Introduction
Glioblastoma (GBM) is the most prevalent malignant
primary brain tumor in adults, constituting 54% of all glioma cases
(1). Despite standard treatment
involving surgery followed by concurrent chemoradiotherapy and
adjuvant chemotherapy, the curative effect remains limited, with a
median patient survival time of only ~15 months (2). Consequently, there is a pressing
need to explore the biological nature of GBM, decipher the
signaling pathways underpinning tumor progression, and develop
therapeutic strategies targeting driver factors to improve disease
management and patient lifespan.
Pseudogenes, a distinct class of long non-coding
RNAs (lncRNAs), have sequences that are dysfunctional copies of
protein-coding genes. Yet, burgeoning evidence suggests their
involvement in various biological processes, and their role in
significant functions (3).
Generally, pseudogenes can be transcribed into antisense RNA to
meddle with coding genes or act as competitive endogenous RNA by
binding to microRNAs (miRNAs) (4). Numerous studies have indicated that
pseudogene dysregulation may contribute to disease development,
including tumors. For instance, the pseudogene PTENP1 can exert a
growth-suppressive function by regulating cellular levels of PTEN
through competitive miRNA binding, but its gene locus is
selectively lost in human cancer (5). A network made up of numerous miRNAs
and several pseudogenes, all originating from a single parent gene,
can be controlled through various mechanisms. Disruption or
dysregulation of this intricate network can lead to the onset and
progression of cancer, such as the case with an FTH1 pseudogene
observed in prostate cancer (6).
Genomic gains and aberrant expression of BRAFP1, which serves as a
ceRNA sponge for miRNAs targeting BRAF, can elicit its oncogenic
activity and induce lymphoma (7).
The enhancement of mitochondrial fission is facilitated by RACGAP1P
via a mechanism that depends on its competitive interaction with
miR-345-5p, counteracting its parent gene, RACGAP1. This process
culminates in the activation of dynamin-related protein 1 (Drp1),
and advances the invasion and metastasis of breast cancer (8). Despite previous revelations about
the prognostic value of two pseudogene signatures in glioma
cohorts, understanding of pseudogene expression patterns, functions
and regulatory mechanisms in glioma remains limited (9,10).
Only a handful of tumor-promoting pseudogenes have been identified
in glioma: Hypoxia-induced PDIA3P1, which facilitates mesenchymal
transition through the PDIA3P1/miR-124-3p/RELA axis; LGMNP1, which
elevates LGMN expression by sponging miR-495-3p; and ANXA2P2, which
competes with miR-9 against LDHA to modulate aerobic glycolysis
progression and tumor cell proliferation (11-13).
Pseudogene UBDP1, residing on chromosome 6p22.1 and
spanning 271 bp, exhibits a high degree of sequence homology with
its coding gene UBD, also known as FAT10. This ubiquitin-like
regulatory protein with a proteasome degradation signal (14) is overexpressed in a variety of
solid tumors, including those of the breast, stomach, colon, liver
and pancreas. It also promotes the invasion and metastasis of
hepatocellular carcinoma (15),
the metastasis of osteosarcoma (16), breast cancer invasion (17) and the chemotherapy resistance of
non-small cell lung cancer (18).
Its overexpression and oncogenic activity have been identified in
glioma as well (19,20). Yet, the interaction and role of
the UBDP1-UBD binary system in glioma progression remain largely
unexplored.
The present study identified a novel pseudogene,
UBDP1, which is upregulated in GBM and associated with a poor
patient prognosis. Further in vitro and in vivo
investigations demonstrated that both UBDP1 and UBD foster GBM
proliferation, migration and invasion. UBDP1 serves as an
endogenous sponge for miR-6072, obstructing its interaction with
UBD, subsequently enhancing UBD's oncogenic capabilities and
promoting tumor advancement. These findings illustrated the
intricate pseudogene regulatory network within GBM and highlight a
potential therapeutic target.
Materials and methods
Clinical samples
Tumor specimens were obtained from patients who were
initially diagnosed with GBM and underwent their first surgical
procedure between December 2016 and May 2020 at Shanghai Changzheng
Hospital, Naval Medical University (Shanghai, China). The median
age of patients was 49 (range, 34-65), comprising 15 men and 15
women. Normal brain tissues were obtained from craniocerebral
trauma patients who received intracranial decompression therapy
between January 2018 and May 2020 at the aforementioned hospital.
The median age of patients was 41 years (range, 29-57). The present
study was approved (approval no. CZEC2018-032) by the Independent
Ethics Committee of Shanghai Changzheng Hospital (Shanghai, China).
Written informed consent was obtained from all patients or their
legal guardians indicating their understanding of the risks and
benefits.
Microarray analysis
lncRNA microarray data were obtained from the
authors' previous study (GEO dataset: GSE51146) (21). Differentially expressed lncRNAs
were identified by using Volcano Plot filtering. The threshold of
upregulated and downregulated lncRNAs was P<0.05 and a fold
change >2.
Cell lines, authentication and culture
conditions
Human cell lines U87MG (RRID: CVCL_0022), U251MG
(RRID: CVCL_0021) and 293T (DSMZ no. ACC 635) were purchased from
the Cell Bank at the Chinese Academy of Sciences (Shanghai, China).
In order to authenticate these cell lines, STR profiling was
conducted in the year 2020; the identities of these cell lines were
confirmed and authenticated. The U87 cell line used in the present
study is most probably an ATCC-originating version of the U87 MG
cell line. All these cells were cultured in Dulbecco's modified
Eagle's medium (DMEM) supplemented with 10% FBS (both from Hyclone;
Cytiva) and 5% CO2 at 37°C.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from tissues and cells using
TRIzol reagent (Wuhan Servicebio Technology Co., Ltd.). RNA was
reverse transcribed to cDNA using RevertAid First Strand cDNA
Synthesis kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. qPCR was performed using FastStart
Universal SYBR Green Master (Roche Diagnostics) on the RT-PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were as follows: The procedure involved 40
cycles, which consisted of denaturation at 98°C for 10 sec,
annealing at 58°C for 25 sec and extension at 68°C for 30 sec.
GADPH was marked as an internal control. The primers used were as
follows: GAPDH forward, 5′-GGA AGC TTG TCA TCA ATG GAA ATC-3′ and
reverse, 5′-TGA TGA CCC TTT TGG CTC CC-3′; UBD forward, 5′-CAA TGC
TTC CTG CCT CTG TGT-3′ and reverse, 5′-GGG TAA GGT GGA TGG TCT TCT
CT-3′; and UBDP1 forward, 5′-TGG CTG CTA AAA TGG AGT GAA GA-3′ and
reverse, 5′-AGG TGA GGT GGA TGG TCT TCT T-3′. The expression levels
were determined using the 2−ΔΔCq method (22).
Fluorescence in situ hybridization
(FISH)
The cells were fixed in 4% paraformaldehyde for 15
min at room temperature, washed with PBS, treated with pepsin (1%
in 10 mM HCl), and dehydrated with 70, 85 and 100% ethanol. The
cells were then air-dried and incubated in a hybridization buffer
containing the FISH probe for 5 min at 73°C in a water bath. The
hybridization was performed for 12 h at 37°C. After washing and
dehydrating the cell slides, they were counterstained with DAPI
(2.5 μg/ml). The RNA FISH probe for UBDP1 was designed and
synthesized by Shanghai GenePharma Co., Ltd., and its sequence was
5′-FAM-CCT ACC TTC TTC ACT CCA TTT TAG CAG CCA-FAM-3′.
Plasmid construction
The gene sequences and target sequences of UBD and
UBDP1 were constructed into the GV657/GV248 vector plasmid
(Shanghai GeneChem Co., Ltd.) by PCR amplification and double
enzyme digestion (restriction enzymes, New England BioLabs, Inc.).
Thus, GV657/UBD-Overexpression, GV657/UBDP1-Overexpression,
GV248/UBD-Knockdown and GV248/UBDP1-Knockdown plasmids were
obtained. GV657/GV248 vector plasmids (Shanghai GeneChem Co., Ltd.)
were used as negative control (NC). The sequences of PCR
amplification primers were as follows: UBD overexpression forward,
5′-CGG AAT TCA TGG CTC CCA ATG CTT CCT-3′ and reverse, 5′-CGG GAT
CCT CAC CCT CCA ATA CAA TAA CAT GCC A-3′; UBD knockdown forward,
5′-CCG GCG AGA CTA AGA CGG GTA TAA TCT CGA GAT TAT ACC CGT CTT AGT
CTC GTT TTT G-3′ and reverse, 5′-AAT TCA AAA ACG AGA CTA AGA CGG
GTA TAA TCT CGA GAT TAT ACC CGT CTT AGT CTC G-3′; UBDP1
overexpression forward, 5′-CGG AAT TCG TTG GTG ATA CCT ACT TTC ACT
GAG-3′ and reverse, 5′-CGG GAT CCG CAT CTC TCT ACC CCT GGG-3′; and
UBDP1 knockdown forward, 5′-CCG GTC TGG TGG AAA CAT GTG ATC AAG CTT
CAT CAC ATG TTT CCA CCA GAT TTT TG-3′ and reverse, 5′-AAT TCA AAA
ATC TGG TGG AAA CAT GTG ATG AAG CTT GAT CAC ATG TTT CCA CCA
GA-3′.
The gene sequences and target sequences of miR-6072
and miR-6818-3p were constructed into the GV251/GV249 vector
plasmid (Shanghai GeneChem Co., Ltd.) by PCR amplification and
double enzyme digestion (restriction enzymes, New England BioLabs,
Inc.). Thus, the GV251/miR-6072-Overexpression,
GV251/miR-6818-3p-Overexpression, GV249/miR-6072-Inhibition and
GV249/miR-6818-3p-Inhibition plasmids were obtained. GV251/GV249
vector plasmids (Shanghai GeneChem Co., Ltd.) were used as NC. The
sequences of PCR amplification primers were as follows: MiR-6072
overexpression forward, 5′-TGT GGA AAG GAC GCG GGA TCA GAT GCA CAG
GAC TGG GCA C-3′ and reverse, 5′-CAG CGG TTT AAA CTT AAG CTA AAA
AAT AAG AAC TAC TCT ATG-3′; miR-6072 inhibition forward, 5′-GCT AAA
AAT CCT CAT CAC ACT GCA CCT TAG G-3′ and reverse, 5′-ATC CCT AAG
GTG CAG TGT GAT GAG GAT TTT T-3′; miR-6818-3p overexpression
forward, 5′-ACG GGC CCT CTA GAC TCG AGT GTT GGT TGT GTA AGA TTT
C-3′ and reverse, 5′-TTA AAC TTA AGC TTG GTA CCT ACT GAC TGT ACC
AGA TGC-3′; and miR-6818-3p inhibition forward, 5′-GCT AAA AAT TGT
CTC TTG TTC CTC ACA CAG G-3′ and reverse, 5′-ATC CCT GTG TGA GGA
ACA AGA GAC AAT TTT T-3′. The vectors and DNA fragments were then
connected using T4 ligase from New England BioLabs, Inc.
Cell transfection
U87 and U251 cell lines were cultured to a density
of 2×105 cells for transfection purposes. Transfection
was performed using 50 pmol of either miRNA mimics or inhibitors.
Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.) was
employed as the transfection reagent, and 5 μl was
administered to each well of a 6-well plate, as per the
manufacturer's instructions. To prepare the transfection mix,
Lipofectamine® 3000 was first diluted in Opti-MEM Medium
(Gibco; Thermo Fisher Scientific, Inc.) and thoroughly mixed.
Separately, a master mix of the DNA was prepared by diluting the
desired amount of DNA also in Opti-MEM Medium and mixing until
being homogeneous. The diluted DNA was then combined with the
previously diluted Lipofectamine® 3000, using a 1:1
ratio, to form a DNA-lipid complex. This complex was allowed to
incubate for 20 min at room temperature (37°C) to facilitate the
formation of a stable DNA-lipid complex. Subsequently, the complex
was administered to the cultured cells. Following transfection, the
cells were incubated at 37°C for a period of 48 h prior to
conducting any subsequent experiments. For lentiviral transduction,
the vectors (GL132/GV248; Shanghai GenePharma Co., Ltd.) were
co-transfected with helper in the 2nd generation transfection
system into 293T cells. The 293T cells were cultured in DMEM
supplemented with 10% FBS at a temperature of 37°C and a
CO2 concentration of 5%. Following a 48-h incubation
period at 37°C, the supernatant of the 293T cells were collected
and the lentivirus was concentrated by subjecting to centrifugation
at 25,000 x g for 2 h at 4°C. The harvested lentiviral vectors were
then introduced into the target GBM cells (U87/U251) for
transduction. Subsequently, U87/U251 cell lines were plated into a
6-well plate and the cells were cultured until they reach 80%
confluence. To generate stable cell lines, GBM cells were
transduced with these lentiviral vectors in a milieu containing
polybrene (Shanghai GenePharma Co., Ltd.) at a concentration of 5
μg/ml. Then, lentivirus was added and co-cultured with the
cells at 37°C for 24 h (Multiplicity of infection, 10). After that,
the medium was replaced, and then culture continued in a 5%
CO2 and 37°C incubator for another 48 h. Following a
72-h incubation period, cells underwent a selection process using 2
μg/ml of puromycin (Shanghai GeneChem Co., Ltd.), over a
course of 3 days.
Western blot analysis
Cell lysates were prepared with RIPA buffer (Thermo
Fisher Scientific, Inc.). The proteins were separated using 10%
SDS-PAGE and then transferred to PVDF membranes (EMD Millipore),
and a total of 25 μg protein were loaded per lane. After 2 h
of blocking using 5% milk at room temperature, membranes were
incubated overnight at 4°C with different primary antibodies (FAT10
Rabbit mAb; 1:1,000; cat. no. 76194; Cell Signaling Technology,
Inc.). The membranes were then incubated at room temperature for 1
h with secondary antibodies (Anti-rabbit IgG HRP-linked antibody;
1:10,000; cat. no. 7074; Cell Signaling Technology, Inc.; RRID:
AB_2099233), and the bands were detected using a chemiluminescence
imager (Syngene) and analyzed with Image Lab™ Software (version
5.1; Bio-Rad Laboratories, Inc.). GAPDH (1:1,000; cat. no.
sc-47724; Santa Cruz Biotechnology, Inc.) served as the
control.
Cell proliferation assay
The proliferation ability of the cells was tested
using the Cell Counting Kit-8 assay (CCK-8; Shanghai Yeasen
Biotechnology Co., Ltd.), according to the manufacturer's
instructions. Cells were seeded in a 96-well plate at a density of
1,000-2,000 each and cultured at 37°C. At the indicated time points
(0, 24, 48, 72 and 96 h), 10 μl CCK-8 was added to the plate
for 4 h. The optical density at 450 nm was measured consecutively
using a microplate reader (Bio-Rad Laboratories, Inc.). Each
experiment was performed thrice.
Cell migration and invasion assays
The migration assays were performed using the
uncoated plates in Transwell chambers (8-μm diameter pores;
Corning, Inc.), while Matrigel-coated plates were used for invasion
assays at 37°C for 1 h. The upper chambers contained
2×104 cells/well in serum-free medium, while FBS with
10% serum was loaded into the lower chamber. After 24 h of
incubation at 37°C with 5% CO2 in the humidified
incubator, the cells were stained with 0.4% crystal violet at room
temperature for 10 min. The remaining cells in the upper chamber
were removed with cotton swabs, while the migratory or invasive
cells in the lower chamber were counted under a light
microscope.
Bioinformatic analysis
The miRanda tool (http://www.microRNA.org/) was used to predict binding
sites between miRNAs, UBD mRNA, and UBDP1, with Score and Energy
thresholds of >140 and <-20, respectively. The screened
miRNAs targeting UBD and UBDP1 were subsequently matched against
databases, including miRWalk 3.0 (http://mirwalk.umm.uni-heidelberg.de/), miRDB
(https://mirdb.org/), ENCORI (https://rnasysu.com/encori/), microRNA (https://mirbase.org/), Tarbase (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index)
and miRNet (https://www.mirnet.ca/). Then, the
miRNA(s) shared by the maximum number of databases and binding with
maximum similarity to the UBD mRNA and UBDP1 sequences were
identified.
Luciferase reporter assay
The dual luciferase reporter plasmids [GM-1013FL02 +
UBDP1 mutant (MT)/wild-type (WT), GM-1013FL02 + UBD MT/WT] were
designed and synthesized by Shanghai GenePharma Co., Ltd.
Co-transfection was conducted with Lipofectamine® 3000
for 48 h. The activities of the firefly and Renilla
luciferases were analyzed using a dual luciferase assay kit (cat.
no. E1910; Promega Corporation), according to the manufacturer's
instructions. The assay was conducted independently in
triplicate.
Tumor xenograft model
Nude female BALB/cA (RRID: MGI:2160349) mice (age, 4
weeks-old) were purchased from the Shanghai Jihui Laboratory Animal
Care Co., Ltd. The nude mice were kept in separate ventilated cages
within a controlled environment that was free from pathogens, and
they were provided with free access to food and water. The mice
were subjected to a 12/12-h light/dark cycle, with the temperature
maintained between 20-25°C and humidity levels ranging from 50-80%.
At the time of tumor cell injection, each mouse weighed ~18±1.75 g
and was 6 weeks-old. They were divided into three groups (n=9 in
each group). The mice were subjected to anesthesia through
intraperitoneal injection of ketamine (100 mg/kg) and xylazine (20
mg/kg). Subsequently, they were secured in a stereotaxic frame
(Kopf Instruments) and placed on a heating pad to ensure
temperature maintenance. After careful sterilization, 2 μl
PBS containing 1×107 U87 cells were injected using a
Hamilton micro-syringe in the mice's parietooccipital median region
with the depth 2-3 mm, the whole injection period lasting for 2
min. After injection, the health and behavior of every nude mouse
was monitored on a daily basis. The tumor burden in the mice began
to adversely affect their quality of life, causing severe
difficulties in fundamental activities such as drinking, eating and
moving, thereby severely diminishing their comfort. Consequently,
to minimize any further distress, all mice were compassionately
euthanized using an intraperitoneal injection of pentobarbital (150
mg/kg). This was followed by a secondary verification of
euthanasia, employing cervical dislocation to confirm the cessation
of life. In total, 60 days after implantation, all mice were
euthanized by intraperitoneal injection of pentobarbital and
cervical dislocation. Cardiac arrest was then employed to confirm
death by the examination of pulse palpation. The overall survival
time from tumor implantation to the death of mice was then
recorded. Their whole brains were removed when the mice were
euthanized and immediately processed for histological evaluation.
In the present investigation, the maximum tumor diameter was found
to be <2 cm, and the maximum tumor volume was <2,000
mm3. All mice experiments were performed according to
the Institutional Guidelines for the Care and Use of Laboratory
Animals of the Naval Medical University and were approved by the
Animal Care and Experimental Committee of Naval Medical University
(approval no. CZEC2018-032; Hefei, China).
Immunohistochemistry (IHC)
The IHC assay was performed as previously described
(23). The primary antibody used
was Ki-67 Rabbit mAb (1:100; cat. no. 9027; Cell Signaling
Technology, Inc.). The tumor tissues sections (5.0-μm-thick)
were stained with hematoxylin and eosin (cat. no. G1003; Wuhan
Servicebio Technology Co., Ltd.) at room temperature for 5 and 7
min, respectively.
Statistical analysis
All statistical analyses were performed using
GraphPad Prism 6.0 (Dotmatics) and SPSS (RRID:SCR_002865) 18.0
software (IBM Corp.). The unpaired Student's t-test was used to
compare the two groups. A one-way ANOVA test followed by Turkey's
post hoc test was performed to analyze data among multiple groups.
Pearson correlation analysis was applied to determine the linear
relationship between the expression of different genes. The
survival curves were calculated by the Kaplan-Meier method, and a
log-rank test detected the difference. Univariate and multivariate
survival analyses used the Cox hazard regression model; univariate
analysis factors with P<0.1 were included in the multivariate
analysis. Data were presented as the mean ± standard deviation
(SD). P<0.05 was considered to indicate a statistically
significant difference.
Results
UBDP1 and UBD are concomitantly highly
expressed in GBM and correlate with a poor prognosis
From the authors' previous microarray data
(GSE51146), differentially expressed lncRNAs in GBM were searched
and eight upregulated and 29 downregulated pseudogenes were
identified (Fig. 1A and Table SI). Among these dysregulated
pseudogenes, UBDP1 was selected due to its significant expression
shifts and unexplored functions in glioma (Fig. 1B). RT-qPCR was then utilized to
assess UBDP1 expression in 30 GBM and 15 normal brain tissue
samples, confirming a substantial upregulation of UBDP1 in GBM
(Fig. 1C). Furthermore, analysis
of the UBDP1 gene disclosed a pseudogene with 75% sequence
similarity to UBD mRNA, which was also found to be significantly
overexpressed in GBM (Fig. 1C),
with its levels exhibiting a strong positive correlation with UBDP1
(Fig. 1D).
To elucidate the clinical significance of UBDP1 in
GBM, clinical data from patients with GBM were collected, and
patients were categorized based on their UBDP1 expression level
(high and low) (Table I). When
analyzed using the Kaplan-Meier curve and Cox hazard regression
model, elevated UBDP1 expression significantly associated with
shorter overall survival times (Fig.
1E) and presented as a potential independent risk factor
(Tables II and SII). Consistently, high expression of
UBD also associated with poor patient prognosis (Fig. 1F). These findings suggested that
both UBDP1 and UBD could potentially drive GBM progression.
 | Table IClinicopathological characteristics
and expression level of UBDP1 of 30 patients with glioblastoma. |
Table I
Clinicopathological characteristics
and expression level of UBDP1 of 30 patients with glioblastoma.
| Clinicopathological
characteristics | Number of
patients
(n=30) (%) | Expression level of
UBDP1
|
|---|
High
(n=15) | Low
(n=15) |
|---|
| Sex | | | |
| Male | 15 (50) | 5 | 10 |
| Female | 15 (50) | 10 | 5 |
| Age | | | |
| ≤45 | 9 (30) | 2 | 7 |
| >45 | 21 (70) | 13 | 8 |
| Tumor size
(cm) | | | |
| <4 | 15 (50) | 11 | 4 |
| ≥4 | 15 (50) | 4 | 11 |
| Resection
degree | | | |
| Total | 26 (87) | 11 | 15 |
| Subtotal | 4 (13) | 4 | 0 |
|
Radio-chemotherapy | | | |
| Yes | 23 (77) | 11 | 12 |
| No | 7 (23) | 4 | 3 |
| IDH1 mutation | | | |
| Yes | 2 (7) | 0 | 2 |
| No | 28 (93) | 15 | 13 |
| UBD | | | |
| High | 15 (50) | 10 | 5 |
| Low | 15 (50) | 5 | 10 |
 | Table IICox multivariate analysis of factors
associated with overall survival of patients with glioblastoma. |
Table II
Cox multivariate analysis of factors
associated with overall survival of patients with glioblastoma.
| Variable | Hazard ratio | 95% confidence
interval | P-value |
|---|
| UBDP1 (low vs.
high) | 4.114 | 1.484-11.405 | 0.007 |
| Age (≤45 vs.
>45) | 6.649 | 2.060-21.459 | 0.002 |
| Tumor size (<4
cm vs. ≥4 cm) | 7.021 | 1.829-26.945 | 0.009 |
| Resection degree
(Total vs. Subtotal) | | | 0.106 |
| Radio-chemotherapy
(Yes vs. No) | 0.173 | 0.042-0.704 | 0.014 |
| IDH1 mutation (Yes
vs. No) | | | 0.175 |
UBDP1 and UBD promote proliferation,
migration and invasion of glioma cells
To investigate the biological roles of UBDP1 and UBD
in glioma, stable UBDP1- and UBD-overexpressing or knocking down
cell lines [U87 and U251 (RRID: CVCL_1G29) cells] were established,
validating their expressions using RT-qPCR and western blotting.
Then, a CCK-8 assay was performed to determine the effect of UBDP1
and UBD on proliferation. Overexpression of UBDP1 and UBD increased
the proliferation potential of glioma cells, whereas the knockdown
of UBDP1 and UBD significantly impeded this capacity (Fig. 2A and B). Furthermore, Transwell
and Matrigel assays were conducted to examine whether UBDP1 and UBD
are involved in glioma migration and invasion. As indicated in
Fig. 2C-F, upregulation of UBDP1
and UBD significantly accelerated glioma cell migration and
invasion compared with the control group. By contrast, knockdown of
UBDP1 and UBD decreased the number of migratory and invasive cells.
These results demonstrated the tumor-promoting capacities of UBDP1
and UBD in glioma cells.
UBDP1 and UBD aggravate the malignancy of
glioma in vivo
To determine the in vivo function of UBDP1
and UBD, U87 cells, either overexpressing UBDP1 or UBD or serving
as a NC, were injected into the brains of nude mice. As expected,
elevated levels of UBDP1 and UBD associated with reduced overall
survival in xenograft mice relative to the NC group (Fig. 3B). To access tumor characteristics
in vivo, additional five mice from each group were
euthanized two weeks post-injection. H&E staining of xenograft
sections revealed that mice bearing UBDP1 or UBD overexpressing U87
cells exhibited significantly enhanced tumor growth than mice in
control group (Fig. 3A).
Consistently, an IHC assay confirmed higher Ki-67 levels in
xenograft samples overexpressing UBDP1 or UBD compared with the
control group (Fig. 3C and D).
Collectively, these results suggested that both UBDP1 and UBD
contribute to the oncogenic progression of glioma.
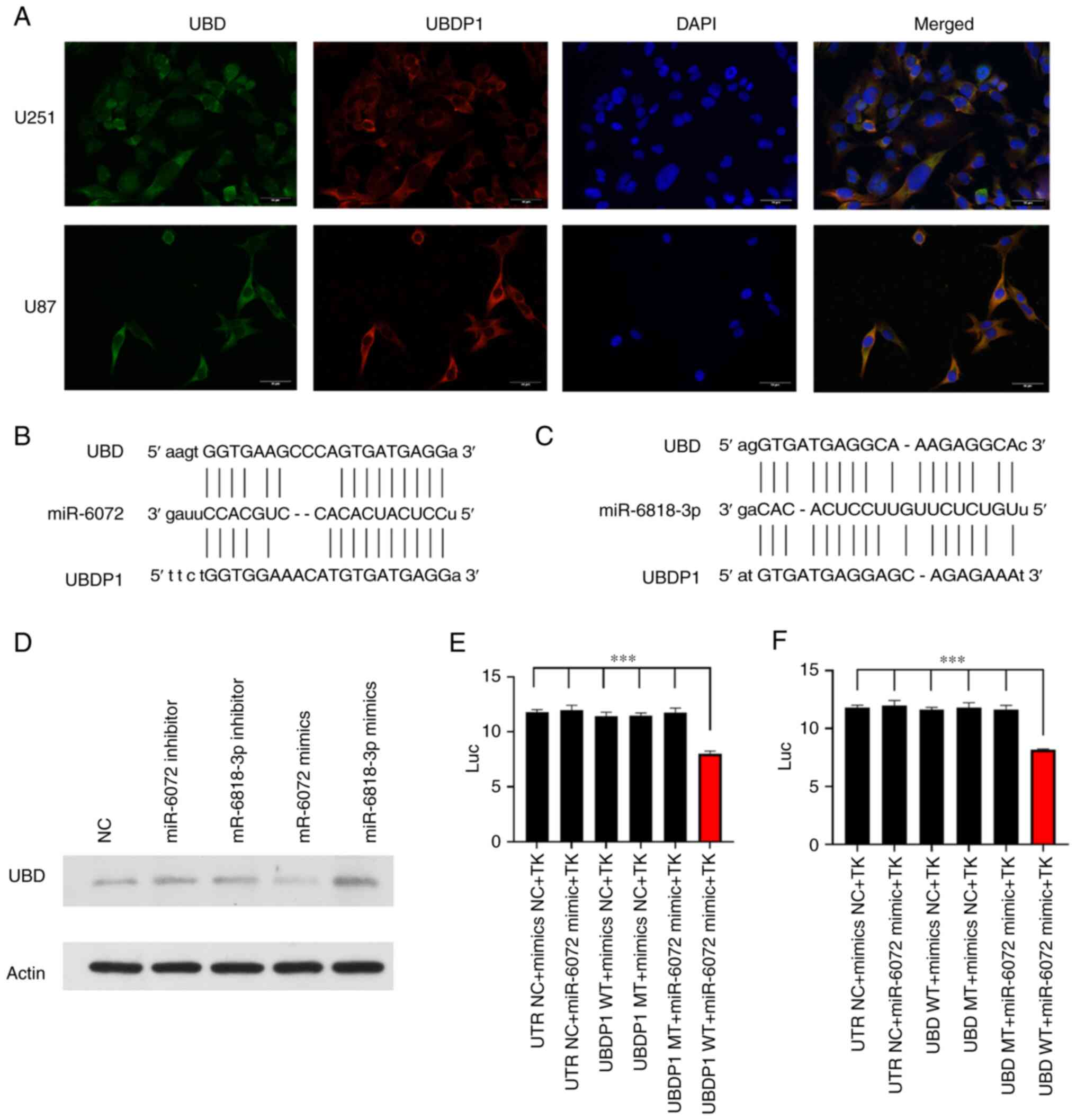
Identification of the common miRNAs
shared between UBD and UBDP1
The localization of UBDP1 in glioma cells was
investigated using FISH and immunofluorescence assays. As
demonstrated in Fig. 4A, UBDP1,
similar to UBD, was predominantly found in the cytoplasm of U251
and U87 cells, suggesting its potential role as a miRNA sponge
(Fig. 4A) (24,25). Several potential miRNA targets for
UBDP1 were predicted using online databases (Table SIII) and two targets, miR-6072
and miR-6818-3p, were found to interact with both UBDP1 and UBD
mRNA (Fig. 4B and C). To evaluate
the influence of these miRNAs on UBD expression, 293T cells were
transfected with miRNA mimics or inhibitors, and alterations in UBD
protein levels were assessed via western blotting. Compared with
the control, miR-6072 mimics were associated with a reduction in
UBD levels, whereas miR-6072 inhibitors resulted in increased UBD
expression. However, miR-6818-3p mimics presented conflicting
results (Fig. 4D).
Dual-luciferase reporter gene assays further confirmed that both
UBDP1 and UBD were direct targets of miR-6072 (Fig. 4E and F). These findings suggested
that miR-6072 plays a pivotal role in the competitive endogenous
RNA network involving UBDP1 and UBD.
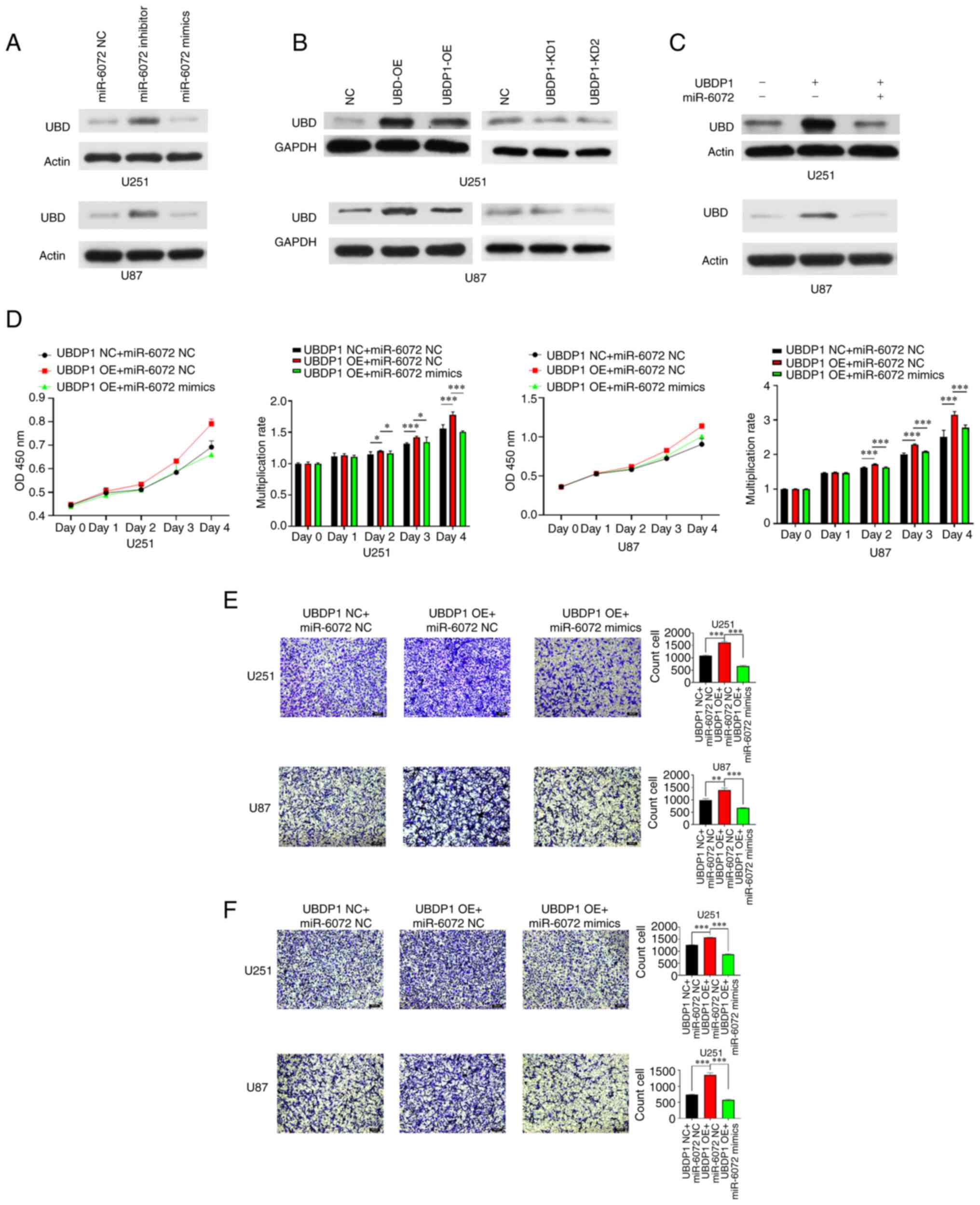
UBD is markedly activated through UBDP1
competitively binding to miR-6072 in glioma
To verify the hypothesis that UBDP1 functions as a
competitive endogenous RNA, sequestering UBD from miRNA-6072
induced degradation, expression of UBD was analyzed in U87 and U251
cells transfected with either a miRNA-6072 mimic or an inhibitor.
The results revealed that overexpression of miRNA-6072
significantly decreased the level of UBD in both glioma cells,
whereas its inhibition increased the UBD expression (Fig. 5A). Additionally, the UBD
expression was detected in U87 and U251 cells transfected with
UBDP1 overexpression or knockdown vector and it was found that the
level of UBD was markedly elevated in UBDP1-overexpressing glioma
cells and notably decreased in UBDP1-knockdown cells (Fig. 5B). Moreover, co-transfection of
UBDP1 and miR-6072 demonstrated that miR-6072 reversed the
protein-level upregulation of UBD mediated by UBDP1 (Fig. 5C). UBDP1 was overexpressed in U87
and U251 glioma cells to determine the role of the
UBD-miR-6072-UBDP1 network in glioma progression. Transfection with
miR-6072 mimics counteracted the increased proliferative ability of
UBDP1-overexpressing cells (Fig.
5D). Similarly, miR-6072 mimics diminished the enhanced
invasion and migration capabilities prompted by UBDP1 (Fig. 5E and F). These results confirmed
that UBDP1 can competitively bind to miR-6072.
Discussion
Pseudogenes can competitively bind to miRNAs,
thereby alleviating their inhibition of the expression of their
parental genes and forming a regulatory network to promote tumor
progression. In the present study, the upregulated pseudogene UBDP1
was identified in GBM, whose high expression was associated with
poor prognosis. Further experiments demonstrated that UBDP1
increases the level of its oncogenic partner UBD by sponging
miRNA-6072, thereby reinforcing the malignant phenotypes of glioma
cells. The present findings revealed a tumor-promoting role of the
UBDP1 and its functioning network with miR-6072 and UBD in glioma
progression.
The UBD gene, located in the 6q21.3 region of the
chromosome, is unique in encoding a ubiquitin-like protein that
directly targets substrates for proteasomal degradation (26). UBD dysregulation has been reported
in various cancers and can promote tumor progression through
multiple pathways (27). Prior
research has shown that UBD, via its ubiquitin-like domain,
interacts with the spindle assembly checkpoint MAD2, thereby
affecting mitotic regulation and contributing to tumor growth and
malignancy (14). Furthermore,
UBD forms a complex with translation elongation factor eEF1A1,
competing with ubiquitin for binding, thus stabilizing eEF1A1
expression to foster tumor proliferation (28). High expression levels of MAD2 and
eEF1A1 have been detected in gliomas, contributing to proliferation
and survival of tumor cells (29-31). It is hypothesized that MAD2 and
eEF1A1 may act as downstream to UBD in a protein-protein
interaction manner during glioma progression.
Moreover, previous studies demonstrated that UBD
influences gene expression at the transcription level. Findings
indicated UBD suppresses the transcriptional activity of tumor
suppressor p53, without significantly affecting its protein levels
(32). The inactivation of p53, a
phenomenon linked to commonly occurring cellular signaling pathways
in glioma genesis, is exacerbated by UBD overactivation (33). UBD and p53 can form a regulatory
loop, maintaining equilibrium between UBD and p53 levels which is
crucial for the precise regulation of p53. When UBD is
overexpressed, it suppresses the transcriptional activity of p53,
which in turn, accelerates tumor development and progression
(32). Interestingly, another
study drew a different conclusion regarding UBD's overexpression
leading to WISP1 protein/mRNA expression discrepancy (34). By stabilizing β-catenin, UBD
overexpression increases WISP1 mRNA expression. While UBD
stabilizes substrates simultaneously which increases WISP1 protein
degradation, and thereby promoting the proliferation of
hepatocellular carcinoma. However, coordinated expression of WISP1
protein and mRNA has been observed in glioma (35). WISP1, as a downstream effector of
the Wnt/β-catenin pathway, is closely associated with glioma
malignancy, promotes glioma cell proliferation,
epithelial-mesenchymal transition, glioma stem cells maintenance
and tumor-supportive macrophages in GBM (35-37). It is reasonable to assume that UBD
upregulates WISP1 in glioma cells without suppressing its protein
level, mediating oncogenic functions of Wnt/β-catenin signaling and
contributing to glioma progression. The mechanisms underlying the
UBDP1/miRNA-6072/UBD regulatory network in glioma need to be
further investigated for verification.
A xenograft GBM model was established in nude mice
using the U87 cell line. The present in vivo studies
revealed a significant correlation between elevated levels of UBDP1
and UBD, decreased overall survival, and increased Ki-67 levels in
xenograft mice compared with the control group. These findings
strongly implicate the involvement of both UBDP1 and UBD in the
oncogenic progression of GBM. However, previous research (38) has brought to light the origin of
the U87 cell line. According to a previous study, the U87 cell
line, while of central nervous system origin, is considered a
bona fide human GBM cell line with an unknown patient
origin, rather than the original GBM cell line established in 1968
at the University of Uppsala. Considering this revelation, it is
imperative that future investigations replicate the current
experiments using alternative stable GBM cell lines. This
replication would serve to validate and reinforce the present
findings, thereby bolstering reliability and generalizability.
The competing endogenous RNA hypothesis suggests
that RNA molecules regulate gene expression by acting as miRNA
sponges. This principle, widely demonstrated in cancer research,
signifies a communication link between pseudogenes and their
parental protein-coding genes. The present study indicated that
miRNA-6072 can target both UBD and UBDP1, suggesting that UBDP1
partly regulates UBD expression by competitively binding with
miRNA-6072, thereby influencing glioma progression. Additionally,
emerging evidence shows that pseudogenes can affect their parental
genes in various ways, either positively or negatively. For
instance, pseudogenes can interact with proteins and localize to
the promoters of their parental genes, which regulate target gene
expression. Antisense RNA (asRNA) generated from pseudogenes can
combine with sense-stranded mRNA from a homologous parent gene,
affecting mRNA stability (39). A
previous study demonstrated that PTENP1, a PTEN pseudogene, encodes
an alpha asRNA isoform that localizes to the PTEN promoter,
epigenetically modulating PTEN transcription by recruiting DNMT3a
and EZH2. The beta asRNA isoform interacts with PTENP1, affecting
its stability and miRNA sponge activity (40). In another study, pseudogene
DUXAP10 was revealed to interact with PRC2 and LSD1, repressing
LATS1 expression at the transcriptional level. It also binds with
HuR, maintaining the stability of β-catenin mRNA and increasing its
protein levels at the post-transcriptional level (41). As the understanding of pseudogenes
has deepened, further mechanistic research is required to explore
the potential effects of UBDP1 on UBD activity in glioma.
Numerous studies have elucidated the pivotal
influence of tumor immune cell infiltration on the prognosis and
therapeutic outcomes for cancer patients. As the role of the immune
system in cancer development and progression becomes increasingly
recognized, immunotherapy has witnessed rapid advancements.
Effective immunotherapy against tumors necessitates sufficient
immune cell infiltration within the tumor microenvironment. UBD
exhibits heterogeneous expression across human tissues, high
expression level was observed within immune system organs such as
lymph nodes, thymus and spleen (42,43). The expression of UBD mRNA in
organs involved in lymphocyte development, maturation and activity
suggests a crucial role in the maturation process of lymphocytes
(44,45). Under normal conditions, UBD is
induced during the maturation process of dendritic cells triggered
by toll-like receptor ligands, enhancing the potential for antigen
presentation and stimulating T cells, thereby playing a key role in
immune defense (46).
A recent study also indicated that pro-inflammatory
cytokines such as interferon-γ, tumor necrosis factor-α and
interleukin-6 can robustly stimulate FAT10 mRNA and protein levels
across all tissues, potentially triggering widespread changes in
cellular processes (47).
Additionally, constitutive stimulation by these cytokines may
promote DNA damage and aberrant tissue healing, creating a
conducive environment for tumorigenesis.
It is crucial to acknowledge that the immune
landscape of gliomas is not solely influenced by T cell
populations. The absence of a broader immune cell profiling is a
limitation of the present study. Other immune cells, including
macrophages, natural killer cells and B cells, also contribute
significantly to the immune dynamics within gliomas. Their roles,
while not explored in the present study, could provide a more
comprehensive understanding of the immune contexture and
therapeutic responses. It is proposed by the authors that future
research should expand on these findings to include a wide array of
immune cells, which will help elucidate the complex interplay
between UBD expression and the immune environment in gliomas.
In summary, the expression of UBD may affect
lymphocytes at tumor sites, altering the efficacy of immune
responses. The complex interplay between UBD expression, GBM
progression and lymphocyte infiltration provides a promising avenue
for future research, particularly in the evaluation of
immunological markers. Prospective studies are encouraged to delve
into the molecular mechanisms underlying UBD-mediated immune
modulation, aiming to identify novel therapeutic targets. Advancing
understanding of UBD's role in glioma immunology paves the way for
the development of more precise and effective immunotherapeutic
strategies, potentially transforming the clinical management of
gliomas.
In conclusion, UBDP1 is upregulated in GBM and has
the potential to serve as a prognostic biomarker for patients.
Moreover, UBDP1 may enhance glioma progression by competitively
binding with miRNA-6072 against its parental gene, UBD. These
findings suggested that blocking the UBDP1/miRNA-6072/UBD network
may represent a novel therapeutic approach for glioma.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FH performed experimental operations and data
analyses. FH, HW, ZG and TH wrote the manuscript. ZG and CC
contributed significantly in data analyses and manuscript revision.
JC and HW conceived and designed the study. TH, QH, ZG, YL, PM and
XZ carried out the study and collected important background
information. HW and JC confirm the authenticity of all the raw
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All methods in the present study were carried out in
accordance with relevant guidelines and regulations and reported in
accordance with ARRIVE guidelines for the reporting of animal
experiments. Animal experiments were approved (approval no.
CZEC2018-032) by the Animal Care and Experimental Committee of
Naval Medical University (Shanghai, China). Human studies were
approved (approval no. CZEC2018-032) by the Independent Ethics
Committee of Naval Medical University (Shanghai, China). Written
informed consents to participate in the present study were obtained
from all patients or their legal guardians prior to study
commencement.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant nos. 81902538, 82272715, 82272904 and
81872072), the Shanghai Sailing Program (grant no. 19YF1448200) and
the Shanghai Basic Research Program (grant no. 19JC1415000).
References
|
1
|
Taphoorn MJB, Dirven L, Kanner AA,
Lavy-Shahaf G, Weinberg U, Taillibert S, Toms SA, Honnorat J, Chen
TC, Sroubek J, et al: Influence of treatment with tumor-treating
fields on health-related quality of life of patients with newly
diagnosed glioblastoma: A secondary analysis of a randomized
clinical trial. JAMA Oncol. 4:495–504. 2018.
|
|
2
|
Stupp R, Taillibert S, Kanner A, Read W,
Steinberg D, Lhermitte B, Toms S, Idbaih A, Ahluwalia MS, Fink K,
et al: Effect of tumor-treating fields plus maintenance
temozolomide vs maintenance temozolomide alone on survival in
patients with glioblastoma: A randomized clinical trial. JAMA.
318:2306–2316. 2017.
|
|
3
|
Pink RC and Carter DRF: Pseudogenes as
regulators of biological function. Essays Biochem. 54:103–112.
2013.
|
|
4
|
Lou W, Ding B and Fu P: Pseudogene-derived
lncRNAs and Their miRNA sponging mechanism in human cancer. Front
Cell Dev Biol. 8:852020.
|
|
5
|
Poliseno L, Salmena L, Zhang J, Carver B,
Haveman WJ and Pandolfi PP: A coding-independent function of gene
and pseudogene mRNAs regulates tumour biology. Nature.
465:1033–1038. 2010.
|
|
6
|
Chan JJ, Kwok ZH, Chew XH, Zhang B, Liu C,
Soong TW, Yang H and Tay Y: A FTH1 gene:Pseudogene:microRNA network
regulates tumorigenesis in prostate cancer. Nucleic Acids Res.
46:1998–2011. 2018.
|
|
7
|
Karreth FA, Reschke M, Ruocco A, Ng C,
Chapuy B, Léopold V, Sjoberg M, Keane TM, Verma A, Ala U, et al:
The BRAF pseudogene functions as a competitive endogenous RNA and
induces lymphoma in vivo. Cell. 161:319–332. 2015.
|
|
8
|
Zhou D, Ren K, Wang M, Wang J, Li E, Hou
C, Su Y, Jin Y, Zou Q, Zhou P and Liu X: Long non-coding RNA
RACGAP1P promotes breast cancer invasion and metastasis via
miR-345-5p/RACGAP1-mediated mitochondrial fission. Mol Oncol.
15:543–559. 2021.
|
|
9
|
Gao KM, Chen XC, Zhang JX, Wang Y, Yan W
and You YP: A pseudogene-signature in glioma predicts survival. J
Exp Clin Cancer Res. 34:232015.
|
|
10
|
Wang Y, Liu X, Guan G, Xiao Z, Zhao W and
Zhuang M: Identification of a five-pseudogene signature for
predicting survival and its ceRNA network in glioma. Front Oncol.
9:10592019.
|
|
11
|
Wang S, Qi Y, Gao X, Qiu W, Liu Q, Guo X,
Qian M, Chen Z, Zhang Z, Wang H, et al: Hypoxia-induced lncRNA
PDIA3P1 promotes mesenchymal transition via sponging of miR-124-3p
in glioma. Cell Death Dis. 11:1682020.
|
|
12
|
Liao K, Qian Z, Zhang S, Chen B, Li Z,
Huang R, Cheng L, Wang T, Yang R, Lan J, et al: The LGMN pseudogene
promotes tumor progression by acting as a miR-495-3p sponge in
glioblastoma. Cancer Lett. 490:111–123. 2020.
|
|
13
|
Du P, Liao Y, Zhao H, Zhang J, Muyiti
Keremu and Mu K: ANXA2P2/miR-9/LDHA axis regulates Warburg effect
and affects glioblastoma proliferation and apoptosis. Cell Signal.
74:1097182020.
|
|
14
|
Theng SS, Wang W, Mah WC, Chan C, Zhuo J,
Gao Y, Qin H, Lim L, Chong SS, Song J and Lee CG: Disruption of
FAT10-MAD2 binding inhibits tumor progression. Proc Natl Acad Sci
USA. 111:E5282–E5291. 2014.
|
|
15
|
Yuan R, Wang K, Hu J, Yan C, Li M, Yu X,
Liu X, Lei J, Guo W, Wu L, et al: Ubiquitin-like protein FAT10
promotes the invasion and metastasis of hepatocellular carcinoma by
modifying β-catenin degradation. Cancer Res. 74:5287–5300.
2014.
|
|
16
|
Ma C, Zhang Z, Cui Y, Yuan H and Wang F:
Silencing Fat10 inhibits metastasis of osteosarcoma. Int J Oncol.
49:666–674. 2016.
|
|
17
|
Zou Y, Ouyang Q, Wei W, Yang S, Zhang Y
and Yang W: FAT10 promotes the invasion and migration of breast
cancer cell through stabilization of ZEB2. Biochem Biophys Res
Commun. 506:563–570. 2018.
|
|
18
|
Xue F, Zhu L, Meng QW, Wang L, Chen XS,
Zhao YB, Xing Y, Wang XY and Cai L: FAT10 is associated with the
malignancy and drug resistance of non-small-cell lung cancer. Onco
Targets Ther. 9:4397–4409. 2016.
|
|
19
|
Yuan J, Tu Y, Mao X, He S, Wang L, Fu G,
Zong J and Zhang Y: Increased expression of FAT10 is correlated
with progression and prognosis of human glioma. Pathol Oncol Res.
18:833–839. 2012.
|
|
20
|
Dai B, Zhang Y, Zhang P, Pan C, Xu C, Wan
W, Wu Z, Zhang J and Zhang L: Upregulation of p-Smad2 contributes
to FAT10-induced oncogenic activities in glioma. Tumour Biol.
37:8621–8631. 2016.
|
|
21
|
Yan Y, Zhang L, Jiang Y, Xu T, Mei Q, Wang
H, Qin R, Zou Y, Hu G, Chen J and Lu Y: LncRNA and mRNA interaction
study based on transcriptome profiles reveals potential core genes
in the pathogenesis of human glioblastoma multiforme. J Cancer Res
Clin Oncol. 141:827–838. 2015.
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
|
|
23
|
Zhou J, Wang H, Hong F, Hu S, Su X, Chen J
and Chu J: CircularRNA circPARP4 promotes glioblastoma progression
through sponging miR-125a-5p and regulating FUT4. Am J Cancer Res.
11:138–156. 2021.
|
|
24
|
He H, Wang Y, Ye P, Yi D, Cheng Y, Tang H,
Zhu Z, Wang X and Jin S: Long noncoding RNA ZFPM2-AS1 acts as a
miRNA sponge and promotes cell invasion through regulation of
miR-139/GDF10 in hepatocellular carcinoma. J Exp Clin Cancer Res.
39:1592020.
|
|
25
|
Cui Y, Yi L, Zhao JZ and Jiang YG: Long
noncoding RNA HOXA11-AS functions as miRNA sponge to promote the
glioma tumorigenesis through targeting miR-140-5p. DNA Cell Biol.
36:822–828. 2017.
|
|
26
|
Zhang JY, Wang M, Tian L, Genovese G, Yan
P, Wilson JG, Thadhani R, Mottl AK, Appel GB, Bick AG, et al: UBD
modifies APOL1-induced kidney disease risk. Proc Natl Acad Sci USA.
115:3446–3451. 2018.
|
|
27
|
Aichem A and Groettrup M: The
ubiquitin-like modifier FAT10 in cancer development. Int J Biochem
Cell Biol. 79:451–461. 2016.
|
|
28
|
Liu X, Chen L, Ge J, Yan C, Huang Z, Hu J,
Wen C, Li M, Huang D, Qiu Y, et al: The ubiquitin-like protein
FAT10 stabilizes eEF1A1 expression to promote tumor proliferation
in a complex manner. Cancer Res. 76:4897–4907. 2016.
|
|
29
|
Wu D, Wang L, Yang Y, Huang J, Hu Y, Shu
Y, Zhang J and Zheng J: MAD2-p31comet axis deficiency
reduces cell proliferation, migration and sensitivity of
microtubule-interfering agents in glioma. Biochem Biophys Res
Commun. 498:157–163. 2018.
|
|
30
|
Scrideli CA, Carlotti CG Jr, Okamoto OK,
Andrade VS, Cortez MAA, Motta FJN, Lucio-Eterovic AK, Neder L,
Rosemberg S, Oba-Shinjo SM, et al: Gene expression profile analysis
of primary glioblastomas and non-neoplastic brain tissue:
Identification of potential target genes by oligonucleotide
microarray and real-time quantitative PCR. J Neurooncol.
88:281–291. 2008.
|
|
31
|
Biterge-Sut B: Alterations in eukaryotic
elongation factor complex proteins (EEF1s) in cancer and their
implications in epigenetic regulation. Life Sci.
238:1169772019.
|
|
32
|
Choi Y, Kim JK and Yoo JY: NFκB and STAT3
synergistically activate the expression of FAT10, a gene
counteracting the tumor suppressor p53. Mol Oncol. 8:642–655.
2014.
|
|
33
|
Wang H, Xu T, Jiang Y, Xu H, Yan Y, Fu D
and Chen J: The challenges and the promise of molecular targeted
therapy in malignant gliomas. Neoplasia. 17:239–255. 2015.
|
|
34
|
Yan J, Lei J, Chen L, Deng H, Dong D, Jin
T, Liu X, Yuan R, Qiu Y, Ge J, et al: Human leukocyte antigen F
locus adjacent transcript 10 overexpression disturbs WISP1 protein
and mRNA expression to promote hepatocellular carcinoma
progression. Hepatology. 68:2268–2284. 2018.
|
|
35
|
Jing D, Zhang Q, Yu H, Zhao Y and Shen L:
Identification of WISP1 as a novel oncogene in glioblastoma. Int J
Oncol. 51:1261–1270. 2017.
|
|
36
|
Tao W, Chu C, Zhou W, Huang Z, Zhai K,
Fang X, Huang Q, Zhang A, Wang X, Yu X, et al: Dual role of WISP1
in maintaining glioma stem cells and tumor-supportive macrophages
in glioblastoma. Nat Commun. 11:30152020.
|
|
37
|
He L, Zhou H, Zeng Z, Yao H, Jiang W and
Qu H: Wnt/β-catenin signaling cascade: A promising target for
glioma therapy. J Cell Physiol. 234:2217–2228. 2019.
|
|
38
|
Allen M, Bjerke M, Edlund H, Nelander S
and Westermark B: Origin of the U87MG glioma cell line: Good news
and bad news. Sci Transl Med. 8:354re32016.
|
|
39
|
Chen X, Wan L, Wang W, Xi WJ, Yang AG and
Wang T: Re-recognition of pseudogenes: From molecular to clinical
applications. Theranostics. 10:1479–1499. 2020.
|
|
40
|
Johnsson P, Ackley A, Vidarsdottir L, Lui
WO, Corcoran M, Grandér D and Morris KV: A pseudogene
long-noncoding-RNA network regulates PTEN transcription and
translation in human cells. Nat Struct Mol Biol. 20:440–446.
2013.
|
|
41
|
Xu Y, Yu X, Wei C, Nie F, Huang M and Sun
M: Over-expression of oncigenic pesudogene DUXAP10 promotes cell
proliferation and invasion by regulating LATS1 and β-catenin in
gastric cancer. J Exp Clin Cancer Res. 37:132018.
|
|
42
|
Lee CGL, Ren J, Cheong ISY, Ban KHK, Ooi
LLPJ, Yong Tan S, Kan A, Nuchprayoon I, Jin R, Lee KH, et al:
Expression of the FAT10 gene is highly upregulated in
hepatocellular carcinoma and other gastrointestinal and
gynecological cancers. Oncogene. 22:2592–2603. 2003.
|
|
43
|
Lukasiak S, Schiller C, Oehlschlaeger P,
Schmidtke G, Krause P, Legler DF, Autschbach F, Schirmacher P,
Breuhahn K and Groettrup M: Proinflammatory cytokines cause FAT10
upregulation in cancers of liver and colon. Oncogene. 27:6068–6074.
2008.
|
|
44
|
Buerger S, Herrmann VL, Mundt S, Trautwein
N, Groettrup M and Basler M: The ubiquitin-like modifier FAT10 is
selectively expressed in medullary thymic epithelial cells and
modifies T cell selection. J Immunol. 195:4106–4116. 2015.
|
|
45
|
Canaan A, Yu X, Booth CJ, Lian J, Lazar I,
Gamfi SL, Castille K, Kohya N, Nakayama Y, Liu YC, et al:
FAT10/diubiquitin-like protein-deficient mice exhibit minimal
phenotypic differences. Mol Cell Biol. 26:5180–1589. 2006.
|
|
46
|
Basler M, Buerger S and Groettrup M: The
ubiquitin-like modifier FAT10 in antigen processing and
antimicrobial defense. Mol Immunol. 68:129–132. 2015.
|
|
47
|
Kandel-Kfir M, Garcia-Milan R, Gueta I,
Lubitz I, Ben-Zvi I, Shaish A, Shir L, Harats D, Mahajan M, Canaan
A and Kamari Y: IFNγ potentiates TNFα/TNFR1 signaling to induce
FAT10 expression in macrophages. Mol Immunol. 117:101–109.
2020.
|