Introduction
Esophageal cancer is one of the most common
malignant tumors; in 2020, 604,100 new cases and 544,076 deaths
were reported worldwide (1-3).
Esophageal cancer is divided into esophageal squamous cell
carcinoma (ESCC) and esophageal adenocarcinoma. ESCC forms from
esophageal squamous epithelium, which accounts for the majority of
esophageal cancer in China; esophageal adenocarcinoma is a
malignant tumor that occurs from glands, with a relatively low
proportion (4). ESCC is highly
invasive, and undergoes lymph node metastasis. Patients with early
esophageal cancer lack typical clinical symptoms and signs, whereas
the middle and late stages of the malignancy are associated with
esophageal obstruction, lesion infiltration and metastasis, all of
which have a detrimental effect on both the quality of life and the
survival prognosis of patients (5). The main treatment methods for ESCC,
including surgery, chemotherapy, radiotherapy and targeted
therapies, have been widely promoted. The 5-year overall survival
rate of patients with esophageal cancer is <30.3% (6). Even with improvements that have been
made in terms of surgery and the development of multimodal
therapies, the clinical outcome for patients remains unfavorable
(7,8). Therefore, an exhaustive analysis of
the origin of ESCC is necessary to identify novel therapeutic
targets (9-14), and the underlying molecular
mechanisms associated with the origin and invasion of ESCC also
need to be investigated (15).
Ferroptosis is an oxidative, iron-dependent type of
regulated cell death that is distinguished by excessive lipid
peroxidation and iron accumulation, which results in oxidative
damage to the cell membrane (16).
Ferroptosis has been linked to tumor growth and therapeutic
responses in various types of cancer (17). Tumor growth is dependent on the
presence of iron, a trace element (18). In animal studies, increased iron
accumulation was found to promote the occurrence of ferroptosis
(19). Excess iron promotes the
subsequent onset of lipid peroxidation through the iron-dependent
Fenton reaction to generate reactive oxygen species (ROS) and to
activate iron-containing enzymes, such as lipoxygenase (20,21).
Glutathione peroxidase 4 (GPX4), an intracellular antioxidant
enzyme, functions as a fundamental repressor of ferroptosis in
cancer cells by directly reducing phospholipid hydroperoxide to
hydroxyphospholipid (22). The
presence of lipid radicals and the depletion of glutathione (GSH)
are among the biochemical phenomena that underlie ferroptosis
(23,24). GPX4 catalyzes the reduction of
lipid hydroperoxides into lipid alcohols by utilizing GSH as a
reducing cofactor. This important antioxidant system (GSH-GPX4)
functions to protect cells from ferroptosis. By contrast, acyl-CoA
synthetase long-chain family member 4 (ACSL4) catalyzes the
esterification of arachidonoyl and adrenoyl into
phosphatidylethanolamines, and therefore has a role as an important
contributor to ferroptosis (25,26).
p53-mediated transcriptional inhibition of SLC7A11 (one of two
subunits of system xc−) has been shown to promote
ferroptosis in cancer cells (16).
Ferroptosis, therefore, serves a complex and a highly
context-dependent role in tumor biology and therapy.
Transgelin (TAGLN; also known as SM22), an
actinbinding protein, is mainly associated with cytoskeleton
remodeling (27). In several types
of tumor, such as prostate cancer, osteosarcoma and breast cancer,
the expression level of TAGLN has been shown to be reduced, or even
entirely deleted (28-30). An association between the
expression of TAGLN and the occurrence, development and invasion of
tumors has also been reported in several studies (31-33).
A previous study showed that TAGLN may be involved in the migration
of epithelial cells by interacting with actin or promoting podosome
formation, and the downregulation of TAGLN expression could inhibit
tumorigenicity and tumor growth in vivo, producing enhanced
cytotoxic effects of metabolite anticancer drugs on pancreatic
cancer cells (33). A recently
published study demonstrated that cytoskeletal remodeling and
malignant progression in colorectal cancer is regulated through the
interaction of TAGLN with cartilage oligomeric matrix protein; this
interaction was found to occur during epithelial-mesenchymal
transition (EMT) (34). In
addition, a previous publication by our research group demonstrated
that the malignant progression of ESCC could be prevented by TAGLN
through impeding EMT (35).
However, to the best of our knowledge, to date relatively few
studies have investigated the function of TAGLN in different types
of tumors, or its association with ferroptosis.
Therefore, the present study aimed to examine the
influence that TAGLN has on the invasive, metastatic and
proliferative capabilities of ESCC cell lines through the
regulation of ferroptosis. The function and underlying molecular
mechanism of TAGLN in the incidence, development, invasion and
metastasis of ESCC cells is also highlighted, and the potential
therapeutic applications of TAGLN with respect to the
identification and treatment of esophageal cancer are
discussed.
Materials and methods
Clinical sample collection and
information
Esophageal tissue samples from 25 patients with ESCC
(age, 64.1±7.49) and 10 esophageal tissues from healthy controls
(age, 63.58±8.76) were acquired from Tianjin Medical University
General Hospital (Tianjin, China) between January 2021 and December
2022. Prior to the surgery, none of the patients had received any
cancer treatment therapies. The last follow-up date was 31 October
2021. The 8th American Joint Committee on Cancer staging method was
used to categorize the tumor stage and grade of the tissue samples
(36). The present study was
conducted according to the principles set out in The Declaration of
Helsinki of the World Medical Association. Written informed consent
was provided by each patient, and the Ethics Committee of Tianjin
Medical University General Hospital approved the study (ethics no.
IRB2021-WZ-134). The clinicopathological characteristics of the
patients are presented in Table
I.
 | Table IData were analyzed by χ2
to investigate the relationship between TAGLN expression and
clinicopathological characteristics. |
Table I
Data were analyzed by χ2
to investigate the relationship between TAGLN expression and
clinicopathological characteristics.
| Clinicopathological
characteristic | TAGLN expression
|
|---|
| Cases, n=25 | Negative | Positive | P-value |
|---|
| Sex | | | | |
| Male | 23 | 11 | 12 | 0.953 |
| Female | 2 | 1 | 1 | |
| Age, year | | | | |
| <65 | 14 | 6 | 8 | 0.562 |
| ≥65 | 11 | 6 | 5 | |
| Tumor grade | | | | |
| T1 + T2 | 4 | 0 | 4 | 0.036 |
| T3 + T4 | 21 | 12 | 9 | |
| Lymphatic
invasion | | | | |
| N0 | 7 | 1 | 6 | 0.035 |
| N1-N3 | 18 | 11 | 7 | |
|
Differentiation | | | | |
| High +
moderate | 12 | 6 | 6 | 0.8475 |
| Low | 13 | 6 | 7 | |
| AJCC stage | | | | |
| I-II | 5 | 0 | 5 | 0.016 |
| III-IV | 20 | 12 | 8 | |
| Size, cm | | | | |
| <3 | 13 | 5 | 8 | 0.32 |
| ≥3 | 12 | 7 | 5 | |
| Tumor site | | | | |
| Upper and
median | 14 | 6 | 8 | 0.562 |
| Lower | 11 | 6 | 5 | |
Immunohistochemical analysis
Paraformaldehyde (4%) was used to fix the tissue
specimens at room temperature for 24 h. The fixed paraffin-embedded
tissue sections (4-µm thick) were subsequently dewaxed using
xylene and hydrated through graded concentrations of ethanol in
water. Sodium citrate buffer (0.01 mol/l) was used for antigen
retrieval and the endogenous peroxidase activity was blocked by
Enhanced Endogenous Peroxidase Blocking Buffer (cat. no. P0100A;
Beyotime Institute of Biotechnology) at 37°C for 20 min. The
primary antibodies used for the immunohistochemical analysis were
anti-TAGLN (1:100; cat. no. ab233971; Abcam), anti-GPX4 (1:200;
cat. no. DF6701; Affinity Biosciences, Ltd.), anti-ACSL4 (1:400;
cat. no. ab155282; Abcam), anti-p53 (1:800; cat. no. 60283-2-Ig;
ProteinTech Group, Inc.), anti-cyclooxygenase-2 (COX2; 1:200; cat.
no. abp51035; Abbkine Scientific Co., Ltd.), anti-transferrin (TF;
1:400; cat. no. 17435-1-AP; ProteinTech Group, Inc.) and
anti-ferritin heavy chain 1 (FTH1; 1:800; cat. no. abp54401;
Abbkine Scientific Co., Ltd.). These primary antibodies were added
to the tissue sections and incubated at 4°C overnight. Following a
washing step using Immunol Staining Wash Buffer, Beijing Beyotime
company, the tissue sections were incubated for 30 min at 37°C with
the following HRP-conjugated secondary antibodies: Goat anti-mouse
(1:500; cat. no. ab6789; Abcam) and goat anti-rabbit (1:1,000; cat.
no. ab6721; Abcam). Horseradish peroxidase was then added, and the
samples were further incubated at 37°C for 20 min. The sections
were scanned using a PRECICE 510B digital slice scanning system
(Unic Technologies, Inc.), and observed by iViewer software
(version 7.2.7.2; Unic Technologies, Inc.). The staining results
were graded according to the staining intensity and percentage of
positive cells. The cell staining intensity was scored as follows:
0, non-staining; 1, light yellow; 2, brownish yellow; and 3,
yellowish brown. The percentages of positive cells were scored as
follows: 0, <5%, 1, 5-25%; 2, 26-50%; 3, 51-75%; and 4, >75%.
The positive intensity is the product of two scores: 0-2, negative;
3-5, weakly positive; 6-8, positive; and 9-12, strongly positive.
Two pathologists assessed the findings independently, and the
average values of five fields of view were used.
TAGLN expression analysis
The mRNA expression levels of TAGLN between the ESCC
samples and normal tissues were compared using data acquired from
the Gene Expression Omnibus (GEO) records (GSE161533, GSE45670 and
GSE100942; http://www.ncbi.nih.gov/geo).
Gene Set Enrichment Analysis (GSEA)
GSEA was completed using ESCC data obtained from The
Cancer Genome Atlas (TCGA) project to identify the signaling
pathways and biological phenomena that exert a role in the
advancement of ESCC. The RNA-seq transcriptome data of ESCC
patients in fragment per kilobase of transcript per million mapped
reads format were extracted from TCGA. The average TAGLN transcript
level was used to divide the patients with ESCC into high- and
low-expression groups. For each assessment, gene set permutations
were set to 1,000. The pathways enriched for each phenotype were
classified according to the false discovery rate, nominal P-value
and normalized enrichment score (NES). The DESeq algorithm for
differential gene expression analysis was used, and the
significance of DEGs was evaluated by P<0.05 and |log FC|>1
(where FC is fold change) and were used as screening criteria. The
R programming language was then used to generate volcano plots of
the DEGs and to conduct clustering analysis. Volcano plots were
generated using ggplot2, a data visualization package in the R
programming language. Specifically, the geom_point function was
used to plot the log FC values against the negative logarithm of
the adjusted P-values. Similarly, heatmaps were created using the R
package heatmap, using the heatmap.2 function to display the
relative expression levels of genes across different conditions. To
investigate the biological roles of DEGs, biological process (BP)
gene ontology (GO) enrichment analysis and molecular function (MF)
GO term enrichment analysis based on the GO database (http://geneontology.org; accessed on April 5, 2022)
was performed. Kyoto Encyclopedia of Genes and Genomes (KEGG)
database (https://www.kegg.jp/kegg; accessed on
April 5, 2022) analysis was performed for further assessment of the
signaling pathways.
Culture of human ESCC cells
The human ESCC cell lines (KYSE-150 and Eca-109)
were obtained from The Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences. Cells were cultured in RPMI-1640
medium containing 10% FBS (Gibco®; Thermo Fisher
Scientific, Inc.) and maintained at 37°C in an atmosphere of 5%
CO2.
Cell transfection
For cell transfection experiments, the Eca-109 and
KYSE-150 cells were added at a density of 5×105
cells/well into a 6-well plate. When confluency was 70-90%, cells
were transfected using Lipofectamine 3000™ Transfection Reagent
(Thermo Fisher Scientific, Inc.) following the manufacturer's
guidelines. The TAGLN-overexpression plasmid (cat. no. HG14991-NY;
TAGLN cDNA ORF Clone; human, pCMV3-N-HA as vector; Sino Biological)
and an empty control plasmid (vector-NC; cat. no. CV017; pCMV3-N-HA
as negative control vector; Sino Biological) were transfected into
the KYSE-150 and Eca-109 cells. The final transfection
concentration of plasmid was 100 nmol/l. After 6-8 h of culture in
10% serum medium without penicillin-streptomycin at 37°C, the
culture was replaced to 10% serum complete medium for 24-48 h.
To avoid off-target results, two different
gene-specific siRNAs against TAGLN (si-TAGLN; one from Santa Cruz
Biotechnology, Inc. (cat. no. sc-44163) and the other from Shanghai
GenePharma Co., Ltd. (sense, 5′-GCU GAA GAA UGG CGU GAU UTT-3′;
antisense, 5′-AAU CAC GCC AUU CUU CSG CTT-3′), and the
corresponding siRNA negative control (si-NC; cat no. sc-37007;
Santa Cruz Biotechnology, Inc.) were transfected into KYSE-150 and
Eca-109 cells, as aforementioned (using Lipofectamine 3000™; the
final transfection concentration of siRNAs was 10 nmol/l). After
culturing the cells at 37°C in 10% serum medium without
penicillin-streptomycin for 6-8 h, the medium was replaced with
complete medium containing 10% FBS (cat. no. 10099141C; Gibco;
Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin
(cat. no. 15140122; Gibco; Thermo Fisher Scientific, Inc.), and the
cells were incubated for an additional 24-48 h prior to the
follow-up experiments.
Experimental cells were separated into the following
groups according to the nature of the transfections: si-NC,
si-TAGLN-1, si-TAGLN-2, vector-NC (empty plasmid) and TAGLN
overexpression plasmid. Subsequently, the cells were examined by
Cell Counting Kit 8 (CCK-8) viability assay, EdU-labeling
proliferation assay, colony formation assay, wound healing assay,
and Transwell invasion and migration assays within 72 h after the
completion of cell transfection.
CCK-8 assay
KYSE-150 and Eca-109 cells were seeded into a
96-well plate at a density of ~5,000 cells/well and cultured at
37°C. After 24, 48 and 72 h, 10 µl CCK-8 reagent (Beyotime
Institute of Biotechnology) was added to each well, and the cells
were incubated at 37°C for 1 h. A Tecan Infinite M200 Pro
microplate analyzer (Tecan Group, Ltd.) was used to measure the
absorbance of each well at 450 nm to calculate cell viability. Each
experiment was repeated three times.
EdU-labeling proliferation assay
KYSE-150 and Eca-109 cells were seeded into 12-well
plates at a density of 5×104 cells/well and incubated in
RPMI-1640 medium containing 10% FBS at 37°C. The BeyoClick™ EdU-488
kit (Beyotime Institute of Biotechnology) was used to determine
cell proliferation by examining DNA synthesis in the cells. EdU
reagent (1 ml at 10 µM) was added to the cells and incubated
at 37°C for 60 min. The cells were fixed with 4% paraformaldehyde
at room temperature for 15 min, and then treated with 1% Triton
X-100 at room temperature for 15 min. Subsequently, 200 µl
Click reaction solution was added to each well, and the cells were
incubated in dark at room temperature for 30 min. Nuclei were
stained with Hoechst (Beyotime Institute of Biotechnology). A Leica
DM5000B fluorescence micro-scope (Leica Microsystems GmbH) was used
to examine and capture images of the cells. Images were analyzed
using LAS AF software v2.6.0.7266 (Leica Microsystems GmbH). The
percentages of EdU-positive (DNA-replicating) and Hoechst-positive
cells (total cells) were determined.
Colony formation assay
KYSE-150 and Eca-109 cells were seeded into a 6-well
plate, at a range of densities between 2×102 and
10×102 cells/well, and cultivated in RPMI-1640 medium
containing 10% FBS at 37°C for 24 h. The cells were treated with
various concentrations of Tanshinone I (Tan-I; Selleck Chemicals),
0, 1.2, 2.4, 4.8 and 9.6 µg/ml, prior to being resuspended
in RPMI-1640 medium containing 10% FBS and cultured at 37°C for 15
days in an atmosphere containing 5% CO2 to stimulate
colony formation; the medium containing 10% FBS was replaced every
three days. The plate was washed using cold PBS and fixed with 4%
paraformaldehyde solution. The fixed colonies were subsequently
stained with 0.1% crystal violet for 30 min at room temperature.
The number of visible colonies, those with a diameter >0.05 mm,
was calculated using FIJI/ImageJ software (v2.1.0/1.53c; National
Institutes of Health).
Wound healing assay
To prevent cell proliferation upon migration, the
cells were allowed to rest overnight in serum-free medium prior to
wound scratching. Digested cells were harvested and cultured (at a
density of 2×105 cells/well). Once the confluency of the
cells had reached to 100%, wounds were made using a pipette tip.
The wound closures were subsequently measured at 0, 12, 24 and 36 h
to assess the degree of cell migration using a Nikon ECLIPSE Ts2
microscope (Nikon Corporation) and calculated using FIJI/ImageJ
software (v2.1.0/1.53c; National Institutes of Health). Experiments
were performed three times.
Transwell invasion and migration
analysis
Matrigel™ (BD Biosciences) was diluted in RPMI-1640
medium containing 10% FBS after dissolution overnight at 4°C. The
mixture (50 µl) was added to the upper and the lower
compartments of Costar® 24-well plates (cat. no. 3422;
Corning, Inc.), which were then incubated at 37°C for 30 min.
Eca-109 and KYSE-150 cells in the exponential growth phase were
collected; serum-free medium was used to wash and resuspend the
cells to 5×104 cells/ml. The bottom part of the Matrigel
insert was merged in RPMI-1640 medium with 20% FBS, and cells were
gently added to the upper compartment. Culture plates were then
incubated at 37°C with 95% relative humidity and in the presence of
5% CO2 for 24 h, after which, the upper Transwell
chambers were removed. The cells on the lower side of the insert
were stained with 0.1% crystal violet for 15-30 min at room
temperature, and images were captured for cell counting. The number
of cells was counted in 10 fields, which were randomly selected
using a Nikon ECLIPSE Ts2 microscope (Nikon Corporation), and the
average numbers of cells were recorded. The Transwell migration
assay was conducted as described above for the Transwell invasion
experiment, but without Matrigel. Each experiment was repeated
three times.
Xenograft tumor model
Four groups of female BALB/c nude mice (aged 4-6
weeks; n=24/group) were acquired from the SPF Beijing Biotechnology
Co., Ltd. The mice were kept under pathogen-free conditions and
provided sterile food and water ad libitum. The mice were
raised in an environment with constant temperature of 20-26°C,
moderate humidity of 50-60% and a 12-h light/dark cycle. A total of
5×106 cells in 100 µl PBS containing the
TAGLN-overexpression vector, vector-NC, si-TAGLN or si-NC were
subcutaneously inoculated into the right flank of the mice. Tumor
growth was observed every 4 days. Following the procedure outlined
in previous studies (37,38), transfection experiments were
performed 10, 13 and 16 days after the injection of the tumor cells
(designated as day 0). On days 10 and 13, 14 µg vector-NC or
TAGLN-overexpression plasmids were combined with 21.7 µl
Lipofectamine 3000™ Transfection Reagent (Thermo Fisher Scientific,
Inc.); in addition, 434 pmol si-NC or si-TAGLN was mixed with 50
µl siRNA-Oligofectamine (Thermo Fisher Scientific, Inc.) to
form a complex. The solutions are mixed with 5% dextrose water to a
total volume of 100 µl. Following a period of incubation
(10-15 min), an insulin syringe with a permanently attached needle
was used to inject these reagent mixtures above into the tumor. On
day 16, double doses of the siRNA and plasmid complexes were
injected, and the amount of Oligofectamine was increased
accordingly. This procedure was performed in accordance with the
guidelines of the Animal Ethics Committee of Tianjin Medical
University. The weight of the tumors in mice was not allowed to
exceed 10% of their body weight, and the average tumor diameter
never exceeded 15 mm. Mice and tumors were examined every 2-3 days;
when the tumor exceeded 15 mm in diameter, the mice were
euthanized. In case of ulceration, infection or necrosis, the
experiment was terminated, and the mice were euthanized at that
stage. On the 18th day of the experiment, one of the mice in the
TAGLN knockdown group died; H&E staining was performed on their
lungs and liver, and it was found that they had systemic
metastasis. The remaining mice survived until the end of the
experiment. At the end of the experiment, the mice were sacrificed
and the tumors were removed. Mice were euthanized using the
CO2 method: Mice were placed into a cage, and 100%
CO2 was introduced at a flow rate of 30% CO2
volume displacement/min. The CO2 added to the chamber
was allowed to reach equilibrium in the gas mixture to quickly
realize the goal of animal unconsciousness and reduce animal pain.
Animal death was confirmed by cardiac arrest, respiratory arrest,
animal stiffness and dilated pupils. A 4% paraformaldehyde solution
was used to fix the tumors at room temperature for 12-24 h, which
were then submerged in paraffin and sectioned (4-µm thick)
for later observation. The animal study was approved by the Ethics
Committee of Tianjin Medical University General Hospital (ethics
no. IRB2019-DWFL-414).
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
TRIzol™ (Invitrogen; Thermo Fisher Scientific, Inc.)
was used to extract total RNA from the cells. cDNA was synthesized
using a FastKing RT Kit with gDNase (cat. no. KR116; Tiangen
Biotech Co., Ltd.) and amplified by qPCR using Applied Biosystems™
PowerUp™ SYBR™ Green Master Mix (cat. no. A25780; Thermo Fisher
Scientific, Inc.), precisely following the manufacturer's
instructions (Thermo Fisher Scientific, Inc.). The forward and
reverse primer sequences were as follows: GAPDH forward, 5′-CCC TTC
ATT GAC CTC AAC TAC ATG G-3′, and reverse 5′-CAT GGT GGT GAA GAC
GCC AG-3′; TAGLN forward, 5′-GGT GGA GTG GAT CAT AGT GC-3′, and
reverse 5′-ATG TCA GTC TTG ATG ACC CCA-3′; GPX4 forward, 5′-ATG AAG
ATC CAA CCC AAG GG-3′, and reverse 5′-AGG TCC TTC TCT ATC ACC
AGG-3′; ACSL4 forward 5′-CCA AAG AAC ACC ATT GCC ATC-3′ and reverse
5′-AGC CTC AGA TTC ATT TAG CCC-3′; and p53 forward 5′-CCT CAG CAT
CTT ATC CGA GTG G-3′, and reverse 5′-TGG ATG GTG GTA CAG TCA GAG
C-3′. The following PCR thermocycling conditions were used: 95°C
for 30 sec, 95°C for 8 sec and 60°C for 32 sec for 40 cycles; then
95°C for 1 min, 60°C for 30 sec and 95°C for 30 sec. The relative
expression of each of gene was calculated using the
2−ΔΔCq method. Each sample was run in duplicate, and
each experiment was repeated three times.
Western blot analysis
Tissues and cells were lysed using RIPA buffer
(Beyotime Institute of Biotechnology) for total protein extraction.
The BCA assay was used to quantify the total protein concentration.
Proteins (40 µg) were separated using 12.5% SDS-PAGE gel,
and the proteins were then transferred to a PVDF membrane using the
wet (submersion) method (1 kDa/min). Subsequently, the membrane was
blocked using 10% skimmed milk for 1 h at room temperature. After
washing with TBS with 0.1% Tween-20, the membrane was incubated
with the primary antibodies anti-TAGLN, anti-β-actin (cat. no.
TA-09 ZSGB-BIO; OriGene Technologies, Inc.), anti-GAPDH (cat. no
TA-08; ZSGB-BIO; OriGene Technologies, Inc.), anti-GPX4 and
anti-ACSL4 antibodies (also see the Immunohistochemical
analysis subsection above for further details) overnight at
4°C. Note that the dilution ratio of the antibodies for western
blot analysis was 1:1,000. The membrane was then washed and
incubated with secondary antibodies, namely HRP-conjugated goat
anti-rabbit IgG (H+L) (cat. no. ZB-2301; ZSGB-BIO; ZSGB-BIO;
OriGene Technologies, Inc.) and goat anti-mouse IgG (H+L) (cat. no.
ZB-2305; ZSGB-BIO; ZSGB-BIO; OriGene Technologies, Inc.) for 1 h at
room temperature. The proteins were visualized using an Enhanced
Chemiluminescence (ECL) Western Blotting Substrate (cat. no.
PE0010; Beijing Solarbio Science & Technology Co., Ltd.).
Densitometric analysis was conducted using Image Lab software
version 6.1 (Bio-Rad Laboratories, Inc.); each experiment was
repeated three times.
Lipid peroxidation malondialdehyde (MDA)
assay
Lipid peroxidation was detected in Eca-109 and
KYSE-150 cells using a Lipid Peroxidation MDA Assay Kit (cat. no.
BC0025; Beijing Solarbio Science & Technology Co., Ltd.),
following the manufacturer's instructions. The cultured and
transfected cells were collected and then subjected to ultrasonic
fragmentation (power, 200 W; ultrasound 3 sec; interval, 10 sec;
repeated 30 times), and the supernatant is centrifuged. MDA
detection working solution was added and the mixture was incubated
for 60 min in a 100°C water bath; subsequently, then the mixture
was centrifuged 8,000 × g at 4°C for 10 min and 200 µl
supernatant was added to each well of a 96-well plate for detection
using a microplate analyzer to measure the absorbance of both the
protein (at wavelength 450 nm) and MDA (at wavelengths of 532 and
600 nm).
GSH assay
GSH was measured in Eca-109 and KYSE-150 cells using
a Reduced Glutathione (GSH) Colorimetric Assay kit (cat. no.
E-BC-K030-M; Elabscience Biotechnology, Inc.), following the
manufacturer's instructions.
ROS assay
The intracellular level of ROS was calculated by
means of the fluorescent 2,7-dichlorofluorescein diacetate probe
(Nanjing Jiancheng Bioengineering Institute), which was added to
serum-free medium at a final concentration of 10 µM. The
probe and Eca-109 and KYSE-150 cells (5×106) were then
incubated for 30 min in the dark. The cells were washed twice with
PBS to remove excess probe, and the fluorescence intensity was
measured using a Tecan Infinite M200 Pro microplate analyzer (Tecan
Group, Ltd.); the optimal excitation wavelength was 488 nm, and the
optimal emission wavelength was 525 nm). Images of the cells were
captured under a fluorescence microscope.
Iron and total iron assay
The level of Fe2+ or total iron in
5×106 transfected Eca-109 and KYSE-150 cells was
measured using the of Sigma-Aldrich Iron Assay kit (cat. no.
MAK025; Merck KGaA) and a Tecan Infinite M200 Pro microplate
analyze, following the manufacturer's instructions.
Fluorescence detection of Iron ions in
living cells
Transfected Eca-109 and KYSE-150 cells
(5×104) were inoculated in black polystyrene 96-well
plates (cat. no. CLS3916; Corning, Inc.) and incubated overnight at
37°C in a 5% CO2 incubator. The culture medium was removed and the
cells were washed three times with serum-free RPMI-1640 medium.
FerroOrange (cat. no. F374; Dojindo Laboratories, Inc.) was added
at a 1 µmol/l working solution and the cells were incubated
at 37°C in a 5% CO2 incubator for 30 min. Immediately
following incubation, the cells were observed under a fluorescent
microscope (Leica Microsystems GmbH).
Co-immunoprecipitation (Co-IP) assay
Co-IP was performed according to the instructions
offered for each reagent in the process. Eca-109 cells
(1×107) were lysed with Cell Lysis Buffer (cat. no
P0013; Beyotime Institute of Biotechnology) and centrifuged at
13,000 × g for 15 min at room temperature. The anti-p53 antibody
(1:100; cat. no. 10442-11-AP; Proteintech Group, Inc.), anti-TAGLN
antibody (1:100; cat. no. ab233971; Abcam) and anti-rabbit IgG
antibody (negative control) were incubated with 40 µg
Protein A+G-Sepharose beads (cat. no. P2105; Beyotime Institute of
Biotechnology) at 4°C for 1 h. Each 1,000 µl extracted
cellular protein sample was mixed with 40 µl of magnetic
bead antibody mix and incubated for 2 h at room temperature. At the
end of the incubation, the mixtures were placed on a magnetic stand
for 10 sec to ensure separation, and the supernatant was removed.
The beads were washed three times with a TBS, and 40 µg
magnetic beads was added to 100 µl of protein loading buffer
in a metal bath heated at 95°C for 5 min, and then placed on the
magnetic rack to remove the supernatant, leaving the target protein
to be analyzed by western blotting.
Immunofluorescence co-localization
assay
TAGLN- overexpressing Eca-109 cells
(1×104) were seeded into 24-well plates and fixed for 10
min using 4% paraformaldehyde at room temperature. PBS containing
0.1% Triton X-100 was added for cell membrane permeabilization, and
the cells were incubated for 10 min at room temperature. Then PBST
containing 1% BSA (cat. no. A8020; Beijing Solarbio Science &
Technology Co., Ltd.) was used for blocking at room temperature for
1 h. The cells were subsequently incubated with the primary
antibodies, anti-TAGLN (1:100; cat. no. ab233971; Abcam), anti-p53
(1:100; cat. no. 10442 11 AP; Proteintech Group, Inc) and
anti-HSPB1 (1:500; cat. no. ab109376; Abcam) and then carefully
washed three times with PBS. Goat anti-,mouse IgG H&L (Alexa
Fluor® 488-conjugated; 1:200; ab150113; Abcam) and
donkey anti-rabbit IgG H&L (Alexa Fluor®
647-conjugated; 1:200; ab150075; Abcam) was used to stain the
secondary antibodies. Nuclei were stained with DAPI (cat. no.
S2110; Beijing Solarbio Science & Technology Co., Ltd.). Images
of the cells were obtained using a laser-scanning confocal
microscope (Leica Microsystems GmbH).
Comprehensive analysis of protein
interactions
The protein-protein interaction (PPI) network
information for TAGLN was downloaded from the Search Tool for the
Retrieval of Interacting Genes/Proteins (STRING) website (version
11.0; https://string-db.org). Using the FerrDb
dataset (version 1; http://www.zhounan.org/ferrdb), a total of 259
ferroptosis-associated genes were identified. Genes involved in the
TAGLN and ferroptosis pathways were illustrated using a Venn
diagram.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 8.0 software (GraphPad Software; Dotmatics). All measurement
data are shown as the mean ± standard deviation. Comparisons
between two groups were analyzed using a two-tailed unpaired
t-test, and multiple groups were analyzed by one-way ANOVA followed
by Tukey's post hoc test for further pairwise comparisons.
Correlation analysis was performed using Pearson's correlation
method, and the Kaplan-Meier followed by log-rank method was used
to detect relevant associations between the expression levels of
TAGLN and the subsistence prognosis of patients with ESCC.
P<0.05 was considered to indicate a statistically significant
difference.
Results
TAGLN expression level is lower in ESCC
and is positively correlated with the prognosis of patients with
ESCC
The respective endoscopic presentations of the
normal esophageal tissue and ESCC are shown in Fig. 1A. The changes in TAGLN expression
level in normal esophageal tissues and in ESCC tissues were
distinguished using immunohistochemical analysis (Fig. 1B). The results showed that relative
TAGLN expression was lower in ESCC compared with normal tissues
(Fig. 1C). Statistical analysis
confirmed that no significant associations were identified between
the relative expression level of TAGLN in ESCC and age, sex, lymph
node metastasis, tumor grade, tumor size, differentiation or tumor
site of the patients (Table I).
However, the relative expression of TAGLN was significantly
associated with tumor grade, lymphatic invasion and AJCC stage
(Table I). Moreover, the
expression of ferroptosis-associated proteins, ACSL4 and GPX4, in
ESCC and normal tissues was confirmed using immunohistochemistry,
and the differences were found to be statistically significant
(Fig. 1B and C). The correlation
analyses were based on IHC stating scores; the expression of GPX4
was negatively correlated with TAGLN expression (Fig. 1E), whereas that of ACSL4 was
positively correlated with TAGLN expression (Fig. 1F). The expression level of TAGLN
was statistically higher in ESCC tissues at the early stages of
cancer (stage IIa) compared with the advanced stages (stage III)
(Fig. 1D); the difference between
the expression of TAGLN in stage IIa and in the control groups was
not statistically significant, whereas the differences between the
expression of TAGLN in stage III groups and in the control group
was identified as statistically significant. The differences
between the expression of ACSL4 and GPX4 in ESCC stages IIa and
stages III were found not to be statistically significant (Fig. 1D); however, the differences between
the expression of ACSL4 and GPX4 in these two ESCC groups compared
with expression in the control group were statistically
significant.
 | Figure 1TAGLN expression in ESCC and its
association with ESCC prognosis. (A) Relative endoscopic
presentation of normal esophagus and that of patients with ESCC.
(B) Immunohistochemistry was used to detect the expression of
TAGLN, GPX4 and ACSL4 in normal esophageal tissues and in ESCC
stages I-II and III-IV tumoral tissues. (C) Statistical analysis of
TAGLN, GPX4 and ACSL4 expression in normal esophagus and ESCC
tissues. Data are presented as the mean ± SD of three independent
experiments. (D) Statistical analysis of the expression of TAGLN,
GPX4 and ACSL4 in normal esophagus, stage I-II and III-IV ESCC
tissues. Data are presented as the mean ± SD of three independent
experiments. *P<0.05, as determined by two-tailed
unpaired Student's t-test. Correlation between TAGLN and (E) GPX4
and (F) ACSL4 expression. (G) Survival curve of patients with ESCC
with high and low TAGLN expression levels in cancer tissues. AJCC,
American Joint Committee on Cancer; ACSL4, acyl CoA synthetase long
chain family member 4; EE, esophageal epithelium; EG, esophageal
gland; ESCC, esophageal squamous cell carcinoma; ns, not
significant; SM, smooth muscle; TAGLN, transgelin. |
The cut-off value used to separate patients into
high and low expression groups was 3.7 (median value) according to
IHC staining score. Kaplan-Meier analysis of the data demonstrated
a significant association between the survival prognosis of
patients with ESCC and the relative expression levels of TAGLN;
that is, patients with high expression of TAGLN had a longer
overall survival rate compared with patients with a low expression
of TAGLN (Fig. 1G).
Subsequently, the expression levels of the
ferroptosis-associated proteins p53, COX2, TF and FTH1 in ESCC and
normal tissues were detected by immunohistochemical analysis. The
differences in the expression of p35, COX2, TF and FTH1 between the
ESCC and normal tissues were found to be statistically significant
(Fig. 2A and B). The differences
in the expression of p53, COX2, TF and FTH1 between ESCC stage III
and normal tissues were found to be statistically significant. The
differences in the expression of p53 and TF between ESCC stage IIa
and normal tissues were not statistically significant, whereas the
differences in the expression of COX2 and FTH1 between ESCC stage
IIa and normal tissues were found to be statistically significant.
The differences between the expression levels of p53, COX2, TF and
FTH1 in ESCC stage IIa and stages III, however, were found not to
be statistically significant (Fig.
2C). FTH1 expression was negatively correlated with TAGLN
expression, whereas TF expression was positively correlated with
TAGLN expression (Fig. 2D).
However, the correlations between p53 and COX2 expression and TAGLN
expression were found not to be statistically significant.
 | Figure 2Ferroptosis-related protein
expression levels in ESCC and their correlation with TAGLN. (A)
Expression levels of p53, COX2, TF and FTH1 detected by
immunohistochemistry in normal esophagus, stage I-II and III-IV
ESCC tumor tissues. (B) Statistical analysis of the expression of
p53, COX2, TF and FTH1 in normal esophagus and cancer tissues. Data
are presented as the mean ± SD of three independent experiments.
(C) Statistical analysis of the expression of p53, COX2, TF and
FTH1 in normal esophagus, stage I-II and III-IV ESCC tumor tissues.
Data are presented as the mean ± SD of three independent
experiments. *P<0.05 and **P<0.01, as
determined by two-tailed unpaired Student's t-test. (D) Correlation
between p53, COX2, TF and FTH1 expression and TAGLN expression in
normal esophagus and cancer tissues. AJCC, American Joint Committee
on Cancer; COX2, cyclooxygenase 2; EE, esophageal epithelium; EG,
esophageal gland; ESCC, esophageal squamous cell carcinoma; FTH1,
ferritin heavy chain 1; ns, not significant; SM, smooth muscle;
TAGLN, transgelin; TF, transferrin. |
TAGLN expression level is decreased in
ESCC and is associated with various cancer cell signaling
pathways
TAGLN expression level is decreased in ESCC and is
associated with various cancer cell signaling pathways (39). The GEO databank is a publicly
accessible genomics data repository for high-throughput sequencing.
The databank contains sequence and array based gene profile data,
and was used in this study to profile the expression of genes in
ESCC. Three datasets, GSE161533, GSE45670 and GSE100942, were used
to examine the differential expression of TAGLN in ESCC and normal
tissues. A significant decrease in the level of TAGLN expression in
ESCC compared with expression in normal tissue was observed in
these datasets (Figs. 3A-C and
S1). Using the median TAGLN mRNA
level as a cutoff value, the patients with ESCC in TCGA were split
into high and low TAGLN subgroups. The signaling pathways and
cellular processes that were significantly associated with the high
TAGLN subgroup compared with the low TAGLN subgroup were identified
by GSEA (Fig. 3D-I). Genes
upregulated in the high TAGLN subgroup were mostly enriched in
'focal adhesion' (NES=2.01; P<0.0001), 'ECM receptor
interaction' (NES=1.94; P=0.002), 'Gap junction' (NES=1.73;
P=0.004), 'TGF-β signaling pathway' (NES=1.65; P<0.05) and 'Cell
adhesion molecules (CAMS)' (NES=1.74; P<0.05). These pathways
have been associated with tumor proliferation, invasion and other
malignant progressions (40,41),
suggesting that TAGLN may serve an important role in ESCC.
 | Figure 3TAGLN expression is lower in ESCC and
is associated with various cancer cell signaling pathways. Three
datasets from the Gene Expression Omnibus, (A) GSE161533, (B)
GSE45670 and (C) GSE100942 were used to validate the differential
expression of TAGLN in ESCC tissues and normal tissues. (D)
Signaling pathways associated with TAGLN expression in ESCC by
KEGG. Genes upregulated in the TAGLN-high subgroup were enriched in
(E) focal adhesion, (F) TGF-β signaling, (G) gap junction, (H) ECM
receptor interaction, and (I) cell adhesion molecules (CAMS). ECM,
extracellular matrix; ESCC, esophageal squamous cell carcinoma;
KEGG, Kyoto Encyclopedia of Genes and Genomes; TAGLN,
transgelin. |
Overexpression of TAGLN attenuates ESCC
cell proliferation, colony formation, invasion and migration
To determine whether TAGLN exerts a role in the
malignant progression of ESCC, TAGLN was overexpressed in Eca-109
and KYSE-150 cell lines. The efficiency of mRNA and protein
overexpression was confirmed using RT-qPCR and western blot
analyses, respectively, and the mRNA and protein expression levels
in TAGLN group were found to be significantly higher compared with
the vector-NC (Fig. 4A and B). The
CCK-8, EdU and colony formation assay results revealed that the
overexpression of TAGLN in ESCC cell lines led to a significant
reduction in the capability of the ESCC cells to proliferate
(Fig. 4C-E). Compared with the
control group, the number of migrating and invading cells was also
significantly decreased (Fig. 4F)
following the overexpression of TAGLN in both cell lines. The
ability of the cells to migrate was also significantly reduced in
the wound healing assay following TAGLN overexpression (Figs. 4G and S2). Taken together, these results
demonstrated that the overexpression of TAGLN may attenuate the
proliferation, colony formation, invasion and migration of the two
ESCC cell lines.
Overexpression of TAGLN suppresses,
whereas TAGLN knockdown promotes tumor growth in a xenograft model
of ESCC
To investigate whether TAGLN serves a role in ESCC
in vivo, a xenograft model was constructed. The inoculation
of the mice with vector-NC or vector-TAGLN transfected Eca-109
cells revealed that TAGLN overexpression caused a marked reduction
in tumor size, volume and weight after one month of growth compared
with the mice inoculated with Eca-109-Vector-NC cells, whereas the
whole-body weight of the mice remained the same (Fig. 5A-D). The TAGLN expression level of
the subcutaneous tumors was subsequently assessed using western
blot, RT-qPCR and immunohistochemical analyses, and the differences
between vector-NC group and vector-TAGLN group were found to be
statistically significant (Fig. 5E, F
and I, respectively). Changes were also observed in the
expression of the ferroptosis marker proteins, including GXP4 and
ACSL4, in the subcutaneous tumors according to RT-qPCR and
immunohistochemical analyses. GPX4 expression decreased, whereas
that of ACSL4 increased, in the TAGLN-overexpression model mice
compared with the control group (Fig.
5G, 5H, J and K). Taken together, these results suggested that
the overexpression of TAGLN may cause a significant suppression of
the progression of ESCC in vivo.
The injection of si-NC- or si-TAGLN-transfected
Eca-109 cells into mice resulted in a significant increase in tumor
volume and weight after one month of growth following TAGLN
knockdown compared with the control Eca-109 cells (Fig. 6A, C and D, respectively), whereas
the whole-body weight of the mice remained unchanged (Fig. 6B). Multiple tumor metastases were
identified (and confirmed by H&E staining) in the livers of
nude mice that were injected with si-TAGLN-transfected Eca-109
cells; the number of metastases was found to be significantly
higher compared with the control group (Fig. 6E and F). The expression level of
TAGLN in the subcutaneous tumors was detected by western blot,
RT-qPCR and immunohistochemical analyses, and the expression was
found to be significantly decreased compared with the control group
(Fig. 6G-I and L). In addition,
changes were also observed in the expression of the ferroptosis
marker proteins, GXP4 and ACSL4, in the subcutaneous tumors using
RT-qPCR and immunohistochemical analyses. According to RT-qPCR,
compared with the control group, the expression of GPX4 was
increased, whereas that of ACSL4 was decreased, following the
knockdown of TAGLN (Fig. 6J and
K). However, according to IHC staining, the difference of GPX4
expression between si-NC and si-TAGLN was not statistically
significant, whereas that of ACSL4 was decreased in si-TAGLN group
compared with the control group, following the knockdown of TAGLN.
(Fig. 6M and N).
Investigation of the mechanism of TAGLN
in inhibiting the malignant progression of ESCC through
ferroptosis
Transcriptome analysis results demonstrated
significant differential normalized transcripts per million and
gene expression in the vector-NC and TAGLN-overexpression Eca-109
cells (Figs. 7A and B, and
S3). To screen the differentially
expressed genes, GO and KEGG enrichment analyses were performed
(Fig. 7C). The analysis of GO
enrichment showed that the BP and MF subontology were
differentially regulated in TAGLN-overexpressing Eca-109 cells. For
GO-BP, this regulation was associated with 'metal ion transport',
'dynamin family protein polymerization related to mitochondrial
fission', 'epithelial cell apoptotic process', 'regulation of amino
acid import across plasma membrane', and 'blood vessel endothelial
cell proliferation in angiogenesis'. The overexpression of TAGLN
induced the regulation of GO-MF gene categories associated with
'dynein intermediate chain binding', 'divalent inorganic cation
transmembrane transporter activity', 'transition metal ion
transmembrane transporter activity', 'ion transmembrane transporter
activity' and 'protein serine/threonine phosphatase activity'. KEGG
analysis showed that the overexpression of TAGLN regulated various
pathways, including 'oxidative phosphorylation', 'transcriptional
misregulation in cancer', 'TGF-β signaling pathway', 'signaling
pathways regulating pluripotency of stem cells', 'pathways in
cancer' and the 'PI3K-Akt signaling pathway'. These results
indicated that TAGLN may contribute to the processes of oxidative
phosphorylation, metal iron ion transport and cell apoptosis,
leading to the occurrence of ferroptosis in ESCC cells and the
inhibition of cancer proliferation, invasion and metastasis,
together with associated signaling pathways. Moreover, the STRING
tool was used to analyze the PPI networks of the TAGLN protein to
determine their interaction in the progression of ESCC (Fig. 7D and E). As the results show, the
crossover gene between PPI network and ferroptosis-related genes
was GPX4, an essential protein of ferroptosis. These data suggested
that TAGLN may regulate ferroptosis.
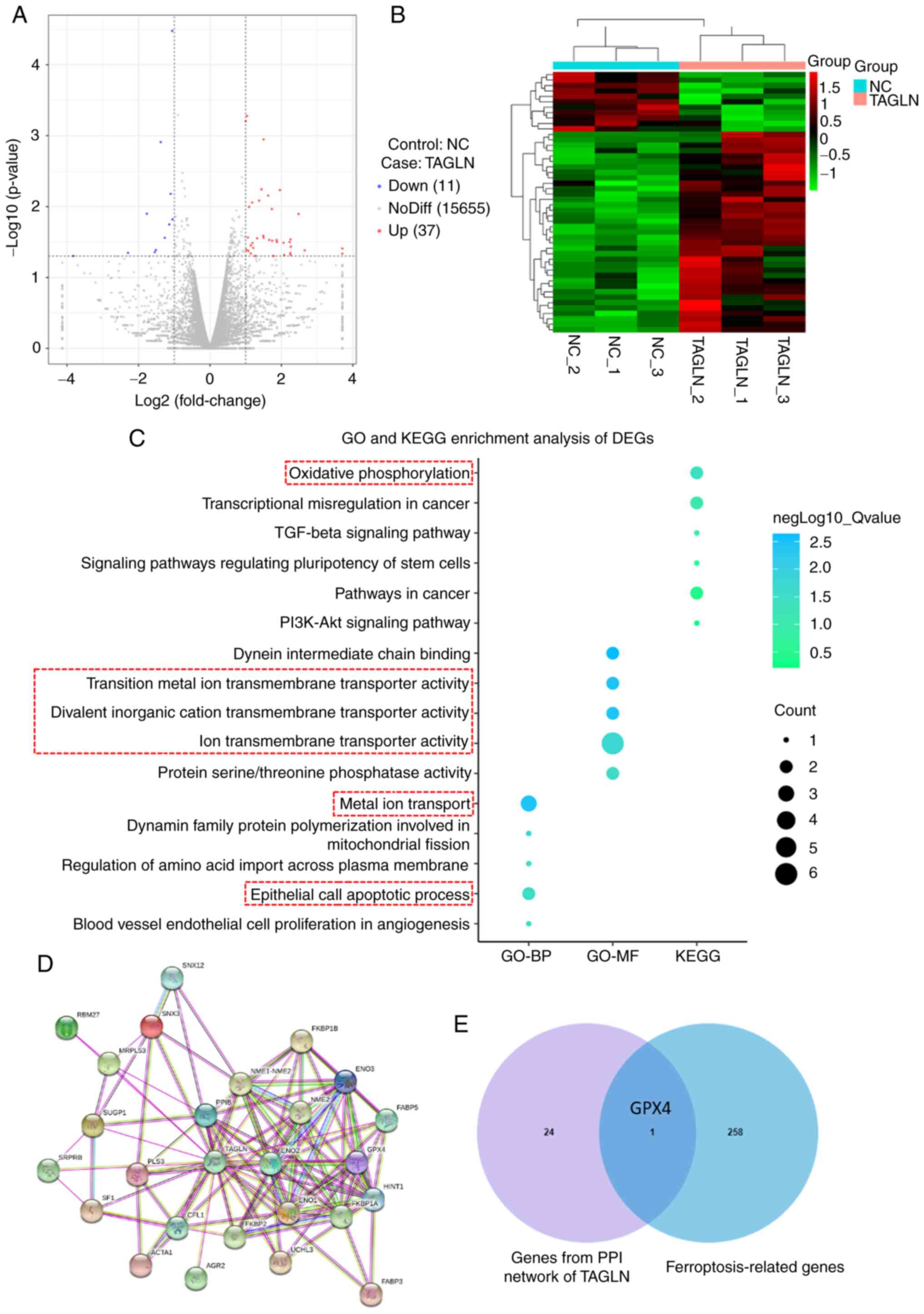 | Figure 7Mechanism of TAGLN inhibition of
esophageal squamous cell carcinoma progression by ferroptosis. DEGs
were identified in Eca-109 cells transfected with vector-NC or
TAGLN-overexpression plasmid. (A) Volcano plot and (B) cluster
heatmap showing the results of differential analyses in the two
groups. (C) Analysis of GO-BP, GO-MF and KEGG enrichment analyses
showed the differential regulation of gene categories induced by
the overexpression of TAGLN. (D) Using STRING tool to analyze the
PPI network of TAGLN protein, p53, one of the main factors that
regulate ferroptosis, was identified from the intersection of the
TAGLN interacting genes. (E) The Venn diagram between genes from
PPI network and ferroptosis-related genes. DEG, differentially
expressed gene; GO, Gene Ontology; GPX4, glutathione peroxidase 4;
KEGG, Kyoto Encyclopedia of Genes and Genomes; NC, negative
control; TAGLN, transgelin. |
Overexpression of TAGLN promotes
ferroptosis and regulates the expression of ferroptosis marker
proteins
Western blot analysis was used to determine the
expression levels of the ferroptosis marker proteins, GPX4 and
ACSL4, following TAGLN overexpression. The protein expression level
of ACSL4 was significantly increased, whereas that of GPX4 was
significantly decreased, in Eca-109 and KYSE-150 cell lines
following TAGLN overexpression compared with the vector-NC group
(Fig. 8A). Furthermore, RT-qPCR
results showed that the mRNA expression level of GPX4 in both ESCC
cell lines was significantly decreased, whereas the expression
level of ACSL4 was significantly increased (Fig. 8B). The overexpression of TAGLN
caused a significant increase in the intracellular concentrations
of MDA and lipid ROS (Fig. 8C and
D, respectively), and a decrease in the concentration of GSH
(Fig. 8E), suggesting that the
overexpression of TAGLN may induce sensitivity to and promote
ferroptosis. Levels of intracellular total iron (Fig. 8F) and Fe2+ iron
(Fig. 8G and H) increased in
Eca-109 and KYSE-150 cells overexpressing TAGLN compared with the
vector-NC transfected groups. Taken together, these results
suggested that TAGLN may serve a crucial oncogenic function in
cancer progression by regulating ferroptosis in ESCC cells.
 | Figure 8Overexpression of TAGLN promotes
ferroptosis and regulates ferroptosis marker proteins expression.
The (A) protein and (B) mRNA expression levels of ferroptosis
marker proteins, GPX4 and ACSL4, were verified by western blotting
(β-actin was used as the internal control) and reverse
transcription-quantitative PCR, respectively. Intracellular
concentrations of (C) MDA and (D) lipid ROS increased, whereas (E)
GSH decreased in Eca-109 and KYSE-150 cells overexpressing TAGLN
compared with the vector-NC groups. Levels of (F) intracellular
total iron, (G) ferrous iron and (H) intracellular Fe2+
iron increased in Eca-109 and KYSE-150 cells overexpressing TAGLN
compared with the vector-NC groups. Data are shown as the mean ± SD
of three independent experiments. *P<0.05 and
**P<0.01, as determined by two-tailed unpaired
Student's t-test. ACSL4, acyl CoA synthetase long-chain family
member 4; CCK-8, Cell Counting Kit-8; fer, ferrostatin; GPX4,
glutathione peroxidase 4; GSH, glutathione; NC, negative control;
ROS, reactive oxygen species; TAGLN, transgelin. |
TAGLN regulates ferroptosis by
interacting with p53
p53, one of the main factors that regulate
ferroptosis, was identified from the intersection of the TAGLN
interacting gene. Therefore, the functional interaction between
TAGLN and p53 was selected for further investigation to unravel the
molecular mechanism and metabolism of the malignancy in ESCC.
RT-qPCR analysis revealed that the mRNA expression level of p53
significantly increased with an upregulation of TAGLN expression in
Eca-109 cells (Fig. 9A). To
investigate a putative interaction of TAGLN with p53, Co-IP and
fluorescence co-localization experiments were conducted in Eca-109
cells (Fig. 9B and C,
respectively). As Fig. 9C shown,
TAGLN and p53 were both found to be abundant in the vector-TAGLN
Eca-109 cell cytoplasm. These results indicated that TAGLN
interacts with p53 and suggested that TAGLN may regulate
ferroptosis through interacting with p53 in ESCC cells. Taken
together, these results suggested that TAGLN inhibits the malignant
progression and regulates ferroptosis by interacting with p53 in
ESCC cells (Fig. 9D).
Discussion
Esophageal cancer has a relatively low 5-year
overall survival rate of ~20% in Europe, USA and China (7); ESCC, the major subtype of esophageal
cancer, has a higher incidence among Eastern Asian and Eastern and
Southern African populations compared with the worldwide population
(42). The aim of the present
study was to suggested potential options for future therapeutic
targets.
TAGLN is an actin-binding protein that stabilizes
actin in vitro and is widely expressed in muscle tissue and
organs, such as GI tract and heart (43-45).
TAGLN may also be associated with cell migration, thereby promoting
an invasive and malignant nature in cells (46). Previous studies have suggested that
TAGLN has tumor-suppressive functions in certain cells that are not
associated with the cytoskeleton per se (47,48).
TAGLN expression was found to be significantly lower in bladder,
breast and renal cell carcinoma tissues compared with matched
normal tissues (49,50). TAGLN expression was shown to be
markedly reduced in colorectal cancer samples compared with normal
colorectal mucosa, and this was associated with poor overall
survival in patients with colorectal cancer (50). In prostate carcinoma cells, TAGLN
was found to block androgen-stimulated cell growth by inhibiting
the binding of an androgen receptor co-activator with its androgen
receptor (51). These studies
indicated that the loss of TAGLN gene expression may be an event in
malignant tumor progression. In our previous study, TAGLN
expression was shown to inhibited ESCC progression, and that,
through inhibiting the occurrence of EMT, it may ultimately be
possible to prevent the malignant progression of ESCC (34). According to results from the
present study, TAGLN may have an important role in the malignant
progression of ESCC. A longer overall survival rate was observed in
patients who had higher levels of TAGLN expression. The level of
TAGLN expression in ESCC tissues may also be beneficial in terms of
clinically evaluating the degree of the malignancy and the
prognosis of patients with ESCC. Thus, another aim of the present
study was to further investigate the role of TAGLN in ESCC and to
explore the underlying mechanism.
According to the GSEA analysis performed in the
present study, TAGLN may be able to influence several pathways that
are linked with ESCC cell functions and tumor characteristics.
Focal adhesions, ECM-receptor interactions, gap junction and CAMs
are the factors/processes that affect tumor invasion. TAGLN can
also affect the NER process that is associated with cancer. The
TGF-β signaling pathway, a tumor immune regulatory pathway, was
also found to be associated with the expression level of TAGLN in
ESCC. Based on these findings, it was possible to speculate that
TAGLN may be a tumor suppressor in ESCC that inhibits the invasion
of ESCC, and possibly the progression of other malignancies,
through tumor immune pathways. Subsequently, in vitro and
in vivo experiments were performed to verify the above
results. The data obtained indicated that increased TAGLN
expression caused an attenuation of the malignancy of Eca-109 and
KYSE-150 cells, whereas the downregulation of TAGLN enhanced the
ability of ESCC cells to proliferate, migrate and invade. The
malignancy of the Eca-109 and KYSE-150 cells was impacted by TAGLN
expression, and these results suggested that the overexpression of
TAGLN in ESCC cell lines both promoted ferroptosis and regulated
the expression of ferroptosis marker proteins.
The results of the transcriptome analysis in the
current study revealed the association between TAGLN expression and
the GO-BP and GO-MF subontologies associated with ion accumulation
and programmed cell death during ferroptosis (52-54).
In addition, the findings further confirmed that TAGLN may
contribute to oxidative phosphorylation, Fe2+ transport
and apoptosis, leading to the occurrence of ferroptosis in ESCC
cells, as well as the inhibition of cancer cell proliferation,
invasion, metastasis and the associated signaling pathways. p53 is
one of the main factors that regulate ferroptosis (16,51).
Subsequently, the STRING tool analysis of the PPI networks of the
TAGLN showed that p53 may interact with TAGLN. RT-qPCR results
confirmed that the p53 expression increased concomitantly with the
overexpression of TAGLN. Moreover, the protein interactions between
p53 and TAGLN in the ESCC cell lines were verified by Co-IP and
immunofluorescence co-localization staining experiments. According
to previous studies (55,56), TAGLN may promotes ferroptosis and
inhibits the malignant evolution of ESCC through interacting with
p53. Tsui et al (55)
reported that TAGLN expression was higher in bladder smooth muscle,
fibroblast and normal epithelial cells compared with carcinoma
cells in vitro. The findings suggested that TAGLN is a p53
upregulated gene; ectopic overexpression of p53 induced TAGLN
expression and, further, TAGLN was shown to inhibit cell
proliferation and invasion in vitro and block tumorigenesis
in vivo. Zhang et al (56) reported that overexpression of TAGLN
resulted in both an increase in the cytoplasmic translocation of
p53 and the upregulation of p53 expression in prostate cancer cell
lines. The interplay between TAGLN and p53 in vivo, and
triggering of the mitochondria-associated apoptotic pathway were
detected in the lymph node carcinoma of the prostate cells
following transfection with TAGLN.
Unlimited lipid peroxidation and a rupture of the
plasma membrane cause ferroptosis (20), which can be stimulated through
extrinsic and intrinsic pathways (57). A blockade of intracellular
antioxidant enzymes (such as GPX4) activates the intrinsic pathway
(16,58). The suppression of cell membrane
transporters, such as the cystine/glutamate transporter (also known
as system xc-), or the stimulation of iron transporters, such as
serotransferrin and lactotransferrin, are two ways in which the
extrinsic pathway can be activated (59). System xc- is comprised of SLC7A11
and SLC3A2 subunits. Both the activity and expression level of
SLC7A11 are negatively regulated by tumor suppressor genes, such as
p53 (60). Ferroptosis in cancer
cells is promoted by the p53-mediated transcriptional inhibition of
SLC7A11, and the ability of p53 to promote apoptosis and
ferroptosis is altered by changes in p53 (mutations or
polymorphisms) (61). Although, in
lung cancer cell lines, the ability of the p533KR
acetylation-defective mutant to induce ferroptosis is retained, it
is not able to induce apoptosis (17). In summary, it is possible to
speculate that TAGLN promotes ferroptosis by interacting with p53
in ESCC.
In conclusion, the present study demonstrated that
TAGLN may prevent the proliferation, migration, invasion and
metastasis of ESCC through regulating the occurrence of
ferroptosis. ESCC is difficult to diagnose at an early stage. At
present, effective treatment methods are lacking, and the prognosis
of ESCC is poor. For these reasons, it is essential to design and
perform additional studies that focus on the incidence, development
and metastasis of ESCC. Recently, TAGLN has been found to have a
role in various types of tumor, although its biological function(s)
and the underlying mechanism(s) remain poorly understood (62). Our previous study demonstrated that
TAGLN inhibited ESCC and regulated EMT in ESCC (34). The present study demonstrated that
the expression level of TAGLN is low in ESCC, and that the
expression level of ESCC is associated with prognostic
characteristics. The expression of TAGLN was significantly
associated with the stage and grade of ESCC, and the overall
survival of patients with ESCC. In this study, the inhibitory
effect of TAGLN on ESCC was verified by more phenotypic experiments
such as EdU, colony formation and nude mice xenograft assays. We
hypothesized that the inhibitory effect of TAGLN on ESCC is not
accomplished by EMT alone. By bioinformatics analysis and the
detection of ferroptosis indicators in ESCC patients, it was found
that TAGLN may regulate ferroptosis through p53 and, thus, inhibit
ESCC. In addition, the number of cases included in this study is
small and the sample size is small, so it has certain limitations.
The sample size will be expanded for further verification in the
future. Finally, the present study has demonstrated that,
regulating ferroptosis may become a new direction of tumor therapy,
and it also suggested that TAGLN may be a therapeutic target for
ESCC.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed in the current
study are available from the corresponding author on reasonable
request. Transcriptome analysis of original data were uploaded to
the NCBI database, and accession to cite for these SRA data is
PRJNA924358 (https://www.ncbi.nlm.nih.gov/sra/PRJNA924358).
Authors' contributions
WZ and BW designed the study. WZ, QC, BY, LZ, CW,
XK, XX, YG, XL, QD, LZ, YL and CW performed the experiments and
analyzed the data. QC, CW and WZ performed the data analysis. QC
and WZ wrote the initial draft of the paper, with contributions
from all authors. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Written informed consent was provided by each
patient, and the Ethics Committee of Tianjin Medical University
General Hospital (Tianjin, China) approved the study (ethics no.
IRB2021 WZ 134). The animal study was approved by the Ethics
Committee of Tianjin Medical University General Hospital (ethics
no. IRB2019-DWFL-414).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was financially supported by The National
Natural Science Foundation of China (grant nos. 82000511, 82170558
and 81900487), Scientific and Technological Projects of Tianjin
(grant no. 21JCQNJC01120), Health Science and Technology Project of
Tianjin (grant no. TJWJ2021QN006), Scientific Research Project of
Tianjin Education Commission (grant no. 2019KJ197), Tianjin Science
and Technology Plan Project (grant no. 21JCQNJC00990), Health
Commission of Shanxi Province Science and Technology Guiding
Project (grant no. 2021XM40), Basic Research Program of Shanxi
Provincial Science and Technology Department (grant no.
20210302123013) and Key Technology Research and Development Project
of Jincheng Science and Technology Bureau (grant no. 20210118).
Abbreviations:
|
ACSL4
|
acyl-CoA synthetase long-chain family
member 4
|
|
Co-IP
|
co-immunoprecipitation
|
|
COX2
|
cyclooxygenase-2
|
|
EMT
|
epithelial-mesenchymal transition
|
|
ESCC
|
esophageal squamous cell
carcinoma
|
|
FTH1
|
ferritin heavy chain 1
|
|
GEO
|
Gene Expression Omnibus
|
|
GO
|
Gene Ontology
|
|
GPX4
|
glutathione peroxidase 4
|
|
GSEA
|
Gene Set Enrichment Analysis
|
|
GSH
|
glutathione
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
MDA
|
malondialdehyde
|
|
ROS
|
reactive oxygen species
|
|
TAGLN
|
transgelin
|
|
TCGA
|
The Cancer Genome Atlas
|
|
TF
|
transferrin
|
References
|
1
|
Ferlay J, Ervik M, Lam F, Colombet M, Mery
L, Piñeros M, Znaor A, Soerjomataram I and Bray F: Global Cancer
Observatory: Cancer Today. International Agency for Research on
Cancer; Lyon: 2020, https://gco.iarc.fr/today. Accessed December 20,
2022.
|
|
2
|
Waters JK and Reznik SI: Update on
management of squamous cell esophageal cancer. Curr Oncol Rep.
24:375–385. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Arnold M, Ferlay J, van Berge Henegouwen
MI and Soerjomataram I: Global burden of oesophageal and gastric
cancer by histology and subsite in 2018. Gut. 69:1564–1571. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fitzmaurice C, Abate D, Abbasi N,
Abbastabar H, Abd-Allah F, Abdel-Rahman O, Abdelalim A, Abdoli A,
Abdollahpour I, Abdulle ASM, et al: Global, regional, and national
cancer incidence, mortality, years of life lost, years lived with
disability, and disability-adjusted life-years for 29 cancer
groups, 1990 to 2017: A systematic analysis for the global burden
of disease study. JAMA Oncol. 5:1749–1768. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pan R, Zhu M, Yu C, Lv J, Guo Y, Bian Z,
Yang L, Chen Y, Hu Z, Chen Z, et al: Cancer incidence and
mortality: A cohort study in China, 2008-2013. Int J Cancer.
141:1315–1323. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lagergren J, Smyth E, Cunningham D and
Lagergren P: Oesophageal cancer. Lancet. 390:2383–2396. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zeng H, Chen W, Zheng R, Zhang S, Ji JS,
Zou X, Xia C, Sun K, Yang Z, Li H, et al: Changing cancer survival
in China during 2003-15: : A pooled analysis of 17 population-based
cancer registries. Lancet Glob Health. 6:e555–e567. 2018.
View Article : Google Scholar
|
|
9
|
Lin DC, Hao JJ, Nagata Y, Xu L, Shang L,
Meng X, Sato Y, Okuno Y, Varela AM, Ding LM, et al: Genomic and
molecular characterization of esophageal squamous cell carcinoma.
Nat Genet. 46:467–473. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu C, Wang Z, Song X, Feng XS, Abnet CC,
He J, Hu N, Zuo XB, Tan W, Zhan Q, et al: Joint analysis of three
genome-wide association studies of esophageal squamous cell
carcinoma in Chinese populations. Nat Genet. 46:1001–1006. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang S, Guo Y, Li Z, Zhang Y, Zhou T, You
W, Pan K and Li W: A systematic review of metabolomic profiling of
gastric cancer and esophageal cancer. Cancer Biol Med. 17:181–198.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen X, Kang R, Kroemer G and Tang D:
Broadening horizons: The role of ferroptosis in cancer. Nat Rev
Clin Oncol. 18:280–296. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu J, Ren L, Li S, Li W, Zheng X, Yang Y,
Fu W, Yi J, Wang J and Du G: The biology, function, and
applications of exosomes in cancer. Acta Pharm Sin B. 11:2783–2797.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen X, Zhou Z, Zhang Z, Zhao C, Li J,
Jiang J, Huang B and Qin Y: Puerarin inhibits EMT induced by
oxaliplatin via targeting carbonic anhydrase XII. Front Pharmacol.
13:9694222022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang WS, SriRamaratnam R, Welsch ME,
Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji
AF, Clish CB, et al: Regulation of ferroptotic cancer cell death by
GPX4. Cell. 156:317–331. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jiang L, Kon N, Li T, Wang SJ, Su T,
Hibshoosh H, Baer R and Gu W: Ferroptosis as a p53-mediated
activity during tumour suppression. Nature. 520:57–62. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta
R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS,
et al: Ferroptosis: An iron-dependent form of nonapoptotic cell
death. Cell. 149:1060–1072. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Torti SV, Manz DH, Paul BT,
Blanchette-Farra N and Torti FM: Iron and cancer. Annu Rev Nutr.
38:97–125. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen X, Yu C, Kang R and Tang D: Iron
metabolism in ferroptosis. Front Cell Dev Biol. 8:5902262020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Stockwell BR, Angeli JP, Bayir H, Bush AI,
Conrad M, Dixon SJ, Fulda S, Gascón S, Hatzios SK, Kagan VE, et al:
Ferroptosis: A regulated cell death nexus linking metabolism, redox
biology, and disease. Cell. 171:273–285. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang WS, Kim KJ, Gaschler MM, Patel M,
Shchepinov MS and Stockwell BR: Peroxidation of polyunsaturated
fatty acids by lipoxygenases drives ferroptosis. Proc Natl Acad Sci
USA. 113:E4966–E4975. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Feng L, Zhao K, Sun L, Yin X, Zhang J, Liu
C and Li B: SLC7A11 regulated by NRF2 modulates esophageal squamous
cell carcinoma radiosensitivity by inhibiting ferroptosis. J Transl
Med. 19:3672021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cao JY and Dixon SJ: Mechanisms of
ferroptosis. Cell Mol Life Sci. 73:2195–2209. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Linkermann A, Skouta R, Himmerkus N, Mulay
SR, Dewitz C, De Zen F, Prokai A, Zuchtriegel G, Krombach F, Welz
PS, et al: Synchronized renal tubular cell death involves
ferroptosis. Proc Natl Acad Sci USA. 111:16836–16841. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Doll S, Proneth B, Tyurina YY, Panzilius
E, Kobayashi S, Ingold I, Irmler M, Beckers J, Aichler M, Walch A,
et al: ACSL4 dictates ferroptosis sensitivity by shaping cellular
lipid composition. Nat Chem Biol. 13:91–98. 2017. View Article : Google Scholar :
|
|
26
|
Kenny EM, Fidan E, Yang Q, Anthonymuthu
TS, New LA, Meyer EA, Wang H, Kochanek PM, Dixon CE, Kagan VE and
Bayir H: Ferroptosis contributes to neuronal death and functional
outcome after traumatic brain injury. Crit Care Med. 47:410–418.
2019. View Article : Google Scholar :
|
|
27
|
Zhong W, Sun B, Gao W, Qin Y, Zhang H,
Huai L, Tang Y, Liang Y, He L, Zhang X, et al: Salvianolic acid A
targeting the transgelin-actin complex to enhance vasoconstriction.
EBioMedicine. 37:246–258. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wen F, Sun X, Sun C, Dong Z, Jia G, Bao W,
Yu H and Yang C: TAGLN is downregulated by TRAF6-mediated
proteasomal degradation in prostate cancer cells. Mol Cancer Res.
19:1113–1122. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xi Y, Liu J and Shen G: Low expression of
IGFBP4 and TAGLN accelerate the poor overall survival of
osteosarcoma. Sci Rep. 12:92982022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sayar N, Karahan G, Konu O, Bozkurt B,
Bozdogan O and Yulug IG: Transgelin gene is frequently
downregulated by promoter DNA hypermethylation in breast cancer.
Clin Epigenetics. 7:1042015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu B, Chen X, Li J, Qu Y, Su L, Peng Y,
Huang J, Yan J, Yu Y, Gu Q, et al: Stromal fibroblasts in the
microenvironment of gastric carcinomas promote tumor metastasis via
upregulating TAGLN expression. BMC Cell Biol. 14:172013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu X, Dong L, Zhang R, Ying K and Shen H:
Transgelin overexpression in lung adenocarcinoma is associated with
tumor progression. Int J Mol Med. 34:585–591. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou L, Zhang R, Zhang L, Sun Y, Yao W,
Zhao A, Li J and Yuan Y: Upregulation of transgelin is an
independent factor predictive of poor prognosis in patients with
advanced pancreatic cancer. Cancer Sci. 104:423–430. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhong W, Hou H, Liu T, Su S, Xi X, Liao Y,
Xie R, Jin G, Liu X, Zhu L, et al: Cartilage oligomeric matrix
protein promotes epithelial-mesenchymal transition by interacting
with Transgelin in colorectal cancer. Theranostics. 10:8790–8806.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang B, Chen Q, Wan C, Sun S, Zhu L, Zhao
Z, Zhong W and Wang B: Transgelin inhibits the malignant
progression of esophageal squamous cell carcinomas by regulating
epithelialmesenchymal transition. Front Oncol. 11:7094862021.
View Article : Google Scholar
|
|
36
|
Rice TW, Kelsen D, Blackstone EH, Ishwaran
H and Hofstetter WL: Esophagus and Esophagogastric Junction.
2017.AJCC Cancer Staging Manual. View Article : Google Scholar
|
|
37
|
Nicchia GP, Stigliano C, Sparaneo A, Rossi
A, Frigeri A and Svelto M: Inhibition of aquaporin-1 dependent
angiogenesis impairs tumour growth in a mouse model of melanoma. J
Mol Med (Berl). 91:613–623. 2013. View Article : Google Scholar
|
|
38
|
Simone L, Gargano CD, Pisani F, Cibelli A,
Mola MG, Frigeri A, Svelto M and Nicchia GP: Aquaporin-1 inhibition
reduces metastatic formation in a mouse model of melanoma. J Cell
Mol Med. 22:904–912. 2018.
|
|
39
|
Boren T, Xiong Y, Hakam A, Wenham R, Apte
S, Wei Z, Kamath S, Chen DT, Dressman H and Lancaster JM: MicroRNAs
and their target messenger RNAs associated with endometrial
carcinogenesis. Gynecol Oncol. 110:206–215. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Najafi M, Farhood B and Mortezaee K:
Extracellular matrix (ECM) stiffness and degradation as cancer
drivers. J Cell Biochem. 120:2782–2790. 2019. View Article : Google Scholar
|
|
41
|
Murphy JM, Rodriguez Y, Jeong K, Ahn EE
and Lim SS: Targeting focal adhesion kinase in cancer cells and the
tumor microenvironment. Exp Mol Med. 52:877–886. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Abnet CC, Arnold M and Wei WQ:
Epidemiology of esophageal squamous cell carcinoma.
Gastroenterology. 154:360–373. 2018. View Article : Google Scholar
|
|
43
|
Lees-Miller JP, Heeley DH and Smillie LB:
An abundant and novel protein of 22 kDa (SM22) is widely
distributed in smooth muscles. Purification from bovine aorta.
Biochem J. 244:705–709. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Camoretti-Mercado B, Forsythe SM, LeBeau
MM, Espinosa R III, Vieira JE, Halayko AJ, Willadsen S, Kurtz B,
Ober C, Evans GA, et al: Expression and cytogenetic localization of
the human SM22 gene (TAGLN). Genomics. 49:452–457. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lawson D, Harrison M and Shapland C:
Fibroblast transgelin and smooth muscle SM22alpha are the same
protein, the expression of which is down-regulated in many cell
lines. Cell Motil Cytoskeleton. 38:250–257. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ueda T, Araki N, Mano M, Myoui A, Joyama
S, Ishiguro S, Yamamura H, Takahashi K, Kudawara I and Yoshikawa H:
Frequent expression of smooth muscle markers in malignant fibrous
histiocytoma of bone. J Clin Pathol. 55:853–858. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gimona M, Kaverina I, Resch GP, Vignal E
and Burgstaller G: Calponin repeats regulate actin filament
stability and formation of podosomes in smooth muscle cells. Mol
Biol Cell. 14:2482–2491. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wei X, Lou H, Zhou D, Jia Y, Li H, Huang
Q, Ma J, Yang Z, Sun C, Meng Y, et al: TAGLN mediated
stiffness-regulated ovarian cancer progression via RhoA/ROCK
pathway. J Exp Clin Cancer Res. 40:2922021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Rao D, Kimler BF, Nothnick WB, Davis MK,
Fan F and Tawfik O: Transgelin: A potentially useful diagnostic
marker differentially expressed in triple-negative and
non-triple-negative breast cancers. Hum Pathol. 46:876–883. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Assinder SJ, Stanton JA and Prasad PD:
Transgelin: An actin-binding protein and tumour suppressor. Int J
Biochem Cell Biol. 41:482–486. 2009. View Article : Google Scholar
|
|
51
|
Thompson O, Moghraby JS, Ayscough KR and
Winder SJ: Depletion of the actin bundling protein SM22/transgelin
increases actin dynamics and enhances the tumourigenic phenotypes
of cells. BMC Cell Biol. 13:12012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yang Z, Chang YJ, Miyamoto H, Ni J, Niu Y,
Chen Z, Chen YL, Yao JL, di Sant'Agnese PA and Chang C: Transgelin
functions as a suppressor via inhibition of ARA54-enhanced androgen
receptor transactivation and prostate cancer cell growth. Mol
Endocrinol. 21:343–358. 2007. View Article : Google Scholar
|
|
53
|
Li J, Cao F, Yin HL, Huang ZJ, Lin ZT, Mao
N, Sun B and Wang G: Ferroptosis: Past, present and future. Cell
Death Dis. 11:882020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li Y, Zeng X, Lu D, Yin M, Shan M and Gao
Y: Erastin induces ferroptosis via ferroportin-mediated iron
accumulation in endometriosis. Hum Reprod. 36:951–964. 2021.
View Article : Google Scholar
|
|
55
|
Tsui KH, Lin YH, Chang KS, Hou CP, Chen
PJ, Feng TH and Juang HH: Transgelin, a p53 and PTEN-upregulated
gene, inhibits the cell proliferation and invasion of human bladder
carcinoma cells in vitro and in vivo. Int J Mol Sci. 20:49462019.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang ZW, Yang ZM, Zheng YC and Chen ZD:
Transgelin induces apoptosis of human prostate LNCaP cells through
its interaction with p53. Asian J Androl. 12:186–195. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tang D and Kroemer G: Ferroptosis. Curr
Biol. 30:R1292–R1297. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lei G, Zhuang L and Gan B: Targeting
ferroptosis as a vulnerability in cancer. Nat Rev Cancer.
22:381–396. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Liu MR, Zhu WT and Pei DS: System Xc(-): A
key regulatory target of ferroptosis in cancer. Invest New Drugs.
39:1123–1131. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Koppula P, Zhuang L and Gan B: Cystine
transporter SLC7A11/xCT in cancer: Ferroptosis, nutrient
dependency, and cancer therapy. Protein Cell. 12:599–620. 2021.
View Article : Google Scholar :
|
|
61
|
Gong D, Chen M, Wang Y, Shi J and Hou Y:
Role of ferroptosis on tumor progression and immunotherapy. Cell
Death Discov. 8:4272022. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhou Y, Bian S, Zhou X, Cui Y, Wang W, Wen
L, Guo L, Fu W and Tang F: Single-cell multiomics sequencing
reveals prevalent genomic alterations in tumor stromal cells of
human colorectal cancer. Cancer Cell. 38:818–828.e5. 2020.
View Article : Google Scholar : PubMed/NCBI
|























