cancer-associated fibroblasts-derived VCAM1 induced
by H. pylori infection facilitates tumor invasion in gastric
cancer. Oncogene. 39:2961–2974. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Neesse A, Michl P, Frese KK, Feig C, Cook
N, Jacobetz MA, Lolkema MP, Buchholz M, Olive KP, Gress TM and
Tuveson DA: Stromal biology and therapy in pancreatic cancer. Gut.
60:861–868. 2011. View Article : Google Scholar
|
|
10
|
Comito G, Giannoni E, Segura CP,
Barcellos-de-Souza P, Raspollini MR, Baroni G, Lanciotti M, Serni S
and Chiarugi P: Cancer-associated fibroblasts and M2-polarized
macrophages synergize during prostate carcinoma progression.
Oncogene. 33:2423–2431. 2014. View Article : Google Scholar
|
|
11
|
Cohen N, Shani O, Raz Y, Sharon Y, Hoffman
D, Abramovitz L and Erez N: Fibroblasts drive an immunosuppressive
and growth-promoting microenvironment in breast cancer via
secretion of Chitinase 3-like 1. Oncogene. 36:4457–4468. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hegab AE, Ozaki M, Kameyama N, Gao J,
Kagawa S, Yasuda H, Soejima K, Yin Y, Guzy RD, Nakamura Y, et al:
Effect of FGF/FGFR pathway blocking on lung adenocarcinoma and its
cancer-associated fibroblasts. J Pathol. 249:193–205. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kojima Y, Acar A, Eaton EN, Mellody KT,
Scheel C, Ben-Porath I, Onder TT, Wang ZC, Richardson AL, Weinberg
RA and Orimo A: Autocrine TGF-beta and stromal cell-derived
factor-1 (SDF-1) signaling drives the evolution of tumor-promoting
mammary stromal myofibroblasts. Proc Natl Acad Sci USA.
107:20009–20014. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kalluri R and Zeisberg M: Fibroblasts in
cancer. Nat Rev Cancer. 6:392–401. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Iwano M, Plieth D, Danoff TM, Xue C, Okada
H and Neilson EG: Evidence that fibroblasts derive from epithelium
during tissue fibrosis. J Clin Invest. 110:341–350. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zeisberg EM, Potenta S, Xie L, Zeisberg M
and Kalluri R: Discovery of endothelial to mesenchymal transition
as a source for carcinoma-associated fibroblasts. Cancer Res.
67:10123–10128. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mishra PJ, Mishra PJ, Humeniuk R, Medina
DJ, Alexe G, Mesirov JP, Ganesan S, Glod JW and Banerjee D:
Carcinoma associated fibroblast like differentiation of human
mesenchymal stem cells. Cancer Res. 68:4331–4339. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Quante M, Tu SP, Tomita H, Gonda T, Wang
SS, Takashi S, Baik GH, Shibata W, Diprete B, Betz KS, et al: Bone
marrow-derived myofibroblasts contribute to the mesenchymal stem
cell niche and promote tumor growth. Cancer Cell. 19:257–272. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jotzu C, Alt E, Welte G, Li J, Hennessy
BT, Devarajan E, Krishnappa S, Pinilla S, Droll L and Song YH:
Adipose tissue derived stem cells differentiate into
carcinoma-associated fibroblast-like cells under the influence of
tumor derived factors. Cell Oncol (Dordr). 34:55–67. 2011.
View Article : Google Scholar
|
|
20
|
Wu MH, Hong HC, Hong TM, Chiang WF, Jin YT
and Chen YL: Targeting Galectin-1 in carcinoma-associated
fibroblasts inhibits oral squamous cell carcinoma metastasis by
downregulating MCP-1/CCL2 expression. Clin Cancer Res.
17:1306–1316. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y, Tang H, Cai J, Zhang T, Guo J,
Feng D and Wang Z: Ovarian cancer-associated fibroblasts contribute
to epithelial ovarian carcinoma metastasis by promoting
angiogenesis, lymphangiogenesis and tumor cell invasion. Cancer
Lett. 303:47–55. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wei L, Ye H, Li G, Lu Y, Zhou Q, Zheng S,
Lin Q, Liu Y, Li Z and Chen R: Cancer-associated fibroblasts
promote progression and gemcitabine resistance via the SDF-1/SATB-1
pathway in pancreatic cancer. Cell Death Dis. 9:10652018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hara M, Nagasaki T, Shiga K and Takeyama
H: Suppression of Cancer-associated fibroblasts and endothelial
cells by Itraconazole in Bevacizumab-resistant gastrointestinal
cancer. Anticancer Res. 36:169–177. 2016.PubMed/NCBI
|
|
24
|
Itoh G, Chida S, Yanagihara K, Yashiro M,
Aiba N and Tanaka M: Cancer-associated fibroblasts induce cancer
cell apoptosis that regulates invasion mode of tumours. Oncogene.
36:4434–4444. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hakala BE, White C and Recklies AD: Human
cartilage gp-39, a major secretory product of articular
chondrocytes and synovial cells, is a mammalian member of a
chitinase protein family. J Biol Chem. 268:25803–25810. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao T, Su Z, Li Y, Zhang X and You Q:
Chitinase-3 like-protein-1 function and its role in diseases.
Signal Transduct Target Ther. 5:2012020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bara I, Ozier A, Girodet PO, Carvalho G,
Cattiaux J, Begueret H, Thumerel M, Ousova O, Kolbeck R, Coyle AJ,
et al: Role of YKL-40 in bronchial smooth muscle remodeling in
asthma. Am J Respir Crit Care Med. 185:715–722. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Berres ML, Papen S, Pauels K, Schmitz P,
Zaldivar MM, Hellerbrand C, Mueller T, Berg T, Weiskirchen R,
Trautwein C, et al: A functional variation in CHI3L1 is associated
with severity of liver fibrosis and YKL-40 serum levels in chronic
hepatitis C infection. J Hepatol. 50:370–376. 2009. View Article : Google Scholar
|
|
29
|
Volck B, Johansen JS, Stoltenberg M,
Garbarsch C, Price PA, Ostergaard M, Ostergaard K, Løvgreen-Nielsen
P, Sonne-Holm S and Lorenzen I: Studies on YKL-40 in knee joints of
patients with rheumatoid arthritis and osteoarthritis. Involvement
of YKL-40 in the joint pathology. Osteoarthritis Cartilage.
9:203–214. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cintin C, Johansen JS, Christensen IJ,
Price PA, Sørensen S and Nielsen HJ: High serum YKL-40 level after
surgery for colorectal carcinoma is related to short survival.
Cancer. 95:267–274. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Francescone RA, Scully S, Faibish M,
Taylor SL, Oh D, Moral L, Yan W, Bentley B and Shao R: Role of
YKL-40 in the angiogenesis, radioresistance, and progression of
glioblastoma. J Biol Chem. 286:15332–15343. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shao R, Hamel K, Petersen L, Cao QJ,
Arenas RB, Bigelow C, Bentley B and Yan W: YKL-40, a secreted
glycoprotein, promotes tumor angiogenesis. Oncogene. 28:4456–4468.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kawada M, Seno H, Kanda K, Nakanishi Y,
Akitake R, Komekado H, Kawada K, Sakai Y, Mizoguchi E and Chiba T:
Chitinase 3-like 1 promotes macrophage recruitment and angiogenesis
in colorectal cancer. Oncogene. 31:3111–3123. 2012. View Article : Google Scholar :
|
|
34
|
Riabov V, Gudima A, Wang N, Mickley A,
Orekhov A and Kzhyshkowska J: Role of tumor associated macrophages
in tumor angiogenesis and lymphangiogenesis. Front Physiol.
5:752014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fan JT, Zhou ZY, Luo YL, Luo Q, Chen SB,
Zhao JC and Chen QR: Exosomal lncRNA NEAT1 from cancer-associated
fibroblasts facilitates endometrial cancer progression via
miR-26a/b-5p-mediated STAT3/YKL-40 signaling pathway. Neoplasia.
23:692–703. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li A, Dubey S, Varney ML, Dave BJ and
Singh RK: IL-8 directly enhanced endothelial cell survival,
proliferation, and matrix metalloproteinases production and
regulated angiogenesis. J Immunol. 170:3369–3376. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Spengler K, Kryeziu N, Große S, Mosig AS
and Heller R: VEGF triggers transient induction of autophagy in
endothelial cells via AMPKα1. Cells. 9:6872020. View Article : Google Scholar
|
|
38
|
Chen CC, Pekow J, Llado V, Kanneganti M,
Lau CW, Mizoguchi A, Mino-Kenudson M, Bissonnette M and Mizoguchi
E: Chitinase 3-like-1 expression in colonic epithelial cells as a
potentially novel marker for colitis-associated neoplasia. Am J
Pathol. 179:1494–1503. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tang H, Sun Y, Shi Z, Huang H, Fang Z,
Chen J, Xiu Q and Li B: YKL-40 induces IL-8 expression from
bronchial epithelium via MAPK (JNK and ERK) and NF-κB pathways,
causing bronchial smooth muscle proliferation and migration. J
Immunol. 190:438–446. 2013. View Article : Google Scholar
|
|
40
|
Maeda Y, Takahashi H, Nakai N, Yanagita T,
Ando N, Okubo T, Saito K, Shiga K, Hirokawa T, Hara M, et al:
Apigenin induces apoptosis by suppressing Bcl-xl and Mcl-1
simultaneously via signal transducer and activator of transcription
3 signaling in colon cancer. Int J Oncol. 52:1661–1673.
2018.PubMed/NCBI
|
|
41
|
Bustin SA: Quantification of mRNA using
real-time reverse transcription PCR (RT-PCR): Trends and problems.
J Mol Endocrinol. 29:23–39. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zarour LR, Anand S, Billingsley KG, Bisson
WH, Cercek A, Clarke MF, Coussens LM, Gast CE, Geltzeiler CB,
Hansen L, et al: Colorectal cancer liver metastasis: Evolving
paradigms and future directions. Cell Mol Gastroenterol Hepatol.
3:163–173. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Melzer C, von der Ohe J, Lehnert H,
Ungefroren H and Hass R: Cancer stem cell niche models and
contribution by mesenchymal stroma/stem cells. Mol Cancer.
16:282017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lee CM, He CH, Nour AM, Zhou Y, Ma B, Park
JW, Kim KH, Dela Cruz C, Sharma L, Nasr ML, et al: IL-13Ralpha2
uses TMEM219 in chitinase 3-like-1-induced signalling and effector
responses. Nat Commun. 7:127522016. View Article : Google Scholar
|
|
45
|
He CH, Lee CG, Dela Cruz CS, Lee CM, Zhou
Y, Ahangari F, Ma B, Herzog EL, Rosenberg SA, Li Y, et al:
Chitinase 3-like 1 regulates cellular and tissue responses via
IL-13 receptor α2. Cell Rep. 4:830–841. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
He CH, Lee CG, Ma B, Kamle S, Choi AMK and
Elias JA: N-glycosylation regulates chitinase 3-like-1 and IL-13
ligand binding to IL-13 receptor α2. Am J Respir Cell Mol Biol.
63:386–395. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen Y, Zhang S, Wang Q and Zhang X:
Tumor-recruited M2 macrophages promote gastric and breast cancer
metastasis via M2 macrophage-secreted CHI3L1 protein. J Hematol
Oncol. 10:362017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Brat DJ, Bellail AC and Van Meir EG: The
role of interleukin-8 and its receptors in gliomagenesis and
tumoral angiogenesis. Neuro Oncol. 7:122–133. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Beard RE, Abate-Daga D, Rosati SF, Zheng
Z, Wunderlich JR, Rosenberg SA and Morgan RA: Gene expression
profiling using nanostring digital RNA counting to identify
potential target antigens for melanoma immunotherapy. Clin Cancer
Res. 19:4941–4950. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kawakami M, Kawakami K, Kasperbauer JL,
Hinkley LL, Tsukuda M, Strome SE and Puri RK: Interleukin-13
receptor alpha2 chain in human head and neck cancer serves as a
unique diagnostic marker. Clin Cancer Res. 9:6381–6388.
2003.PubMed/NCBI
|
|
51
|
Joshi BH, Plautz GE and Puri RK:
Interleukin-13 receptor alpha chain: A novel tumor-associated
transmembrane protein in primary explants of human malignant
gliomas. Cancer Res. 60:1168–1172. 2000.PubMed/NCBI
|
|
52
|
Barderas R, Bartolomé RA,
Fernandez-Aceñero MJ, Torres S and Casal JI: High expression of
IL-13 receptor α2 in colorectal cancer is associated with invasion,
liver metastasis, and poor prognosis. Cancer Res. 72:2780–2790.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Geng B, Pan J, Zhao T, Ji J, Zhang C, Che
Y, Yang J, Shi H, Li J, Zhou H, et al: Chitinase 3-like 1-CD44
interaction promotes metastasis and epithelial-to-mesenchymal
transition through β-catenin/Erk/Akt signaling in gastric cancer. J
Exp Clin Cancer Res. 37:2082018. View Article : Google Scholar
|
|
54
|
Piawah S and Venook AP: Targeted therapy
for colorectal cancer metastases: A review of current methods of
molecularly targeted therapy and the use of tumor biomarkers in the
treatment of metastatic colorectal cancer. Cancer. 125:4139–4147.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ando N, Hara M, Shiga K, Yanagita T,
Takasu K, Nakai N, Maeda Y, Hirokawa T, Takahashi H, Ishiguro H, et
al: Eicosapentaenoic acid suppresses angiogenesis via reducing
secretion of IL-6 and VEGF from colon cancer-associated
fibroblasts. Oncol Rep. 42:339–349. 2019.PubMed/NCBI
|
|
56
|
Togo S, Polanska UM, Horimoto Y and Orimo
A: Carcinoma-associated fibroblasts are a promising therapeutic
target. Cancers (Basel). 5:149–169. 2013. View Article : Google Scholar
|
|
57
|
Roma-Rodrigues C, Mendes R, Baptista PV
and Fernandes AR: Targeting tumor microenvironment for cancer
therapy. Int J Mol Sci. 20:8402019. View Article : Google Scholar :
|





















