Introduction
Liver cancer is the sixth most common cancer type
and the fourth leading cause of death due to cancer globally
(1). The carcinogenesis of the
liver involves multiple mechanisms, such as immune escape, somatic
mutations, abnormal lipid metabolism and aberrant changes in
multiple molecular pathways (2,3). It
is well known that long noncoding RNAs (lncRNAs) have a critical
role in gene regulation (4).
However, only limited studies have investigated the regulatory
roles of lncRNAs and genomic stability (GS). Multiple lncRNAs have
been reported to be abnormally expressed and contribute to
malignant phenotypes in hepatocellular carcinoma (HCC). They
commonly exert their effects by interacting with DNA, RNA or
proteins, or encoding short peptides. For instance, lncRNA
proliferating cell nuclear antigen pseudogene 1 promotes hepatitis
B virus replication and hepatocarcinogenesis by modulating
signalling (5). LncRNA small
nucleolar RNA host gene 6 accelerates the development of HCC by
interacting with heterogeneous nuclear ribonucleoprotein
L/polypyrimidine tract binding protein 1 to facilitate the
destabilisation of SET domain containing 7/leucine zipper
transcription factor like 1 mRNA (6). Significant upregulation of HOX
transcript antisense RNA expression in HCC is associated with poor
prognosis (7). Apart from that,
lncRNAs affect the development of cancer by influencing tumor
metabolism and immunomodulation. Hepatocellular carcinoma
upregulated long non-coding RNA enhances glycolysis and promotes
the proliferation of liver cancer cells through lactate
dehydrogenase A and pyruvate kinase M1/2 (8). LncRNAs may regulate tumor immunity by
directly influencing immune-associated genes (9). Aberrant expression of lncRNAs is
involved in various biological processes of cancers. The competing
endogenous RNA (ceRNA) mechanism is one of the most well-studied
aspects of lncRNA mechanisms. LncRNAs is able to interact with
microRNAs (miRNAs) and inhibit gene silencing caused by miRNAs
(10). By modulating focal
adhesion signaling, ITGB8-AS1 acts as a ceRNA to regulate cell
proliferation and tumor formation (11).
Increasing evidence suggests that GS is inextricably
linked to cancer (12). GS appears
to influence the multistep process of cancer progression through
abnormal DNA repair pathways and dysregulation of genes encoding
homologous recombination proteins (13). Aberrant lncRNAs have an equally
significant impact on GS by regulating key homologous recombination
DNA repair genes, which in turn influences tumorigenesis (14). DNA double-strand break repair is
regulated by lncRNA lnc-RI by stabilising RAD51 Recombinase (RAD51)
as a ceRNA (15). The non-coding
RNA activated by DNA damage forms a topoisomerase complex that is
essential for GS (16). Noncoding
RNA activated by DNA damage regulates GS through Pumilio protein
binding (17). The accumulation of
mutations in somatic cells is linked to GS and development
(18). These studies offer a good
basis for the development of a novel carcinogenic mechanism and
potential therapeutic targets for HCC via lncRNAs and GS.
Therefore, the present study established a
prognostic model related to genomic instability (GI). To verify the
relationship between prognostic lncRNAs and GS, the role of the
lncRNA zinc finger protein, FOG family member 2 antisense 1
(ZFPM2-AS1) in HCC was investigated through basic experiments.
Materials and methods
Data collection
Data on somatic mutation profiles, RNA expression
profiles and clinical characteristics of patients with HCC were
obtained from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). Samples with
missing information on lncRNA and mRNA expression profiles,
clinical characteristics or somatic mutations were excluded. A
total of 343 patients with HCC were enrolled. Table SI lists all of the websites that
were accessed for the present study. Somatic mutations were
analyzed and counted by using the 'maftools' R package (v2.10.5) to
obtain the tumor mutation burden of each patient. Heatmaps, boxplot
graphs, receiver operating characteristic (ROC) analysis with area
under the curve (AUC) and Kaplan-Meier survival curves were
generated in R (v3.5.2). The GSE14520 (19) and GSE76427 (20) datasets were downloaded from the
Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). miRDB (http://www.mirdb.org/), LncBase (http://carolina.imis.athena-innovation.gr/diana_tools/web/)
and TargetScan (http://www.targetscan.org/vert_72/) were applied to
predict candidate binding miRNAs.
Exploration of GI-related lncRNAs
After computing the cumulative number of somatic
mutations per patient and then ranking the patients in decreasing
order, the top 25% (n=93) and bottom 25% (n=90) were designated as
the GI and the GS group, respectively. The 'LIMMA' R package
(v3.38.3) was used to compare the lncRNA expression profiles of the
two groups of patients. A total of 85 significantly differentially
expressed lncRNAs were screened based on the cutoff criteria of
fold change >2 and P<0.05. These 85 lncRNAs were defined as
GI-related lncRNAs.
Tissue samples and cell culture
Tumor and adjacent nontumor tissues from 80 patients
with HCC (age range, 15-91 years; females/males, 4:6) were obtained
from Zhongnan Hospital of Wuhan University (Wuhan, China) and
collected by surgery. Tissue collection dates ranged from April
2014 to May 2020. The exclusion criteria were patients with a
history of local or systemic treatment for HCC or other clinical
disorders. None of the patients in this study received any
chemotherapy or radiation therapy prior to surgery. All patients
provided written informed consent. The present study conformed to
the Declaration of Helsinki. The tissue samples were collected with
the approval of the ethics committee of Zhongnan Hospital (Wuhan,
China; no. KELUN2020100).
Furthermore, five liver cancer cell lines and a
normal liver cell line were used: HepG2, Li-7, HCC-LM3, Huh-7,
Hep3B and THLE-3. HepG2 is a human liver cancer cell line. All cell
lines were maintained in Dulbecco's modified Eagle's medium (DMEM)
or minimum essential medium (MEM) containing 10% foetal bovine
serum (FBS, Gibco; Thermo Fisher Scientific, Inc.). The cell lines
were obtained from the Cell Resource Center of Shanghai Institutes
for Biological Sciences, Chinese Academy of Sciences and cultured
in a humidified incubator (5% CO2, 37°C). All cell lines
were authenticated using short tandem repeat profiling.
DNA damage analysis
To detect DNA damage, an alkaline comet assay was
performed using a reagent kit (ELK Biotechnology) according to the
manufacturer's protocol. ImageJ-OpenComet software [ImageJ, v1.53e;
National Institutes of Health (NIH)] was employed to evaluate the
results of the comet assay. The extent of DNA damage was expressed
as the tail moment, which corresponded to the fraction of the DNA
in the tail of the comet.
RNA extraction and reverse transcription
quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted using the TRIzol method
according to the manufacturer's instructions (Vazyme Biotech Co.,
Ltd.). RT was performed by using an RT kit (cat. no. R223-01;
Vazyme Biotech Co., Ltd.). According to the manufacturer's
protocol, the reverse transcription conditions were as follows:
42°C for 2 min, 50°C for 15 min and 85°C for 5 sec. The product was
used for qPCR. A standard SYBR Green PCR kit (cat. no. Q711-02;
Vazyme Biotech Co., Ltd.) was employed to perform qPCR as described
previously (21). GAPDH served as
an endogenous control. Relative expression was calculated using the
comparative quantification cycle (Cq) method (2−ΔΔCq)
(22). The thermocycling
conditions are provided in Table
SII and the primers in Table
SIII.
Plasmid, microRNA and short hairpin RNA
(shRNA) transfection
For transfection, HCC cells were seeded in T25
flasks one night prior to transfection. When the cell confluence
reached 80%, the cells were transfected with ZFPM2-AS1
overexpression plasmid and a corresponding empty plasmid using
Lipofectamine 3000 (Thermo Fisher Scientific, Inc.). According to
the manufacturer's protocol, 5 µg plasmid, 10 µl
P3000, 10 µl Lipofectamin® 3000 and 500 µl
Opti-MEM Medium (Gibco; Thermo Fisher Scientific, Inc.) were mixed
gently and then incubated at room temperature (RT) for 15 min. The
mixture was then slowly added to HCC cells. After incubating at
37°C for 12 h, the culture medium was subsequently replaced with
fresh DMEM supplemented with 10% FBS. HCC cells were harvested at
48 h after plasmid transfection. Total RNA was extracted and used
to assess the transfection efficiency via RT-qPCR.
ZFPM2-AS1-wild-type (WT)/mutant (MUT) plasmids and X-ray repair
cross complementing 4 (XRCC4)-WT/MUT plasmids were constructed by
Genomeditech. The seed sequences of ZFPM2-AS1 plasmids were TTT GTT
G (WT) and CGG ACC T (MUT). The seed sequences of XRCC4 plasmids
were TTT GTT G (WT) and CGG ACC T (MUT). shRNA, miR-3065-5p primers
and miRNA mimic/inhibitor were designed by Wuhan Qingke
Biotechnology Co., Ltd. HCC-LM3 and Huh7 cells were transfected
with shRNA lentiviral plasmid (pLKO.1-purp). Puromycin (2
µg/ml) was used for sorting positive shRNA cells. TSnanofect
(Wuhan Qingke Biotechnology Co., Ltd) was used for transfection of
miR-3065-5p mimic/inhibitor. The shRNA and miRNA sequences are
presented in Table SIII.
Western blot (WB) analysis
WB was performed using standard methods as described
previously (23). The adherent
cells were washed twice with cold PBS, trypsinized to collect the
cell precipitate and resuspended in PBS. The cell suspension was
then centrifuged at 4°C and 500 × g for 3 min and the cell
precipitate was harvested. Cells were lysed for 15 min on ice in
RIPA buffer (Beyotime Institute of Biotechnology). Cell lysates
were centrifuged at 14,000 × g at 4°C for 10 min to collect the
supernatant. Protein samples (supernatant) were denatured at 95°C
for 10 min. Quantification of protein levels was performed using a
BCA kit (Beyotime Institute of Biotechnology). A total of 50
µg protein was loaded per lane and separated by 10% SDS-PAGE
and transferred to 0.45-µm polyvinylidene difluoride (PVDF)
membranes (Immobilon-P; Millipore) by electro-transblotting, at 300
mA for 1 h. Electro-transblotting was performed by using
transblotting buffer (192 mM glycine, 25 mM TrisBase, 20%
methanol). Subsequently, 5% skimmed milk was used to block PVDF
membranes for 1 h at RT. The membranes were incubated with primary
antibodies overnight at 4°C. They were then washed thrice with
tris-buffered saline/Tween 20 (TBST) for 30 min and incubated with
secondary antibody for 1 h at RT. Finally, the membranes were
washed thrice with TBST for 30 min. The antibodies and antibody
dilution ratios used in this experiment are listed in Table SIV. Protein expression was
identified using enhanced chemiluminescence western blotting
detection reagents (Thermo Fisher Scientific, Inc.) and a
Tanon-5200 Chemiluminescent Imaging System (Tanon Science &
Technology) and densitometrically quantified using Image Lab
software (v5.2; BioRad Laboratories, Inc.). GAPDH served as a
loading control for normalization.
In vivo experiments
Male mice (BALB/c nude mice; age, 5 weeks; body
weight, 18-20 g; Shulaibao Biotech) were housed at 5 per cage under
specific pathogen-free conditions (temperature, 21±2°C; humidity,
40-60%; 12-h light/dark cycle; free access to standard sterile food
and water) and ear-notched for identification. For establishing the
subcutaneous xenograft model in 10 mice, negative control or
HCC-LM3 cells with stable ZFPM2-AS1 knockdown (0.1 ml;
2×106 cells/mouse, 5 mice/group) were subcutaneously
injected into the axillary tissue by using a 27-gauge syringe. To
establish the lung metastasis model in another 10 mice, negative
control or HCC-LM3 cells with stable ZFPM2-AS1 knockdown (0.2 ml;
1×106 cells/mouse, 5 mice/group) were intravenously
injected into the tail vein of nude mice by using a 28-gauge
syringe. The tumor volume (mm3) in each mouse was
measured by a Vernier caliper every 3 days and calculated as
follows: Tumor volume=tumor length x width2/2. Mice were
euthanized following 21 days for the xenograft study and 50 days
for the lung metastasis experiment. The following humane endpoints
were established: Tumor diameter >2.0 cm, weight loss >25%
and poor overall condition. None of the mice reached the humane
endpoints in this study. To reduce suffering, mice were
anesthetized with 2% isoflurane. Mice were then rapidly euthanized
by cervical dislocation. Verification of death included cardiac and
respiratory arrest, lack of reflection and changes in mucosal
color. After subcutaneous tumors were dissected, tumors were
weighed using a digital balance (Mettler), fixed with 4% formalin
(Biosharp) and embedded with paraffin to prepare sections for
histology. No metastatic nodules were found in the abdominal and
thoracic organs of the subcutaneous xenograft mice. After death
verification, lungs of the lung metastasis model mice were
completely dissected. The lungs were fixed with 4% formalin
(Biosharp), embedded in paraffin, cut into sections and stained
with hematoxylin-eosin (H&E) for histopathological evaluation.
Metastatic lung nodules were counted using a microscope (IX51;
Olympus Corporation).
Chromosomal aberration assay
HCC cells with stable ZFPM2-AS1 knockdown were
washed and treated with a demecolcine solution (cat. no. D1925;
Sigma-Aldrich; Merck KGaA) for 2 h. The cells were collected and
suspended in a hypotonic solution (0.075 M KCl) for 20 min at room
temperature. The cells were then fixed in cold Carnoy's fixative
(methanol/acetic acid, 3:1), placed on slides for staining with
Giemsa (cat. no. G146-10; Thermo Fisher Scientific, Inc.) and
examined by a microscope (DM2500; Leica Microsystems GmbH).
Nuclear and cytoplasmic RNA
fractionation
Nuclear and cytoplasmic RNA was extracted using
Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol. RNA was
subjected to RT-qPCR detection. GAPDH and U6 served as the
cytoplasmic and nuclear controls, respectively.
RNA pull-down
The interaction between ZFPM2-AS1 and miR-3065-5p
was validated using a Pierce Magnetic RNA-Protein Pull-Down Kit
(Thermo Fisher Scientific, Inc.). The miR-3065-5p-WT (sequence: TCA
ACA AAA TCA CTG ATG CTG GA)/miR-3065-5p-MUT (sequence: TAC GAG CGA
TCA CTG ATG CTG GA) and control were synthesized and
biotin-labelled by Wuhan Jinkairui Bioengineering Co., Ltd. After
being cultivated for 2 days, HCC cells were lysed. Cell lysates
were then mixed with the biotinylated probes and magnetic beads at
4°C for 1 h. RT-qPCR was used to verify the miR-3065-5p targets
from the pull-down reaction mixture.
Immunohistochemistry (IHC)
For IHC staining, 4 µm-thick slices of mouse
tumor tissue were used. Paraffin sections were placed in an oven at
65°C for 2 h, dewaxed and washed three times with PBS (5 min each
time). Primary antibody incubation was performed overnight at 4°C
and secondary antibody incubation 50 min at 37°C. After labeling
proteins, the staining was visualized using diaminobenzidine
staining followed by hematoxylin counterstaining. Antibody
information and dilution ratios are listed in Table SIV. After alcohol dehydration, a
microscope (IX51; Olympus Corporation) was used to scan the slides
at magnifications of ×400 or ×200. The mean optical density values
of IHC images were determined using the IHC Toolbox plugin for
ImageJ (v1.53e; NIH).
Luciferase reporter assay
A luciferase reporter assay was performed using the
Dual-Luciferase Reporter Assay System (Promega Corp.) with the
pmirGLO dual-luciferase miRNA target expression vector (Promega
Corp.). HCC-LM3 cells were seeded in a 96-well plate and cultured
overnight. They were cotransfected with miR-3065-5p mimics and
ZFPM2-AS1 3′-untranslated region (UTR), XRCC4 3′-UTR, ZFPM2-AS1
3′-UTR-MUT or XRCC4 3′-UTR-MUT. Luciferase activities were measured
using a dual-luciferase reporter gene assay system (Promega Corp.)
and normalised to Renilla activity.
Cell proliferation, apoptosis, invasion
and migration assays, Gene Ontology (GO), Kyoto Encyclopedia of
Genes and Genomes (KEGG) enrichment analyses
The methods for these assays are provided in the
supplementary methods S1.
Statistical analysis
All statistical analyses were performed using SPSS
25.0 software (IBM Corporation), GraphPad Prism 8.0 (GraphPad
Software, Inc.) and R software (version 3.5.2). Categorical
variables were compared using χ2 tests. An unpaired
Student's t-test was used to analyze two independent groups, while
a paired t-test was used for paired samples. Multiple-group
statistical comparisons were performed by one-way ANOVA with
Tukey's multiple-comparisons test. Quantitation of the cell number,
colony number, wound gap closure and WB band integrated density
were performed with ImageJ software (v1.53e; NIH). Kaplan-Meier
survival analysis was performed using the log-rank test. The Renyi
test was performed to generate the P-values when survival curves
crossed over. Cox proportional hazards models were applied to
evaluate the hazard ratios (HR) in uni- and multivariate logistic
regression analyses. All experiments were performed three times and
P<0.05 was considered to indicate statistical significance.
Results
GI-linked lncRNAs are closely related to
prognosis of patients with HCC
The processes performed in the present study are
summarized in the flowchart in Fig.
1A. A total of 85 GI-linked lncRNAs with significant
differences in expression between the high and low mutation groups
were identified. Fig. S1 provides
the results of various bioinformatics analyses. The expression
levels of DNA damage-related proteins were closely associated with
GI-linked lncRNAs. An lncRNA-mRNA coexpression network was then
constructed (Fig. S1A) and genes
coexpressed with these lncRNAs were significantly associated with
GI according to GO and KEGG enrichment analyses (Figs. 1B and S1B). A total of 343 patients with HCC
were randomly allocated into two groups: The training group (n=172)
and the testing group (n=171). There were no significant
differences between these groups in terms of any parameter (all
P>0.05; Table SV). In the
training group, 22 prognostic lncRNAs were screened out by
univariate Cox regression analysis (Fig. S1C). In multivariate Cox regression
analysis, only lung cancer-associated transcript 1 (LUCAT1), MIR210
host gene (MIR210HG) and ZFPM2-AS1 were significant (Fig. 1C). A GI-related lncRNA signature
(GIlncsig) composed of six lncRNAs was established, with the
following formula: (coef, coefficient of each lncRNA; expr,
relative expression level of each lncRNA). The specific parameters
are provided in Table SVI.
Validation of the predictive power of the
model
The patients were divided into high-risk and
low-risk groups based on the median risk score. The GIlncsig had a
good clinical predictive performance in the training cohort
(Fig. S2). In the testing cohort,
the high-risk group had significantly worse outcomes than the
low-risk group (P<0.001; Fig.
1D). The ROC curve analysis indicated good predictive power of
the model [AUC=0.786; Fig. 1E].
The expression of DNA repair-related genes and somatic mutations
were significantly higher in the high-risk group (P<0.05;
Figs. 1F and S1D). Among the DNA repair-related genes,
breast cancer gene 1 (BRCA1), RAD51 and XRCC4 were the most
significant. The expression of prognosis-related lncRNAs differed
significantly between the high- and low-risk groups (Fig. 1G) and the number of somatic
mutations and DNA repair protein expression levels increased with
increasing score (Fig. S1E).
GIlncsig also exhibited a similar clinical predictive performance
in the TCGA group (Fig. S3).
Overall, multivariate Cox regression analysis in each group
indicated GIlncsig to be a prognostic factor independent of other
clinicopathological and mutational parameters (Table SVII). A Kaplan-Meier survival
analysis indicated that the model was applicable to patients of
different ages, pathological grades, genders and stages (Fig. S4). Significantly higher TP53
mutations were found in the high-risk group as compared with those
in the low-risk group (P<0.01; Fig.
1H). The low-risk group with mutant TP53 had the best survival
outcomes, whereas the high-risk group with wild-type TP53 had the
worst survival outcomes (P=0.02; Fig.
1I). The model was then validated in the GEO datasets (GSE14520
and GSE76427) by using survival and ROC curve analyses (Fig. S5A-D).
ZFPM2-AS1 knockdown inhibits tumor
development in vitro and in vivo
As ZFPM2-AS1 in GIlncsig had the largest HR value
and the most significant P-value, ZFPM2-AS1 was selected for
experimental research. the expression level of ZFPM2-AS1 was
evaluated in 80 pairs of tissues (tumor tissues and corresponding
adjacent nontumor tissues) and it was indicated to be highly
expressed in tumor tissues (P<0.001; Fig. 2A). Kaplan-Meier analysis revealed
that patients with high ZFPM2-AS1 expression had poor overall
survival (P<0.001; Fig. 2B).
ZFPM2-AS1 expression was associated with TNM stage, tumor size and
lymphatic metastasis (P<0.05; Table
I) according to the clinicopathological features of patients
with HCC. Furthermore, it was indicated to be an independent
prognostic factor (P=0.04; Table
SVIII). The expression of ZFPM2-AS1 in the liver cancer cell
lines was significantly higher than that in a normal liver cell
line (Fig. 2C). It was indicated
that shRNA#1 and shRNA#2 were able to significantly knock down
ZFPM2-AS1 expression (P<0.001; Fig.
2D). ZFPM2-AS1 knockdown significantly inhibited the
proliferation of HCC-LM3 and Huh7 cells (P<0.001; Fig. 2E and F) and impaired the invasion
and migration of hepatoma cells (P<0.05; Fig. 2G-I). In an in vivo
experiment, a subcutaneous tumor formation assay in nude mice
indicated that ZFPM2-AS1 knockdown significantly inhibited tumor
volume and tumor weight (P<0.05; Fig. 2J). Furthermore, no intrahepatic
metastatic nodules were found according to H&E staining of
mouse liver tissues (Fig. S5E).
After RNA was extracted from the tumor tissue of the mice, RT-qPCR
revealed low expression levels of ZFPM2-AS1 in the ZFPM2-AS1
knockdown group (Fig. 2K). IHC of
mouse tumor tissues suggested that Ki67 expression was decreased in
the ZFPM2-AS1 knockdown group (Fig.
2L). In a separate experiment, ZFPM2-AS1 knockdown also reduced
the formation of pulmonary metastatic nodules (Fig. 2M).
 | Figure 2ZFPM2-AS1 knockdown inhibits hepatoma
cell growth. (A) ZFPM2-AS1 expression was measured by quantitative
PCR in 80 pairs of HCC and corresponding adjacent normal tissues.
(B) Overall survival curves of patients with HCC based on GEPIA
data. (C) Expression of ZFPM2-AS1 in five liver cancer cell lines
and a normal liver cell line. (D) Knockdown efficiency of shRNA in
HCC-LM3 and Huh-7 cells. (E and F) Cell proliferation was
determined using (E) the Cell Counting Kit-8 assay and (F) a plate
colony-formation assay (scale bars, 10 mm). (G and H) The migration
ability of (G) HCC-LM3 and (H) Huh7 cells was detected using
wound-healing experiments (scale bars, 200 µm). (I)
Cell-invasion ability was detected using the Transwell assay (scale
bars, 200 µm). (J) Volume and weight of subcutaneous
xenograft tumors in nude mice. (K) ZFPM2-AS1 was significantly
downregulated in subcutaneous xenograft tumors where
ZFPM2-AS1-knockdown HCC-LM3 cells were injected. (L) IHC analysis
of subcutaneous tumors showed that the expression level of Ki67 was
reduced after ZFPM2-AS1 knockdown (magnification, ×200 and ×400).
The MOD value was used for the quantification of IHC analysis. (M)
ZFPM2-AS1 knockdown inhibited lung metastasis (scale bar in cm).
The arrowhead indicates lung metastasis nodules. H&E staining
results in lung tissue sections of each group (scale bar, 500
µm). The error bars and bars indicate the mean and standard
deviation of at least three independent experiments.
*P<0.05; **P<0.01;
***P<0.001. HCC, hepatocellular carcinoma; HR, hazard
ratio; OD450, optical density at 450 nm; MOD, mean optical density;
shRNA, short hairpin RNA; d, days; IHC, immunohistochemistry;
ZFPM2-AS1, zinc finger protein, FOG family member 2 antisense 1;
H&E, hematoxylin-eosin. |
 | Table IAssociation of ZFPM2-AS1 expression
with demographic and clinicopathological characteristics of
patients with hepatocellular carcinoma (n=80). |
Table I
Association of ZFPM2-AS1 expression
with demographic and clinicopathological characteristics of
patients with hepatocellular carcinoma (n=80).
| Characteristic | Cases, n (%) | ZFPM2-AS1
| P-value |
|---|
| High | Low |
|---|
| Age, years | | | | 0.370 |
| <65 | 42 (52.5) | 19 | 23 | |
| ≥65 | 38 (47.5) | 21 | 17 | |
| Gender | | | | 0.361 |
| Female | 32 (40.0) | 18 | 14 | |
| Male | 48 (60.0) | 22 | 26 | |
| Tumor diameter,
cm | | | | 0.014 |
| <5 | 43 (53.8) | 16 | 27 | |
| ≥5 | 37 (46.2) | 24 | 13 | |
| HBV infection | | | | 0.366 |
| No | 34 (42.5) | 15 | 19 | |
| Yes | 46 (57.5) | 25 | 21 | |
| AFP,
µg/l | | | | 0.369 |
| <400 | 44 (55.0) | 24 | 20 | |
| ≥400 | 36 (45.0) | 16 | 20 | |
| TNM | | | | 0.025 |
| I+II | 38 (47.5) | 14 | 24 | |
| III+IV | 42 (52.5) | 26 | 16 | |
| Lymphatic
invasion | | | | 0.044 |
| No | 43 (53.8) | 17 | 26 | |
| Yes | 37 (46.2) | 23 | 14 | |
| PVTT | | | | 0.818 |
| No | 31 (38.8) | 16 | 15 | |
| Yes | 49 (61.2) | 24 | 25 | |
DNA damage increases after ZFPM2-AS1
knockdown
DNA damage was assessed using the comet assay and it
was indicated that DNA damage in HCC-LM3 cells increased after
ZFPM2-AS1 knockdown (P<0.05; Fig.
3A). Furthermore, ZFPM2-AS1 knockdown increased the number of
chromosomal aberrations (Fig. 3B).
DNA damage has been reported to induce apoptosis and cell-cycle
arrest (24,25). Thus, the effect of ZFPM2-AS1
knockdown on cell apoptosis and the cell cycle was examined. Flow
cytometric analyses suggested that knockdown of ZFPM2-AS1 promoted
apoptosis and contributed to G1-phase arrest (Fig. 3C and D). Next, the expression of
apoptosis- and cell cycle-related genes was examined. ZFPM2-AS1
knockdown significantly upregulated cleaved (CL) caspase 3
expression and downregulated full-length (FL) caspase 3 expression
(apoptosis-related proteins). Furthermore, the expression of P21
was significantly upregulated, but that of cyclin E, cyclin B1,
cyclin-dependent kinase 2 (CDK2), CDK4 and CDK6 (cell cycle-related
proteins) was significantly downregulated after ZFPM2-AS1 knockdown
(Fig. 3E).
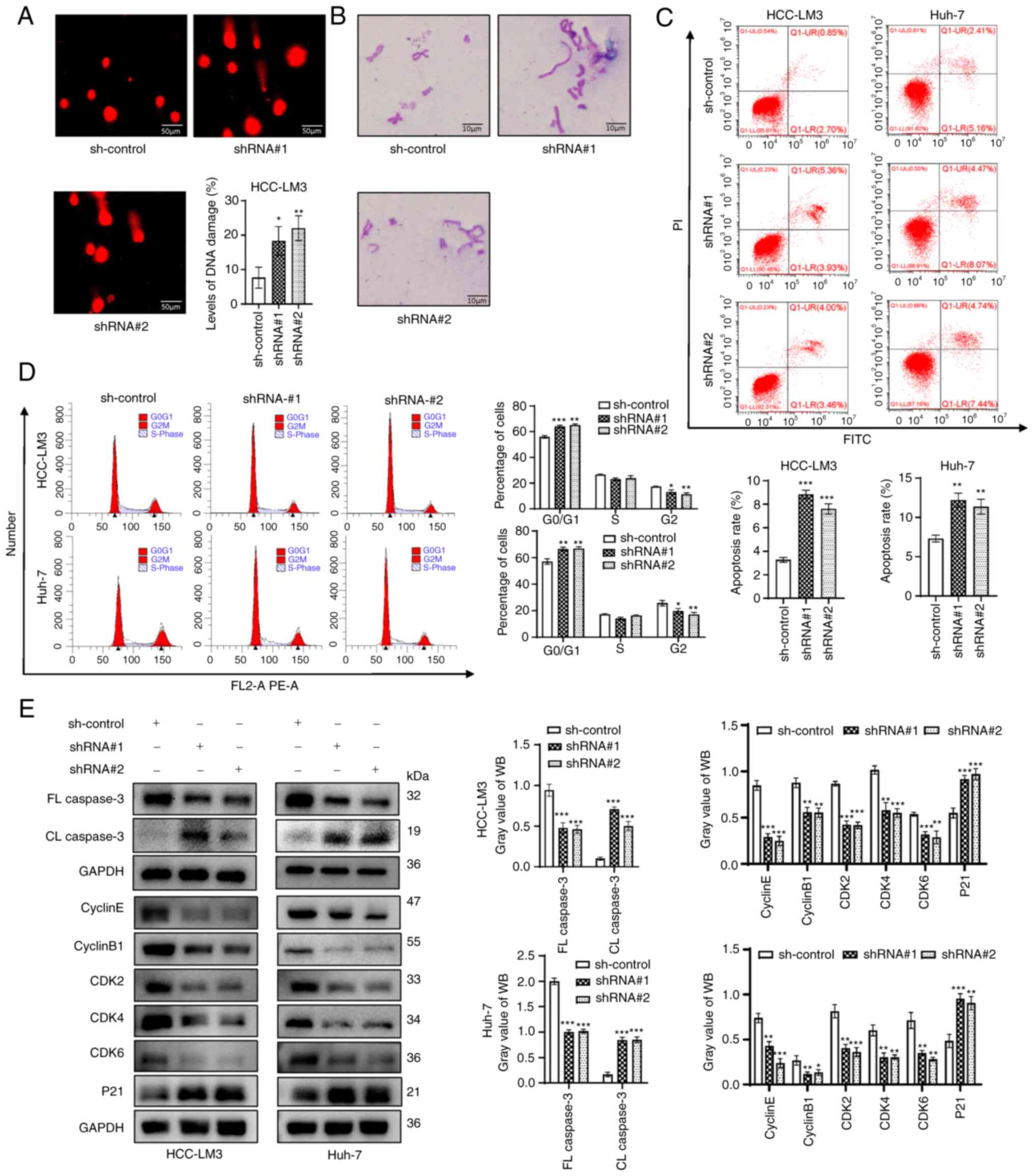 | Figure 3ZFPM2-AS1 knockdown induced DNA
damage in hepatoma cells. (A) Level of DNA damage in HCC-LM3 cells
(scale bar, 50 µm). (B) Distribution of chromosomal
aberrations (scale bar, 10 µm). (C and D) Flow cytometry
indicated that ZFPM2-AS1 knockdown (C) promoted apoptosis and (D)
induced G1 arrest in hepatoma cells. (E) The effect of ZFPM2-AS1
knockdown on apoptosis and cell cycle progression was verified
using WB assays. The error bars and bars indicate the mean and
standard deviation of at least three independent experiments.
*P<0.05; **P<0.01;
***P<0.001. FL, full length; CL, cleaved length; CDK,
cyclin-dependent kinase; shRNA, short hairpin RNA; HCC,
hepatocellular carcinoma; WB, western blot; PI, propidium iodide;
ZFPM2-AS1, zinc finger protein, FOG family member 2 antisense
1. |
ZFPM2-AS1 overexpression promotes
hepatoma-cell growth and DNA repair
A plasmid was constructed to overexpress ZFPM2-AS1
in Hep3B cells, with the empty pcDNA3.1 vector used as a negative
control. The overexpression efficiency of ZFPM2-AS1 was verified by
RT-qPCR (Fig. 4A). It was
confirmed that overexpression of ZFPM2-AS1 promoted the
proliferation, migration and invasion ability of hepatoma cells
(Fig. 4B-E). Flow-cytometric
analysis revealed that the apoptosis rate decreased and the S-phase
population increased after pcDNA/ZFPM2-AS1 overexpression (Fig. 4F and G). Similarly, WB results
revealed that FL caspase3 was upregulated and CL caspase 3 was
downregulated after ZFPM2-AS1 overexpression. In addition,
ZFPM2-AS1 overexpression downregulated P21 and significantly
upregulated cyclin E, cyclin B1, CDK2, CDK4 and CDK6 (Fig. 4H). Cell DNA damage in the ZFPM2-AS1
overexpression group was significantly decreased compared with that
in the control group, according to the comet assay (P<0.01;
Fig. 4I).
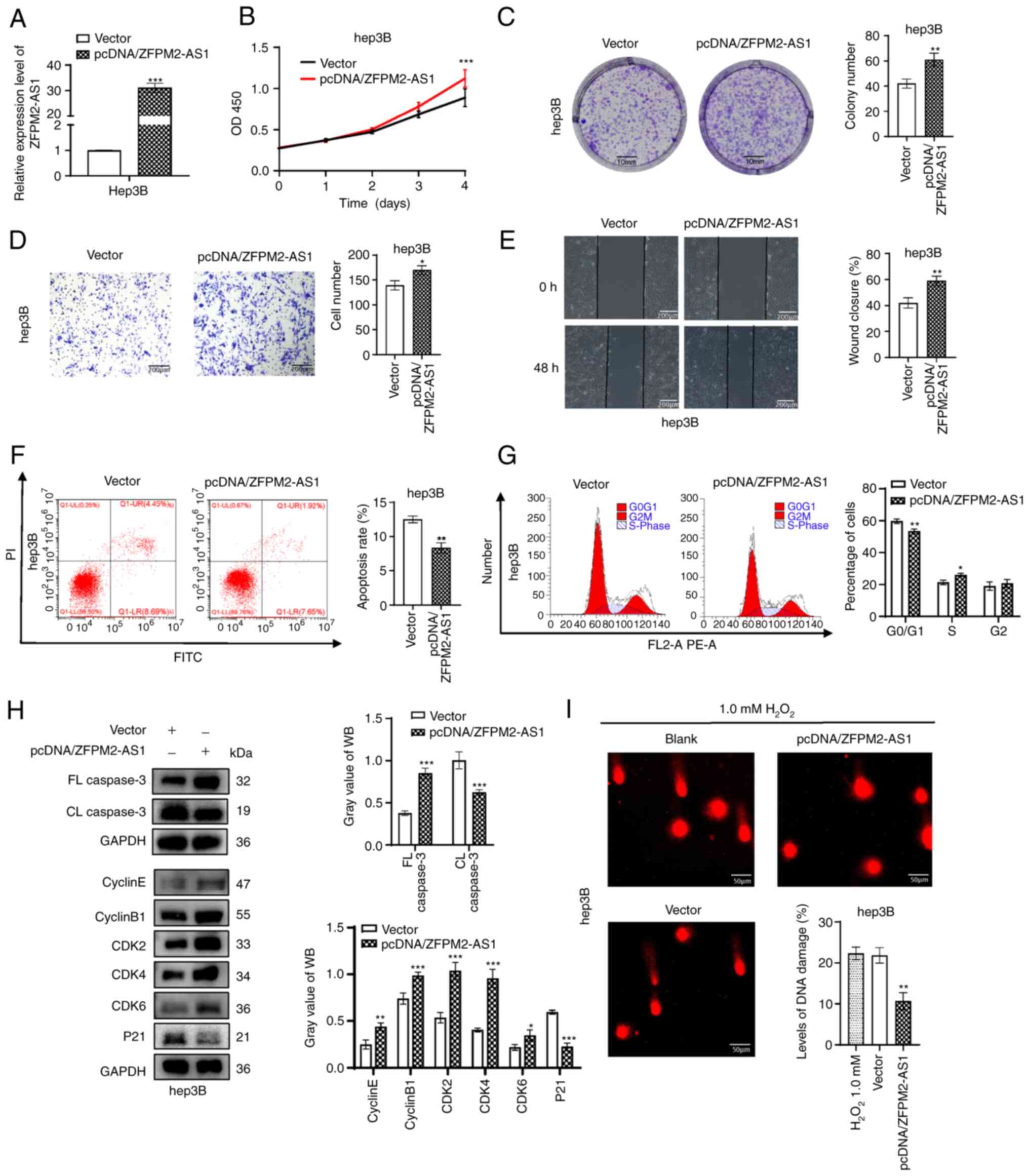 | Figure 4ZFPM2-AS1 overexpression promotes
hepatoma cell progression. (A) Expression of ZFPM2-AS1 in Hep3B
cells after transfection with pcDNA/ZFPM2-AS1. (B) Cell Counting
Kit-8 and (C) plate colony formation assays (scale bars, 10 mm)
indicated that ZFPM2-AS1 overexpression facilitated the
proliferation of Hep3B cells. (D and E) Cell invasion and migration
increased after ZFPM2-AS1 overexpression, as indicated by the (D)
Transwell assay and (E) wound-healing assay, respectively (scale
bars, 200 µm). (F) Apoptosis and (G) the cell cycle were
assessed using flow cytometric analysis. (H) WB analyses revealed
the expression of apoptosis- and cell cycle-related proteins. (I)
An alkaline comet assay indicated that the DNA damage level was
reduced after ZFPM2-AS1 overexpression in Hep3B cells.
H2O2 (1.0 mM) was added to induce cell DNA
damage (scale bars, 50 µm). The error bars and bars indicate
the mean and standard deviation of at least three independent
experiments. *P<0.05; **P<0.01;
***P<0.001. OD, optical density; d, days; FL, full
length; CL, cleaved length; CDK, cyclin-dependent kinase; WB,
western blot; PI, propidium iodide; ZFPM2-AS1, zinc finger protein,
FOG family member 2 antisense 1. |
XRCC4 is influenced by ZFPM2-AS1 and
miR-3065-5p
The GIlncsig indicated that XRCC4, BRCA1 and RAD51
were expressed at higher levels in the high-risk group compared to
the low-risk group. Therefore, it was investigated whether
ZFPM2-AS1 influences the expression levels of these three proteins.
After knockdown of ZFPM2-AS1 in HCC-LM3 and Huh-7 cells, XRCC4 was
significantly downregulated. However, BRCA1 and RAD51 were not
significantly altered (Fig. 5A and
B). When ZFPM2-AS1 was overexpressed in Hep3B cells, the
opposite result was observed (P<0.001; Fig. 5C and D). ZFPM2-AS1 was positively
correlated with XRCC4 in the TCGA dataset (R=0.41, P<0.001;
Fig. 5E). IHC analysis revealed
that ZFPM2-AS1 knockdown downregulated XRCC4 expression in
subcutaneous tumors (Fig. 5F).
Since ZFPM2-AS1 altered XRCC4 at the mRNA and protein levels, it
was hypothesised that ZFPM2-AS1 affected the transcriptional
regulation of XRCC4 via a ceRNA mechanism. Subsequently, five
candidate miRNAs (miR-548aj-3p, miR-548j-3p, miR-186-5p,
miR-3065-5p and miR-548x-3p) were predicted in a screening for
their ability to bind to ZFPM2-AS1 and XRCC4 by miRDB, LncBase and
TargetScan (Fig. 5G). The mRNA
expression of XRCC4 was significantly downregulated when hepatoma
cells were transfected with miR-3065-5p mimics (P<0.001;
Fig. 5H). This result was
consistent with the WB results (P<0.001, Fig. 5I). In addition, a correlation
analysis performed using the TCGA dataset revealed that miR-3065-5p
expression was negatively correlated with XRCC4 and with ZFPM2-AS1
(R=-0.25 and -0.29, respectively, P<0.001; Fig. 5J). Nuclear and cytoplasmic RNA
fractionation assays indicated that ZFPM2-AS1 was primarily
expressed in the tumor cell cytoplasm (Fig. 5K).
 | Figure 5Expression of XRCC4 is affected by
ZFPM2-AS1 and miR-3065-5p. (A and B) The expression levels of three
DNA repair proteins after ZFPM2-AS1 knockdown in HCC-LM3 and Huh-7
cells were detected using (A) RT-qPCR and (B) WB assays. (C and D)
Expression level of XRCC4 after ZFPM2-AS1 overexpression in Hep3B
cells, as detected by (C) RT-qPCR and (D) WB. (E) Pearson
correlation analysis was performed to evaluate the correlation
between ZFPM2-AS1 and XRCC4 in the The Cancer Genome Atlas dataset.
(F) XRCC4 was reduced after ZFPM2-AS1 knockdown, as indicated by
IHC analysis (magnification, ×200 and ×400). Quantification of IHC
analysis of XRCC4 expression. (G) Venn diagram indicating the
predicted target miRNAs of ZFPM2-AS1 and XRCC4 from databases. (H)
mRNA expression of XRCC4 after transfection of miRNA mimics in
HCC-LM3 cells. (I) XRCC4 was significantly reduced by miR-3065-5p,
as indicated by WB assay. (J) Pearson correlation analyses
identified negative correlations between miR-3065-5p and XRCC4 or
between miR-3065-5p and ZFPM2-AS1. (K) Nuclear and cytoplasmic RNA
fractions were isolated from HCC-LM3 cells and analyzed by RT-qPCR.
The error bars and bars indicate the mean and standard deviation of
at least three independent experiments. *P<0.05;
**P<0.01; ***P<0.001; ns, not
significant. WB, western blot; RT-qPCR, reverse
transcription-quantitative PCR; miR, microRNA; shRNA, short hairpin
RNA; NC, negative control; IHC, immunohistochemistry; MOD, mean
optical density; ZFPM2-AS1, zinc finger protein, FOG family member
2 antisense 1; RP, reads per kilobase per million mapped reads;
XRCC4, X-ray repair cross complementing 4; BRCA1, breast cancer
gene 1. |
ZFPM2-AS1 regulates XRCC4 by sponging
miR-3065-5p
To further verify the present hypothesis, binding
sites for miR-3065-5p and XRCC4 were predicted through the
TargetScan database, and XRCC4 mutant plasmids were designed and
synthesized (Fig. 6A).
Furthermore, miR-3065-5p mimics and inhibitor were constructed,
which had good transfection efficiency and significantly regulated
mRNA expression levels of XRCC4 in hepatoma cells (Fig. 6B-D). A luciferase reporter assay
revealed the interaction between miR-3065-5p and XRCC4, while no
significant binding between miR-3065-5p and the XRCC4 mutant was
observed (Fig. 6E). Similarly, the
binding site between miR-3065-5p and ZFPM2-AS1 was predicted and
ZFPM2-AS1 mutant plasmids were constructed (Fig. 6F). A luciferase reporter assay
demonstrated the direct interaction between miR-3065-5p and
ZFPM2-AS1, while miR-3065-5p did not significantly bind to the
ZFPM2-AS1 mutant sequence (Fig.
6G). An RNA pull-down assay confirmed that ZFPM2-AS1 was
significantly enriched by biotinylated miR-3065-5p WT (Fig. 6H). The ZFPM2-AS1 mutant had no
effect on XRCC4, but the miR-3065-5p inhibitor markedly upregulated
XRCC4 at the protein expression level (Fig. 6I). Through RT-qPCR and WB assays,
it was verified that ZFPM2-AS1 regulated XRCC4 expression via
miR-3065-5p (Fig. 6J and K).
Overall, ZFPM2-AS1 acted as a ceRNA to regulate XRCC4 by binding to
miR-3065-5p.
 | Figure 6Mechanisms by which ZFPM2-AS1
regulates XRCC4. (A) Predicted binding sites for miR-3065-5p in the
3′ UTR of XRCC4 and mutations in the binding sites are presented.
(B and C) Expression levels of miR-3065-5p after transfection with
(B) miR-3065-5p mimics and (C) inhibitor. (D) Effect of the
miR-3065-5p mimics and inhibitor on XRCC4 expression. (E) Binding
of miR-3065-5p and XRCC4 was examined using luciferase reporter
assays. (F) Predicted binding sites for miR-3065-5p in the 3′ UTR
of ZFPM2-AS1 and mutations in the binding sites. (G) A luciferase
reporter assay confirmed the direct interaction between miR-3065-5p
and ZFPM2-AS1. (H) RNA pull-down indicated enrichment of ZFPM2-AS1
by application of biotin-labeled miR-3065-5p WT. (I) XRCC4
expression was upregulated by the miR-3065-5p inhibitor, as
detected using WB assays. (J) RT-qPCR assay and (K) WB analysis
indicated that the low expression level of XRCC4 was rescued after
transfection with a miR-3065-5p inhibitor. The error bars and bars
indicate the mean and standard deviation of at least three
independent experiments. **P<0.01;
***P<0.001; ns, not significant. miR, microRNA; WB,
western blot; RT-qPCR, reverse transcription-quantitative PCR; UTR,
untranslated region; WT, wild-type; MUT, mutated; ZFPM2-AS1, zinc
finger protein, FOG family member 2 antisense 1; XRCC4, X-ray
repair cross complementing 4. |
ZFPM2-AS1 regulates XRCC4 to accelerate
HCC progression by competitively binding to miR-3065-5p
To demonstrate that ZFPM2-AS1 promotes HCC by
regulating miR-3065-5p/XRCC4 axis, rescue experiments were
performed (Figs. S6 and 7). The
overexpression efficiency of pcDNA/XRCC4 was examined in HCC-LM3
cells by RT-qPCR and WB (Fig.
S6A). According to CCK-8 and plate colony-formation assays, the
cell proliferation ability was markedly decreased after ZFPM2-AS1
knockdown. However, when co-transducing with miR-3065-5p inhibitor
or co-transfecting with pcDNA/XRCC4, the cell proliferation ability
recovered noticeably again (Fig. 7A
and B). In parallel, the miR-3065-5p inhibitor or pcDNA/XRCC4
reversed the high apoptosis rate and G1-phase arrest caused by
ZFPM2-AS1 knockdown (Fig. 7C and
D). The cell apoptosis and cycle-related protein expression
detected by WB assay further validated the above conclusions
(Fig. 7E). The Transwell and
wound-healing assays demonstrated that the miR-3065-5p inhibitor or
pcDNA/XRCC4 restored the inhibition of cell migration and invasion
ability caused by ZFPM2-AS1 knockdown (Fig. S6B-D). In addition, the promoting
effects of ZFPM2-AS1 knockdown on cell DNA damage were reversed by
transduction with miR-3065-5p inhibitor or transfection with
pcDNA/XRCC4 (Fig. 7F and G).
 | Figure 7Zinc finger protein, FOG family
member 2 antisense 1 facilitates hepatocellular carcinoma growth by
regulating miR-3065-5p-mediated XRCC4. (A and B) Rescue experiments
were conducted using sh-control, shRNA#1+mock, shRNA#1+miR
inhibitor, shRNA#1+vector and shRNA#1+pcDNA/XRCC4 groups, with (A)
Cell Counting Kit-8 and (B) colony-formation assays (scale bars, 10
mm). (C and D) miR-3065-5p inhibitor or pcDNA/XRCC4 rescued (C) the
hepatoma cell cycle and (D) apoptosis as assessed by flow
cytometry. (E) Apoptosis and cell cycle-related proteins were
detected using WB assays. (F and G) The DNA damage level was
rescued after transduction with (F) the miR-3065-5p inhibitor or
(G) transfection with pcDNA/XRCC4 (scale bars, 50 µm). The
error bars and bars indicate the mean and standard deviation of at
least three independent experiments. *P<0.05;
**P<0.01; ***P<0.001. OD, optical
density; d, days; FL, full length; CL, cleaved length; CDK,
cyclin-dependent kinase; WB, western blot; PI, propidium iodide;
shRNA, short hairpin RNA; d, days; miR, microRNA; XRCC4, X-ray
repair cross complementing 4. |
Discussion
Cancer develops from the clonal proliferation of a
single aberrant cell (26). The
cell frequently undergoes irreversible gene mutation and obtains
functions that drive transformation into cancer. Mutations result
from replication errors or DNA damage that is improperly repaired
(27). Therefore, an abnormal DNA
damage repair pathway is closely related to cancer progression.
An in-depth study of lncRNAs suggests that they have
a significant role in cancer development (28,29).
They lack the ability to encode proteins, yet have regulatory
functions. LncRNA ENO1 intronic transcript 1 contributes to cancer
growth by promoting glycolysis (30). Through the stabilization of PD-L1
protein and the degradation of GATA3 protein, lncRNA GATA3-AS1
promotes tumor immune evasion (31). Pivotal lncRNAs function as hinges
between tumor metabolism and immunology (32). However, only a small number of
studies have investigated lncRNAs related to GS. Therefore, the
present study was performed and 85 GI-associated lncRNAs were
identified. To examine the potential value of GI-related lncRNAs, a
model composed of six lncRNAs was developed by Cox regression
analysis. The model was then verified and a favorable prediction
efficiency analysis was performed. The expression of DNA
repair-related genes was significantly higher in the high-risk
group. Among the DNA repair-related genes, BRCA1, RAD51 and XRCC4
were the most significant. The XRCC4, BRCA1 and RAD51 proteins
promote DNA repair (33-36) and have an important role in
maintaining genomic stability (37). Of note, the model still had
excellent prognostic ability in the GEO validation groups.
Therefore, the prognostic model is expected to be adopted by
physicians in clinical applications. A significant proportion of
patients with HCC with TP53 mutations have poor prognosis (38,39).
The combination of GIlncsig and TP53 mutation status will provide a
more accurate prognostic indicator for the diagnosis and treatment
of HCC patients.
Among the six lncRNAs of the GIlncsig, the
literature reports that LUCAT1, ZFPM2-AS1 and MIR210HG promote the
progression of HCC (40-42). The biological functions of
AC004862.1 and AC010205.1 have not been reported and potassium
calcium-activated channel subfamily M regulatory β subunit 2
antisense 1 has not been reported in HCC. None of the six lncRNAs
have been reported to affect GS. In the present study, ZFPM2-AS1
was selected as the focus of subsequent analyses, as it had the
largest coefficient in the GIlncsig and the largest hazard
ratio.
In the present study, ZFPM2-AS1 was indicated to be
significantly upregulated in tumor tissues. ZFPM2-AS1 was
demonstrated to increase the proliferation, migration and invasion
ability of hepatoma cells in vitro and in vivo. The
effects of ZFPM2-AS1 on apoptosis and cell-cycle progression were
also verified using WB assays. Of note, the present study was the
first to indicate that ZFPM2-AS1 knockdown increased DNA damage and
chromosome aberrations in hepatoma cells. CeRNA is an important
regulatory mechanism of lncRNAs (43). LncRNAs may serve as sponges for
miRNAs through identical binding sites and modulate the function of
miRNAs on their target mRNAs (44-46).
The present study proved that ZFPM2-AS1 functions as a ceRNA to
influence miR-3065-5p activity and regulates XRCC4. XRCC4 is an
NHEJ protein that is recruited to DNA damage sites to mediate
double-strand break repair and cell survival (47,48).
In addition, XRCC4 increases the malignancy of breast cancer
(49) but has not been reported in
HCC, to the best of our knowledge. The present study discovered for
the first time that miR-3065-5p is able to regulate DNA damage
repair by mediating XRCC4. From the results of the luciferase
reporter and RNA pull-down assays, it was indicated that
miR-3065-5p directly interacted with ZFPM2-AS1 and XRCC4. Through
rescue experiments, a new axis (ZFPM2-AS1/miR-3065-5p/XRCC4) in HCC
was determined. This axis would be expected to be a key target for
interfering with the progression of HCC in the future.
There are certain limitations to this study.
Although the GIlncSig was applied to test its clinical predictive
value in the TCGA and GEO samples, it requires further validation
in a large cohort of tissue samples from clinical patients. It
holds promise as a useful model in the prognosis prediction of
patients with HCC. The other five lncRNAs in the GIlncsig were not
validated in the initial experiments, and these lncRNAs will be
validated in a follow-up study. Although ZFPM2-AS1 has been
reported numerous times, its relationship with GS has not been
previously reported, to the best of our knowledge. In the present
study, ZFPM2-AS1 was analyzed because it was the most significant
lncRNA in the current model. It was discovered that ZFPM2-AS1 had
the ability to promote lung metastasis of HCC in vivo. In
addition, an entirely novel ceRNA mechanism in HCC was
investigated.
In conclusion, a prognostic model with excellent
predictive performance for HCC was constructed. Through in
vivo and in vitro functional experiments, it was
verified that ZFPM2-AS1 promotes HCC progression and DNA damage
repair. ZFPM2-AS1 was found to act as a ceRNA to promote GS and HCC
proliferation via miR-3065-5p/XRCC4 (Fig. 8). During tumor progression, the
genomic instability of tumors generally increases. However, the
present results indicated that ZFPM2-AS1 facilitated DNA damage
repair by upregulating XRCC4 to promote tumor progression,
suggesting that GS may be a double-edged sword. The underlying
mechanisms should be further investigated. The present study
provides insight into the molecular mechanisms underlying GS and
may offer a potential therapeutic strategy against HCC in the
future.
 | Figure 8Mechanistic map depicting the role of
the ZFPM2-AS1/miR-3065-5p/XRCC4 axis in the progression of HCC.
ZFPM2-AS1 acted as a ceRNA by binding to miR-3065-5p. miR-3065-5p
was loaded into the RISC and guided this complex to the 3′ UTR of
XRCC4 mRNA, leading to suppressed XRCC4 protein expression.
Consequently, ZFPM2-AS1 upregulated XRCC4 by sponging miR-3065-5p
to promote genomic stability and HCC progression. miR, microRNA;
RISC, RNA-induced silencing complex; ceRNA, competing endogenous
RNA; UTR, untranslated region; HCC, hepatocellular carcinoma;
ZFPM2-AS1, zinc finger protein, FOG family member 2 antisense 1;
XRCC4, X-ray repair cross complementing 4. |
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HZ, ZL and JL designed experiments. HZ performed the
bioinformatics analysis. JL, HZ, KX, YZ, and PX performed and
analyzed the data. YZ assisted with the western blot analysis and
methodology. KX concentrated primarily on the cell screening. JL,
HZ and PX wrote the manuscript. JL was a major contributor in
writing the manuscript. HZ, ZL and YY supervised the research. ZL
and YY revised the manuscript critically for important intellectual
content. YY provided the study's facilities. ZL and YY applied for
the funding. JL and HZ confirmed the authenticity of all the raw
data. ZL and YY agreed to be accountable for all aspects of the
work. All authors have read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
This study conformed to the Declaration of Helsinki.
The tissue samples were collected with the approval of the ethics
committee of Zhongnan Hospital (Wuhan, China; no. KELUN2020100).
All of the patients provided written informed consent. All animal
experiments were performed according to ethical guidelines and were
approved by the ethics committee of Zhongnan Hospital (Wuhan,
China; no. ZN2022006).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
This work was supported by the Research Fund of the Health
Commission of Hubei Province (grant no. WJ2021M255); Cancer
Research and Translational Platform Project of Zhongnan Hospital of
Wuhan University (grant no. ZLYNXM202004); Grant from the Key
Research and Development Program of Hubei Province, China (grant
no. 2021BCA114); and the Translational Medicine and
Interdisciplinary Research Joint Fund Project of Zhongnan Hospital
of Wuhan University (grant no. ZNJC201918).
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Calderaro J, Couchy G, Imbeaud S, Amaddeo
G, Letouzé E, Blanc JF, Laurent C, Hajji Y, Azoulay D, Bioulac-Sage
P, et al: Histological subtypes of hepatocellular carcinoma are
related to gene mutations and molecular tumour classification. J
Hepatol. 67:727–738. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ruiz de Galarreta M, Bresnahan E,
Molina-Sánchez P, Lindblad KE, Maier B, Sia D, Puigvehi M, Miguela
V, Casanova-Acebes M, Dhainaut M, et al: β-Catenin activation
promotes immune escape and resistance to Anti-PD-1 therapy in
hepatocellular carcinoma. Cancer Discov. 9:1124–1141. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chaudhary R and Lal A: Long noncoding RNAs
in the p53 network. Wiley Interdiscip Rev RNA. 8. pp.
10.1002/wrna.14102017, View Article : Google Scholar
|
|
5
|
Feng J, Yang G, Liu Y, Gao Y, Zhao M, Bu
Y, Yuan H, Yuan Y, Yun H, Sun M, et al: LncRNA PCNAP1 modulates
hepatitis B virus replication and enhances tumor growth of liver
cancer. Theranostics. 9:5227–5245. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang H, Ma P, Liu P, Guo D, Liu Z and
Zhang Z: lncRNA SNHG6 promotes hepatocellular carcinoma progression
by interacting with HNRNPL/PTBP1 to facilitate SETD7/LZTFL1 mRNA
destabilization. Cancer Lett. 520:121–131. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ishibashi M, Kogo R, Shibata K, Sawada G,
Takahashi Y, Kurashige J, Akiyoshi S, Sasaki S, Iwaya T, Sudo T, et
al: Clinical significance of the expression of long non-coding RNA
HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 29:946–950.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang C, Li Y, Yan S, Wang H, Shao X, Xiao
M, Yang B, Qin G, Kong R, Chen R and Zhang N: Interactome analysis
reveals that lncRNA HULC promotes aerobic glycolysis through LDHA
and PKM2. Nat Commun. 11:31622020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vishnubalaji R, Shaath H, Elango R and
Alajez NM: Noncoding RNAs as potential mediators of resistance to
cancer immunotherapy. Semin Cancer Biol. 65:65–79. 2020. View Article : Google Scholar
|
|
10
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin X, Zhuang S, Chen X, Du J, Zhong L,
Ding J, Wang L, Yi J, Hu G, Tang G, et al: lncRNA ITGB8-AS1
functions as a ceRNA to promote colorectal cancer growth and
migration through integrin-mediated focal adhesion signaling. Mol
Ther. 30:688–702. 2022. View Article : Google Scholar
|
|
12
|
Jeggo PA, Pearl LH and Carr AM: DNA
repair, genome stability and cancer: A historical perspective. Nat
Rev Cancer. 16:35–42. 2016. View Article : Google Scholar
|
|
13
|
Moynahan ME and Jasin M: Mitotic
homologous recombination maintains genomic stability and suppresses
tumorigenesis. Nat Rev Mol Cell Biol. 11:196–207. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wickramasinghe VO and Venkitaraman AR: RNA
processing and genome stability: Cause and consequence. Mol Cell.
61:496–505. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shen L, Wang Q, Liu R, Chen Z, Zhang X,
Zhou P and Wang Z: LncRNA lnc-RI regulates homologous recombination
repair of DNA double-strand breaks by stabilizing RAD51 mRNA as a
competitive endogenous RNA. Nucleic Acids Res. 46:717–729. 2018.
View Article : Google Scholar :
|
|
16
|
Munschauer M, Nguyen CT, Sirokman K,
Hartigan CR, Hogstrom L, Engreitz JM, Ulirsch JC, Fulco CP,
Subramanian V, Chen J, et al: The NORAD lncRNA assembles a
topoisomerase complex critical for genome stability. Nature.
561:132–136. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee S, Kopp F, Chang TC, Sataluri A, Chen
B, Sivakumar S, Yu H, Xie Y and Mendell JT: Noncoding RNA NORAD
regulates genomic stability by sequestering PUMILIO Proteins. Cell.
164:69–80. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
López EH and Palumbi SR: Somatic mutations
and genome stability maintenance in clonal coral colonies. Mol Biol
Evol. 37:828–838. 2020. View Article : Google Scholar
|
|
19
|
Roessler S, Jia HL, Budhu A, Forgues M, Ye
QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX and Wang XW: A
unique metastasis gene signature enables prediction of tumor
relapse in early-stage hepatocellular carcinoma patients. Cancer
Res. 70:10202–10212. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Grinchuk OV, Yenamandra SP, Iyer R, Singh
M, Lee HK, Lim KH, Chow PK and Kuznetsov VA: Tumor-adjacent tissue
co-expression profile analysis reveals pro-oncogenic ribosomal gene
signature for prognosis of resectable hepatocellular carcinoma. Mol
Oncol. 12:89–113. 2018. View Article : Google Scholar
|
|
21
|
Chen X, Ma W, Yao Y, Zhang Q, Li J, Wu X,
Mei C, Jiang X, Chen Y, Wang G, et al: Serum deprivation-response
protein induces apoptosis in hepatocellular carcinoma through
ASK1-JNK/p38 MAPK pathways. Cell Death Dis. 12:4252021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Xia P, Zhang H, Xu K, Jiang X, Gao M, Wang
G, Liu Y, Yao Y, Chen X, Ma W, et al: MYC-targeted WDR4 promotes
proliferation, metastasis, and sorafenib resistance by inducing
CCNB1 translation in hepatocellular carcinoma. Cell Death Dis.
12:6912021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cai J, Wang N, Lin G, Zhang H, Xie W,
Zhang Y and Xu N: MBNL2 Regulates DNA damage response via
stabilizing p21. Int J Mol Sci. 22:7832021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lord CJ and Ashworth A: The DNA damage
response and cancer therapy. Nature. 481:287–294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Martincorena I and Campbell PJ: Somatic
mutation in cancer and normal cells. Science. 349:1483–1489. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Al Zouabi L and Bardin AJ: Stem cell DNA
damage and genome mutation in the context of aging and cancer
initiation. Cold Spring Harb Perspect Biol. 12:a0362102020.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Elguindy MM and Mendell JT: NORAD-induced
Pumilio phase separation is required for genome stability. Nature.
595:303–308. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang Y, He Q, Hu Z, Feng Y, Fan L, Tang
Z, Yuan J, Shan W, Li C, Hu X, et al: Long noncoding RNA LINP1
regulates repair of DNA double-strand breaks in triple-negative
breast cancer. Nat Struct Mol Biol. 23:522–530. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hong J, Guo F, Lu SY, Shen C, Ma D, Zhang
X, Xie Y, Yan T, Yu T, Sun T, et al: F: Nucleatum targets lncRNA
ENO1-IT1 to promote glycolysis and oncogenesis in colorectal
cancer. Gut. 70:2123–2137. 2021. View Article : Google Scholar
|
|
31
|
Zhang M, Wang N, Song P, Fu Y, Ren Y, Li Z
and Wang J: LncRNA GATA3-AS1 facilitates tumour progression and
immune escape in triple-negative breast cancer through
destabilization of GATA3 but stabilization of PD-L1. Cell Prolif.
53:e128552020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang J, Liu F, Wang Y, Qu L and Lin A:
LncRNAs in tumor metabolic reprogramming and immune
microenvironment remodeling. Cancer Lett. 543:2157982022.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen S, Lee L, Naila T, Fishbain S, Wang
A, Tomkinson AE, Lees-Miller SP and He Y: Structural basis of
long-range to short-range synaptic transition in NHEJ. Nature.
593:294–298. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hatchi E, Goehring L, Landini S,
Skourti-Stathaki K, DeConti DK, Abderazzaq FO, Banerjee P, Demers
TM, Wang YE, Quackenbush J and Livingston DM: BRCA1 and RNAi
factors promote repair mediated by small RNAs and PALB2-RAD52.
Nature. 591:665–670. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Feretzaki M, Pospisilova M, Valador
Fernandes R, Lunardi T, Krejci L and Lingner J: RAD51-dependent
recruitment of TERRA lncRNA to telomeres through R-loops. Nature.
587:303–308. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim S, Jin H, Seo HR, Lee HJ and Lee YS:
Regulating BRCA1 protein stability by cathepsin S-mediated
ubiquitin degradation. Cell Death Differ. 26:812–825. 2019.
View Article : Google Scholar :
|
|
37
|
Arnould C, Rocher V, Finoux AL, Clouaire
T, Li K, Zhou F, Caron P, Mangeot PE, Ricci EP, Mourad R, et al:
Loop extrusion as a mechanism for formation of DNA damage repair
foci. Nature. 590:660–665. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hussain SP, Schwank J, Staib F, Wang XW
and Harris CC: TP53 mutations and hepatocellular carcinoma:
Insights into the etiology and pathogenesis of liver cancer.
Oncogene. 26:2166–2176. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Woo HG, Wang XW, Budhu A, Kim YH, Kwon SM,
Tang ZY, Sun Z, Harris CC and Thorgeirsson SS: Association of TP53
mutations with stem cell-like gene expression and survival of
patients with hepatocellular carcinoma. Gastroenterology.
140:1063–1070. 2011. View Article : Google Scholar
|
|
40
|
Lou Y, Yu Y, Xu X, Zhou S, Shen H, Fan T,
Wu D, Yin J and Li G: Long non-coding RNA LUCAT1 promotes
tumourigenesis by inhibiting ANXA2 phosphorylation in
hepatocellular carcinoma. J Cell Mol Med. 23:1873–1884. 2019.
View Article : Google Scholar
|
|
41
|
He H, Wang Y, Ye P, Yi D, Cheng Y, Tang H,
Zhu Z, Wang X and Jin S: Long noncoding RNA ZFPM2-AS1 acts as a
miRNA sponge and promotes cell invasion through regulation of
miR-139/GDF10 in hepatocellular carcinoma. J Exp Clin Cancer Res.
39:1592020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang Y, Li W, Chen X, Li Y, Wen P and Xu
F: MIR210HG predicts poor prognosis and functions as an oncogenic
lncRNA in hepatocellular carcinoma. Biomed Pharmacother.
111:1297–1301. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yoon JH, Abdelmohsen K and Gorospe M:
Posttranscriptional gene regulation by long noncoding RNA. J Mol
Biol. 425:3723–3730. 2013. View Article : Google Scholar
|
|
44
|
Chen S, Xie C and Hu X: lncRNA SNHG6
functions as a ceRNA to up-regulate c-Myc expression via sponging
let-7c-5p in hepatocellular carcinoma. Biochem Biophys Res Commun.
519:901–908. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB,
Yin DD, Kong R, Xia R, Lu KH, Li JH, et al: Lnc RNA HOTAIR
functions as a competing endogenous RNA to regulate HER2 expression
by sponging miR-331-3p in gastric cancer. Mol Cancer. 13:922014.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhao CC, Jiao Y, Zhang YY, Ning J, Zhang
YR, Xu J, Wei W and Kang-Sheng G: Lnc SMAD5-AS1 as ceRNA inhibit
proliferation of diffuse large B cell lymphoma via Wnt/β-catenin
pathway by sponging miR-135b-5p to elevate expression of APC. Cell
Death Dis. 10:2522019. View Article : Google Scholar
|
|
47
|
Ochi T, Blackford AN, Coates J, Jhujh S,
Mehmood S, Tamura N, Travers J, Wu Q, Draviam VM, Robinson CV, et
al: DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku
to promote DNA double-strand break repair. Science. 347:185–188.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gao Y, Ferguson DO, Xie W, Manis JP,
Sekiguchi J, Frank KM, Chaudhuri J, Horner J, DePinho RA and Alt
FW: Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis,
genomic stability and development. Nature. 404:897–900. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wen Y, Dai G, Wang L, Fu K and Zuo S:
Silencing of XRCC4 increases radiosensitivity of triple-negative
breast cancer cells. Biosci Rep. 39:BSR201808932019. View Article : Google Scholar : PubMed/NCBI
|






















