Introduction
Acute myeloid leukemia (AML) is a heterogeneous
clonal disorder of the hematopoietic system that is characterized
by arrest of differentiation in myeloid progenitors (blasts) in
bone marrow and peripheral blood (1,2).
Such cells infiltrate the bone marrow, blood or other tissues,
ultimately leading to hematopoietic failure (2-4).
The incidence of AML increases with age. In 2022, an estimated
20,050 new cases of AML and 11,540 deaths were reported worldwide.
The 5-year survival rate for AML is 30.5%, the lowest 5-year
relative survival rate among all leukemias. Currently, ~40% of
patients with AML under the age of 60 and 15% of aged >60 years
are cured (4-6). Patients with AML are treated
primarily with chemotherapy, called the 7 + 3 regimen (7 days of
cytarabine and 3 days of anthracycline) (7-9).
Nevertheless, these chemotherapeutic agents have limitations of
poor efficacy and high toxicity (10).
Recently, more specific targeted therapies such as
fms-like tyrosine kinase 3 (FLT3) and isocitrate dehydrogenase
(IDH) inhibitor have been developed based on better understanding
of the molecular pathogenesis of AML (11). Mutations in FLT3 genes occur in
one-third of patients with AML. These mutations cause constitutive
activation of signaling, which promotes cell proliferation and
survival and inhibits differentiation. FLT3 mutation in AML is
associated with increased risk of relapse and adverse prognosis
(12,13). FLT3 inhibitors (sorafenib,
midostaurin and quizartinib) have improved overall survival as
single agents and in combination with the 7 + 3 regimen in patients
with FLT3-mutated AML (13).
Another mutation frequently occurring in AML is isocitrate
dehydrogenase (IDH), which has been identified in 15-20% of
patients. Mutations in this gene cause abnormal enzyme activity,
producing a metabolite 2-hydroxyglutarate (2-HG). 2-HG blocks the
differentiation of myeloid precursors and causes uncontrolled
proliferation. IDH1 inhibitor (ivosidenib) and IDH2 inhibitor
(enasidenib) have been used in patients with relapsed or refractory
AML with the IDH mutation and improved median overall survival
(3.3-9.0 and 3.3-9.3 months, respectively) (13,14). CD33 is a cell surface marker
highly expressed on leukemic blasts in almost all patients with AML
(15). Gemtuzumab ozogamicin, a
CD33 antibody conjugated to the cytotoxic drug calicheamicin, was
developed based on molecular studies of AML (16-20), but withdrawn due to lack of
improvement in survival rates and concerns about liver toxicity.
These targeted therapies also have limited therapeutic efficacy
because of the genomic complexity and clonal architecture of AML
(15,21).
Differentiation therapy is a therapeutic method that
stimulates differentiation in undifferentiated cancer cells to
eliminate tumor phenotypes (22,23). One of the hallmarks of AML is
blockade of differentiation, suggesting that induction of
differentiation may be a feasible therapeutic option for AML
subtypes. Differentiation therapy notable improves survival of
patients with acute promyelocytic leukemia (APL) (23), a subtype of AML characterized by t
(15;17) chromosomal translocation, resulting
in fusion of the promyelocytic leukemia (PML) gene with a retinoic
acid receptor (RARα) to form the PML-RARα fusion protein. All
trans-retinoid acid (ATRA) and arsenic trioxide reverse the
differentiation blockade induced by the fusion protein, leading to
APL differentiation and clearance (24,25). However, ATRA and arsenic trioxide
have limited effects in other subtypes of AML (25), necessitating novel agents with low
toxicity and high efficacy for differentiation therapy.
Plants provide a source of drug research and
development. They contain natural bioactive compounds, such as
vitamins, carotenoids, terpenoids, flavonoids, alkaloids, tannins
and minerals, which are associated with anti-oxidant,
anti-microbial, anti-inflammatory and anti-tumor activity (26-28). Plants belonging to the
Corydalis genus exert tumor-suppressive effects.
Tetrahydrocoptisine from C. impatiens has shown
anti-inflammatory activity by inhibiting the NF-κB and MAPK
signaling pathways (29). In
addition, corynoline isolated from C. bungeana Turcz.
decreases pro-inflammatory mediators by regulating the Nrf2/MAPK
pathway (30). Furthermore, C.
edulis Maxim exerts an anti-diabetic effect by increasing
insulin secretion via protein kinase C activation (31). C. yanhusco extract inhibits
cell proliferation in breast cancer and exhibits anti-proliferative
and anti-tumor effects in liver carcinoma cells (32,33).
Alkaloids are bioactive compounds found frequently
in natural herbs that elicit anti-tumor effects in various types of
cancers by inducing DNA damage, cell cycle arrest and apoptosis.
Taxol, camptothecin and vinblastine are among the most widely used
alkaloids to treat cancer (34,35). Alkaloids also exert anti-leukemic
effects in AML (36-40). Securinine is a major alkaloid
isolated from the roots of Securinega suffruticosa. This
compound induces differentiation by activating the DNA damage
response (39). Tetrandrine, an
alkaloid isolated from the roots of Stephaniae tetrandrae,
promotes reactive oxygen species (ROS) accumulation and inhibits
c-myc expression, inducing autophagy and differentiation (40).
The present study investigated the effect of
Corydalis incisa (Thunb.) Pers. (CIP) and norchelerythrine,
an alkaloid isolated from CIP, on differentiation and apoptosis in
AML cells.
Materials and methods
Preparation of plant extracts
Plant extracts (Table
SI) were obtained from the Korea Plant Extract Bank at the
Korea Research Institute of Bioscience and Biotechnology. Each
extract was dissolved in DMSO.
Cell culture and chemicals
Human AML cell lines HL-60, U937 and THP-1 were
purchased from Korean Cell Line Bank (Seoul, Korea) and cultured in
RPMI-1640 medium (Cytiva, cat. no. SH30027.01) supplemented with
10% FBS (HyClone, Cytiva; cat. no. SH30919.03), 1% HEPES (cat. no.
15630-080), 1% penicillin/streptomycin (cat. no. 15140-122) and 1%
L-glutamine (all Gibco, cat. no. 25030-81; Thermo Fisher
Scientific, Inc.) at 37°C in a 5% CO2 incubator. U937
cell line was authenticated using STR profiling by Cosmogenetech
Co., Ltd. Normal bone marrow cells from wild-type C57BL/6 mice were
cultured in RPMI-1640 medium with 10% FBS as aforementioned.
Norchelerythrine and peltatoside were purchased from ChemFaces
(cat. no. CFN92737 and CFN70318). Phorbol-12 myristate-13 acetate
(PMA) (cat. no. P8139) and ROS scavenger N-acetylcysteine (NAC) was
purchased from Sigma-Aldrich (cat. no. A9165; Merck KGaA).
Flow cytometry assay
The induction of AML cell differentiation was
determined by assessing CD11b and CD14 expression on the cell
surface. Briefly, cells were treated with DMSO, CIP (20
μg/ml for 96 h), norchelerythirne (2, 5 or 10 μM for
96 h) or Corydalis speciosa Maxim. (20 μg/ml for 96
h) at 37°C, harvested and washed with PBS, followed by staining
with FITC-conjugated CD11b (cat. no. 101206) or PE-cy7 conjugated
CD11b (both Biolegend, Inc.; cat. no. 101216) or
PercP-Cy5.5-conjugated CD14 (BD Biosciences, cat. no. 550787) for 1
h at 4°C in the dark. To assess the effect of norchelerythrine on
normal hematopoiesis in mice, bone marrow cells were treated with 5
μM norchelerythrine at 37°C for 96 h. Cells were harvested
and washed with PBS, then stained with PE-conjugated CD45
(eBioscience, cat. no. 12-0451-82; Thermo Fisher Scientific, Inc.),
PE-Cy7-conjugated CD11b (BioLegend, Inc.; cat. no. 101216),
APC-conjugated Ly-6G (BD Biosciences, cat. no. 560599) and
FITC-conjugated CD14 (BioLegend, Inc.; cat. no. 123307) for 1 h at
4°C in the dark. The samples were analyzed using a FACSDiva Fusion
Flow Cytometer (BD Biosciences). At least 10,000 cells were
analyzed for each data point. Data analysis was carried out using
FlowJo software version 10.8.1 (Treestar Inc.).
Reverse transcription-quantitative (RT-q)
PCR
Total RNA was isolated from U937 cells using Tri-RNA
reagent (Favorgen, cat. no. FATRR001) and cDNA was synthesized
using the PrimeScript RT reagent kit (Takara Bio, Inc., cat. no.
RR047A) according to the manufacturer's protocol. Equal amounts of
cDNA were used for transcript PCR amplification, which was
performed using TOPreal qPCR PreMIX SYBR Green with low ROX
(Enzynomics Co., Ltd.; cat. no. RT500M). The thermocycling
conditions were as follows: 95°C for 15 min followed by 40 cycles
of 95°C for 15 sec, 59°C for 30 sec and 72°C for 30 sec. Table I lists the primers used. Relative
gene expression was analyzed using the 2-∆∆Cq method (41).
 | Table IPrimers used in reverse
transcription-quantitative PCR. |
Table I
Primers used in reverse
transcription-quantitative PCR.
| Gene | Forward, 5′→3′ | Reverse, 5′→3′ |
|---|
| CSF1R |
GTGGCTGTGAAGATGCTGAA |
CCTTCCTTCGCAGAAAGTTG |
| MAFB |
GCCTGCGCTAATTGTAGGAG |
CGCACTTGAAAGTTGCAAAA |
| ITGAM |
AGAACAACATGCCCAGAACC |
GCGGTCCCATATGACAGTCT |
| CD14 |
CTGCAACTTCTCCGAACCTC |
CCAGTAGCTGAGCAGGAACC |
| Lyz |
GCCAAATGGGAGAGTGGTTA |
ATCACGGACAACCCTCTTTG |
| MMP9 |
TTGACAGCGACAAGAAGTGG |
GCCATTCACGTCGTCCTTAT |
| Myb |
GGCAGAAATCGCAAAGCTAC |
GCAGGGAGTTGAGCTGTAGG |
| Myc |
TTCGGGTAGTGGAAAACCAG |
CAGCAGCTCGAATTTCTTCC |
| TBP |
TATAATCCCAAGCGGTTTGCTGCG |
AATTGTTGGTGGGTGAGCACAAGG |
Cell proliferation assay
Cell proliferation rates were determined using
trypan blue exclusion assay (42). The cells (2×105
cells/well) were seeded into a 24-well plate and treated with CIP
(0, 10 or 20 μg/ml), norchelerythrine (0, 5 or 10 μM)
or peltatoside (0, 500 nM, 1, 2 or 5 μM) at 37°C for 24-120
h. Cells were stained with 0.4% Trypan blue (Gibco; Thermo Fisher
Scientific, Inc.; cat. no. 15250061) for 3 min at room temperature
every day, and the number of viable cells was counted using a
hemocytometer.
Measurement of cell viability
Cell viability was assessed using CellTiter 96
AQueous MTS assay. The cytotoxicity of CIP and Corydalis
speciosa Maxim. extract in human AML cell lines (HL-60 and
THP-1) was tested by seeding cells in a six-well plate at a density
of 4×105/well followed by treatment with CIP or
Corydalis speciosa Maxim. extracts (20 μg/ml) at 37°C
for 5-8 days. The cytotoxicity of CIP was identified by plating
cells in a 96-well plate at a density of 3×104 AML cells
(HL-60 and U937) or 5×105 normal bone marrow cells/well
and treating them with CIP (0, 50, 100, 200 or 300 μg/ml) at
37°C for 24 h. The cytotoxic effect of norchelerythrine and
peltatoside was examined by treating HL-60, U937 and THP-1 cells
with norchelerythrine (0 or 10 μM for 72 or 96 h) or
peltatoside (0, 500 nM, 1, 2 or 5 μM for 48, 72 h or 96 h)
at 37°C. MTS reagent (Promega Corporation; cat. no. G1112) was
added for 4 h at 37°C. The absorbance at 450 nm was measured using
a GloMax Microplate multi-mode reader (Promega Corporation)
(43).
Trypan blue staining
Trypan blue stain was used to determine the cell
apoptosis rate. HL-60 and THP-1 cells were plated in six-well
plates at a density of 4×105 cells/well and treated with
Corydalis incisa (Thunb.) Pers. or Corydalis speciosa
Maxim. extract (20 μg/ml) at 37°C for 5-8 days. The cells
were stained with 0.4% trypan blue (Gibco, Thermo Fisher
Scientific, Inc.; cat. no. 15250061) for 3 min at room temperature.
The number of positively stained cells was counted using a
hemocytometer (44).
Colony forming assay
HL-60 cells (1×104 cells/well) were
seeded in a 12-well plate in methylcellulose (Methocult H4100,
StemCell Technologies) supplemented with 10% FBS and 1%
penicillin/streptomycin, as previously described (43) and treated with CIP (2
μg/ml) at 37°C for 11 days. The colonies containing >50
cells were counted manually 11 days after plating using an Olympus
CX31microscope (Olympus Corporation) at 400× magnification.
Ultra-performance liquid
chromatography-quadrupole time-of-flight mass spectrometry
(UPLC-QTOF-MS) analysis of CIP
CIP was analyzed using Waters AQUITYTM UPLC system
equipped with an XEVO-QTOF mass detector. ACQUITY UPLC BEH C18
column was used, with two mobile phases containing water with 0.1%
formic acid (solvent A) and acetonitrile with formic acid (solvent
B). The mobile phase was delivered at a flow rate of 0.4 ml/min,
and a 1 μl injection volume was used. The elution gradient
was as follows: 1, B 8%; 13, B 8-40%; 2.5, B 40%; 0.5, B 40-100%;
2.5, B 100%; 0.5, B 100-8%; 2 min B 8%. XEVO-QTOF mass detector
featured electrospray ionization. The analysis was conducted in
negative and positive ion modes in the range of 100-1,500 m/z.
N2 was used as the desolvation gas. The desolvation
temperature was set to 350°C at a flow rate of 800 l/h with a
source temperature of 110°C. The capillary and cone voltages were
set to 300 and 40 V, respectively. CIP was prepared (3 mg/ml) in
methanol.
Measurement of intracellular ROS
Intracellular ROS were detected by MitoSOX Red
staining. Following treatment with 10 μM norchelerythrine at
37°C for 48 h, cells were harvested and washed with PBS, followed
by staining with MitoSOX Red (MedChemExpress, cat. no. HY-D1055)
for 20 min at 37°C in the dark. The cells were washed with PBS and
analyzed using a FACSDIVA fusion Flow Cytometer (BD Biosciences).
At least 10,000 cells were analyzed for each data point. Data
analysis was performed using FlowJo software version 10.8.1
(Treestar Inc.).
Western blot analysis
HL-60 and U937 cells were treated with
norchelerythrine (5 or 10 μM at 37°C for 6, 12 or 48 h) and
washed in PBS. The cells lysed in RIPA buffer (Elpis Biotechnology,
cat. no. EBA-1149) containing 1 Na-vanadate, 50 β-glycerophosphate
disodium salt (both Sigma-Aldrich; Merck KGaA; cat. no. G9422), 142
β-mercaptomethanol (Bioworld Technology, Inc.; cat. no. 41300000-1)
and 5 mM EDTA and ProteaseArrest (Thermo Scientific, Inc.; cat. no.
87786). Protein concentrations were measured using BCA assay kit
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. The samples were boiled at 100°C for 10 min, 25
μg proteins loaded on 6 or 15% polyacrylamide gels and
transferred to Immobilon-P transfer membrane. The membranes were
blocked for 1 h at room temperature in 0.1% TBST with 1% BSA (MP
Biomedicals, LLC; cat. no. 160069) and incubated with primary
antibodies overnight at 4°C, washed three times for 5 min in TBST,
and incubated with HRP-conjugated anti-mouse or anti-rabbit
secondary antibodies for 1 h at room temperature. The membranes
were washed three times for 10 min in TBST. The protein bands were
detected using chemiluminescent substrate (EzWestLumi plus; ATTO
corporation) and visualized using the Luminograph II (ATTO
Corporation). The proteins were quantified using ImageJ 1.54 g
(National Institutes of Health). The primary antibodies against p21
(cat. no. #2947S), γH2AX (cat. no. #9718S), Ataxia telangiectasia
mutated (ATM) (cat. no. #2873S) and phosphorylated ATM (all
1:1,000; #5883S) were purchased from Cell Signaling Technology,
Inc. The primary antibody against β-actin (1:5,000; cat. no.
sc-47778) and the anti-mouse secondary antibody (cat. no.
sc-516102, 1:5,000) was obtained from Santa Cruz Biotechnology,
Inc. The anti-rabbit secondary antibody (cat. no. A120-101P,
1:5,000) was purchased from Bethyl Laboratories, Inc. (45).
Patient samples
Pusan National University Hospital (Busan, South
Korea) provided the bone marrow samples from 4 male patients with
AML, collected between August 2023 and November 2023, after
obtaining informed written consent from patients and approval from
the institutional review board of Pusan National University
Hospital (approval no. IRB 2403-010-137). The mean age of the
patients was 55 years (range, 21-80 years). Table SII lists the mutation profiles of
patients.
Generation of CIP-resistant cells
CIP-resistant cell line was generated by seeding
HL-60 cells (2×105 cells/well) in 12-well plates
followed by treatment with DMSO or 2 μg/ml CIP at 37°C for
96 h. Medium was removed, and the cells were cultured for an
additional 48 h in RPMI-1640 medium supplemented with 10% FBS, 1%
HEPES, 1% penicillin/streptomycin and 1% L-glutamine. Subsequently,
the cells were treated with 4, 6, 8, 10, 12, 15, 18 and 20
μg/ml CIP, respectively, in the same manner. This cycle was
repeated for over 6 months. Resistance was confirmed by FACS
analysis of CD11b and cell counting.
QuantSeq-3′ mRNA-sequencing (seq)
Total RNA from HL-60-DMSO and HL-60-CIP cell lines
was extracted using Tri-RNA reagent (Favorgen, FATRR001). DNA was
removed using Ambion AM1906 DNase Treatment and Removal Reagents
(Ambion) according to the manufacturer's instructions. RNA quality
was assessed by Agilent 4200 TapeStation System (Agilent
Technologies, Inc.). QuantSeq-3′mRNA-seq was performed by ebiogen,
Inc. Briefly, RNA library construction was performed using the
Quant-Seq 3′ mRNA-Seq V2 library prep kit FWD with UDI 12 nt Sets
A1-A4, (UDI12A_0001-0384). 384 preps (cat. no. 193.384; Lexogen
GmbH). The loading concentration of the final library was >4 nM
and was measured using Qubit (Thermo Fisher Scientific Inc.).
High-throughput single-end 75 bp sequencing was conducted using
NextSeq 500/550 (Illumina, Inc.) using a NextSeq 500/550 High
Output Kit v2.5 (75 cycles, cat. no. 20024906; Illumina, Inc.).
Gene ontology (GO) and differentially expressed genes (DEG) were
analyzed using ExDEGA version 5.0 (ebiogen, Inc.). Differentially
expressed genes with fold change >2 and P<0.05 were selected.
A heatmap was generated using MultiExperiment Viewer 4.9.0
(sourceforge.net/projects/mev-tm4/).
Statistical analysis
All data are presented as the mean ± SD.
Statistically significant differences were calculated using
non-parametric Mann-Whitney U or unpaired t-test or one-way ANOVA
test with Tukey's post hoc test using Prism version 5.03 software
(GraphPad Software, Inc.; Dotmatics). All experiments were repeated
≥3 times. P<0.05 was considered to indicate a statistically
significant difference.
Results
Extracts of CIP efficiently promote
differentiation of AML
A total of 100 plant extracts native to South Korea
were screened morphologically based on the presence of adherent
cells, after treatment with 20 μg/ml plant extracts for 96
h, which indicated the induction of differentiated cells. Four
extracts that promoted the formation of adherent cells were then
analyzed for CD11b expression, a myeloid differentiation marker,
following exposure to 20 μg/ml plant extracts for 96 h.
Extracts of CIP were most effective in increasing CD11b expression
(Table SI). Therefore, the
present study focused on CIP extracts for further characterization.
U937 AML cell line was exposed to CIP extracts (20 μg/ml) to
confirm the ability of CIP to induce AML differentiation, which
significantly increased CD11b expression (Fig. 1A). In addition, increases in size
(forward scatter) and granularity (side scatter) are commonly
observed as the cells differentiate (46), which was the case following CIP
treatment (Fig. 1B). PMA can
induce differentiation of AML cells and make them adherent and
elongated (47). Co-treatment of
CIP notably increased the number of adherent/elongated cells
compared with PMA alone, suggesting that CIP enhances PMA-induced
differentiation of THP-1 AML cells (Fig. 1C).
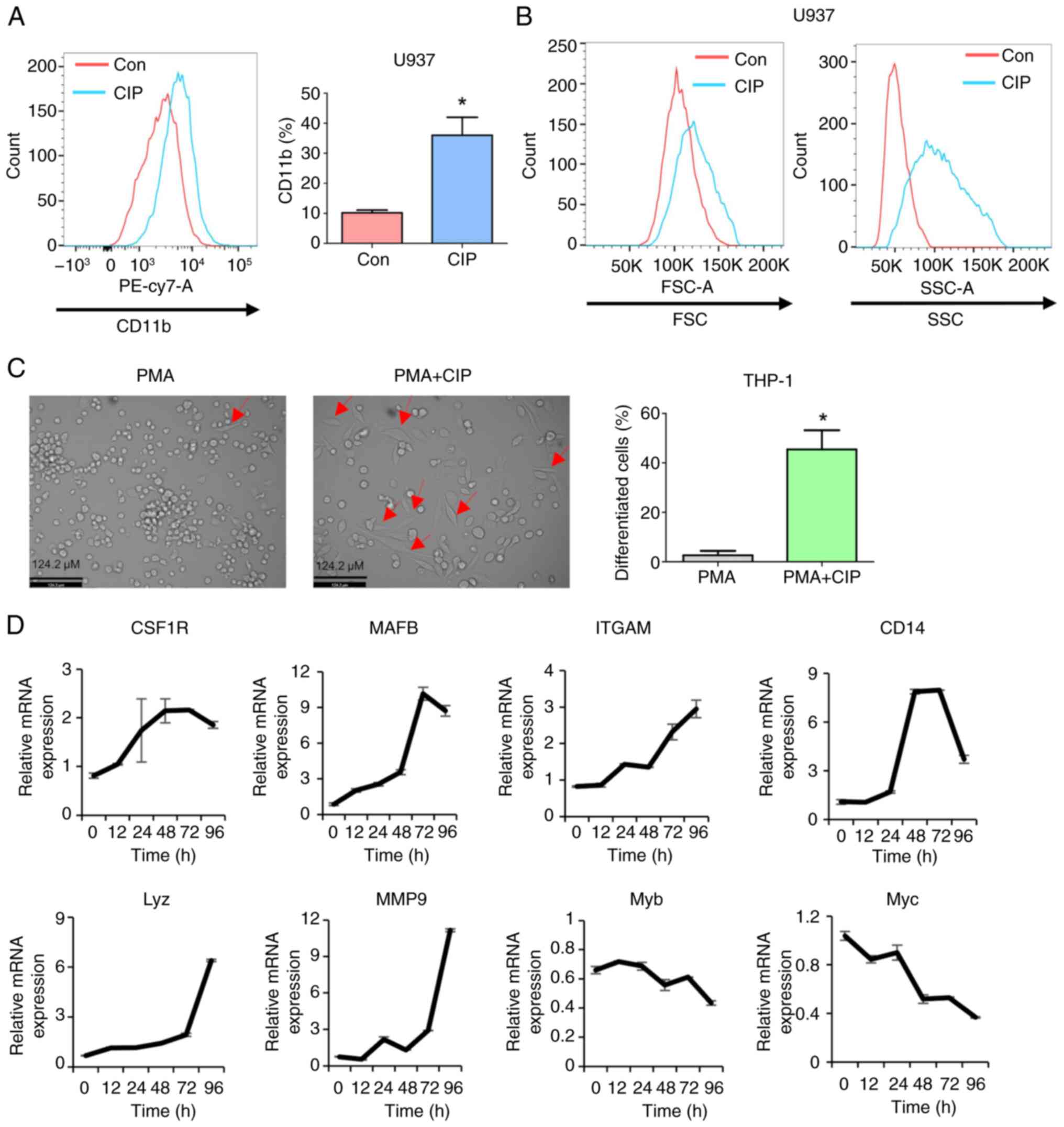 | Figure 1CIP induces differentiation in AML
cells. (A) Fluorescence activated cell sorting analysis of CD11b
expression levels following the CIP treatment (20 μg/ml) for
96 h. (B) FSC and SSC measured by flow cytometry. (C) THP-1 AML
cells were treated with 1 ng/ml PMA in the presence or absence of
CIP (20 μg/ml) for 96 h, followed by microscopic observation
of adherent and elongated cells. (D) Relative expression of CSF1R,
MAFB, ITGAM, CD14, Lyz, MMP9, Myb and Myc measured by reverse
transcription-quantitative PCR. *P<0.05; CIP,
Corydalis incisa (Thumb.) Pers.; AML, acute myeloid
leukemia; FSC, forward scatter; SSC, side scatter; PMA, phorbol-12
myristate-13 acetate; CSF1R, colony stimulating factor 1 receptor;
MAFB, V-maf musculoaponeurotic fibrosarcoma oncogene homolog B;
ITGAM, integrin alpha M; Lyz, lysozyme; Myb, v-myb avian
myeloblastosis viral oncogene homolog; con, control. |
The present study analyzed expression of genes
related to myeloid differentiation using RT-qPCR. Consistent with
the data that CIP efficiently induces AML differentiation, myeloid
differentiation markers (colony stimulating factor 1 receptor,
integrin αM and CD14) were upregulated and negative regulators of
myeloid differentiation (Myb and Myc) were downregulated following
CIP treatment. Furthermore, MAFB, a transcription factor key for
early myeloid and monocytic differentiation (48), exhibited increased expression
following exposure to CIP. In addition, lysozyme and MMP9, genes
associated with differentiated cell functions such as neutrophils
and macrophages, showed elevated expression in response to CIP
treatment. These results collectively showed that CIP effectively
stimulates differentiation in human AML cells (Fig. 1D).
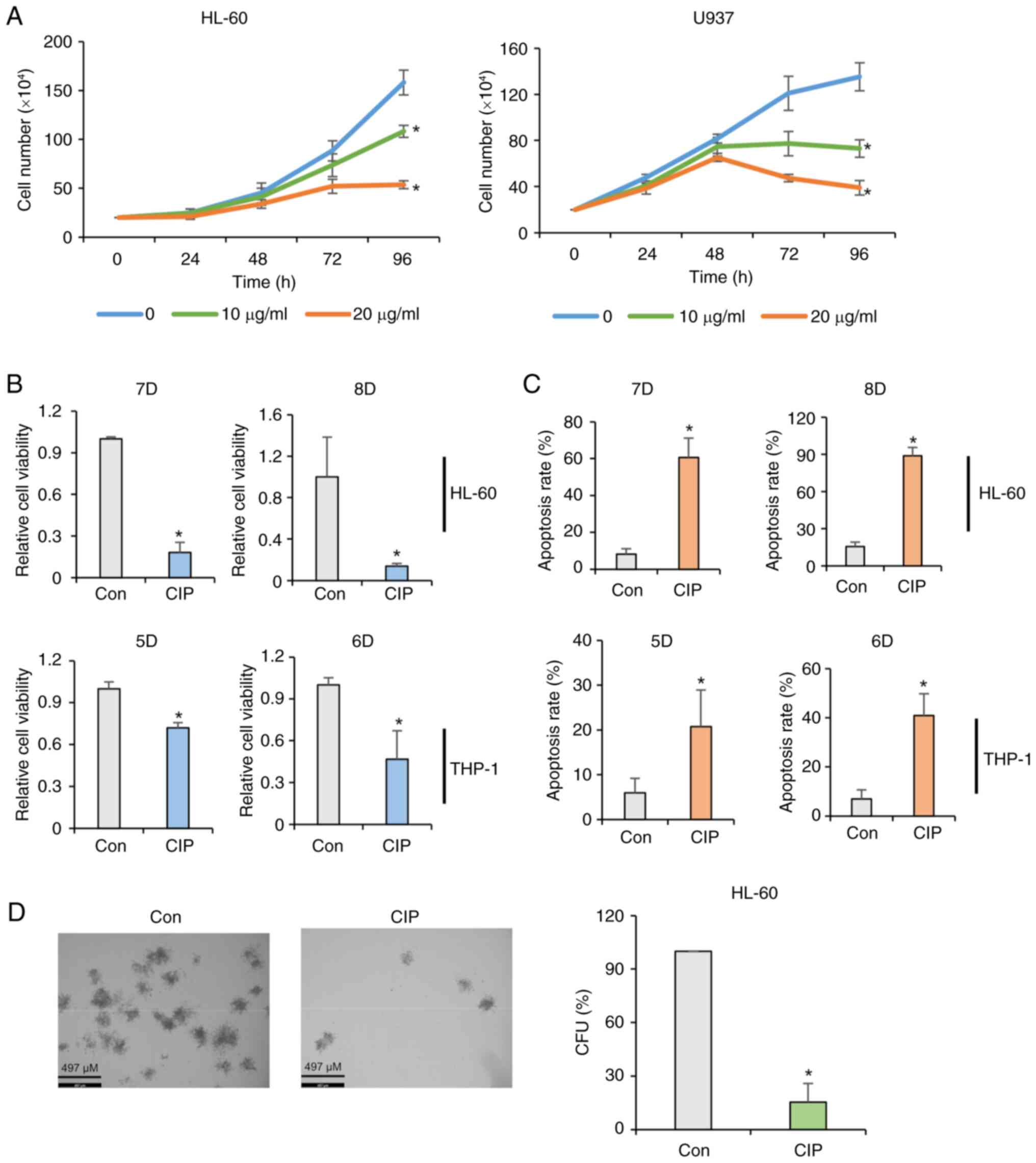
CIP inhibits cell proliferation in
AML
HL-60 and U937 AML cells were exposed to CIP (0, 10
or 20 μg/ml), followed by cell counting every 24 h to
determine differentiation by CIP would affect proliferation in AML.
CIP significantly decreased the cell number in a dose-dependent
manner in HL-60 and U937 cells (Fig.
2A). Consistently, CIP decreased viability and increased the
apoptosis rates in HL-60 and THP-1 cells (Fig. 2B and C). CIP extracts (2
μg/ml) had an inhibitory effect on the colony formation in
AML (Fig. 2D). In addition, CIP
exhibited more toxicity in HL-60 and U937 AML cells than normal
bone marrow cells (Fig. S1),
suggesting that the cytotoxic effect of this extract may be
specific to AML cells with minimal impact on normal cells. These
data collectively indicated that CIP extracts have
anti-proliferative and pro-apoptotic effects by triggering
differentiation in AML cells.
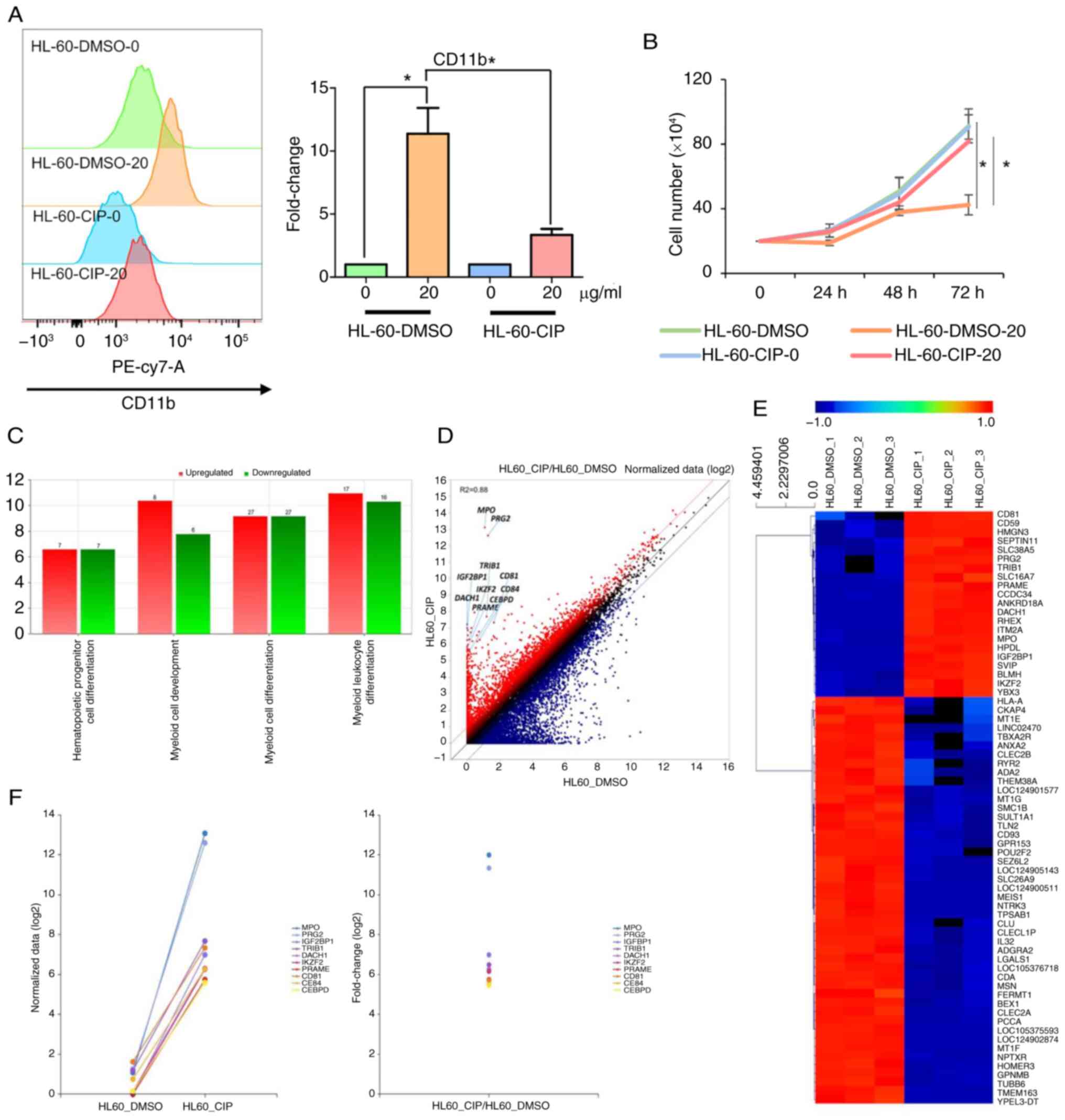
Genome-wide profiling of genes involved
in CIP-induced AML differentiation
CIP-resistant cells were generated to understand
CIP-induced differentiation and identify potential targets of CIP
(49). Human AML cell line HL-60
was treated with DMSO or increasing CIP concentrations (2-20
μg/ml) for 6 months. Following exposure to CIP (20
μg/ml), CD11b expression notably increased in HL-60-DMSO but
increased to a lesser extent in HL-60-CIP cells (Fig. 3A and B). HL-60-DMSO cells were
sensitive to CIP treatment, leading to a decrease in cell number,
while HL-60-CIP cells were resistant to CIP, showing no decrease in
cell number. These data showed that HL-60-CIP cells are resistant
to CIP-induced differentiation and cell cycle arrest (Fig. 3A and B). Furthermore, when
HL-60-CIP cells were cultured in the absence of CIP, they remained
resistant to CIP for 3 weeks, suggesting that resistance was
achieved through stable genetic alteration (Fig. S2).
The genes involved in CIP-induced AML
differentiation were identified at the genome level using Quantseq
3′mRNA-seq using HL-60-DMSO and HL-60-CIP cells. The genes
regulated by CIP were enriched in 'hematopoietic progenitor cell
differentiation', 'myeloid cell development' and 'myeloid cell
differentiation' (Fig. 3C).
RNA-seq analysis showed that genes that promote leukemogenesis and
inhibit myeloid differentiation, such as myeloperoxidase (50), proteoglycan 2 (51), insulin-like growth factor 2
mRNA-binding protein 1 (52),
tribbles homolog 1 (53),
dachshund homolog 1 (54), ikaros
family zinc finger 2 (55),
preferentially expressed antigen in melanoma (56), CD81 (57), CCAAT/enhancer binding protein δ
(58) and CD84 (59), were upregulated in CIP-resistant
HL-60 cells, validating that HL-60-CIP cells are resistant to
differentiation-inducing agents by regulating genes involved in
modulating AML differentiation (Fig.
3D-F).
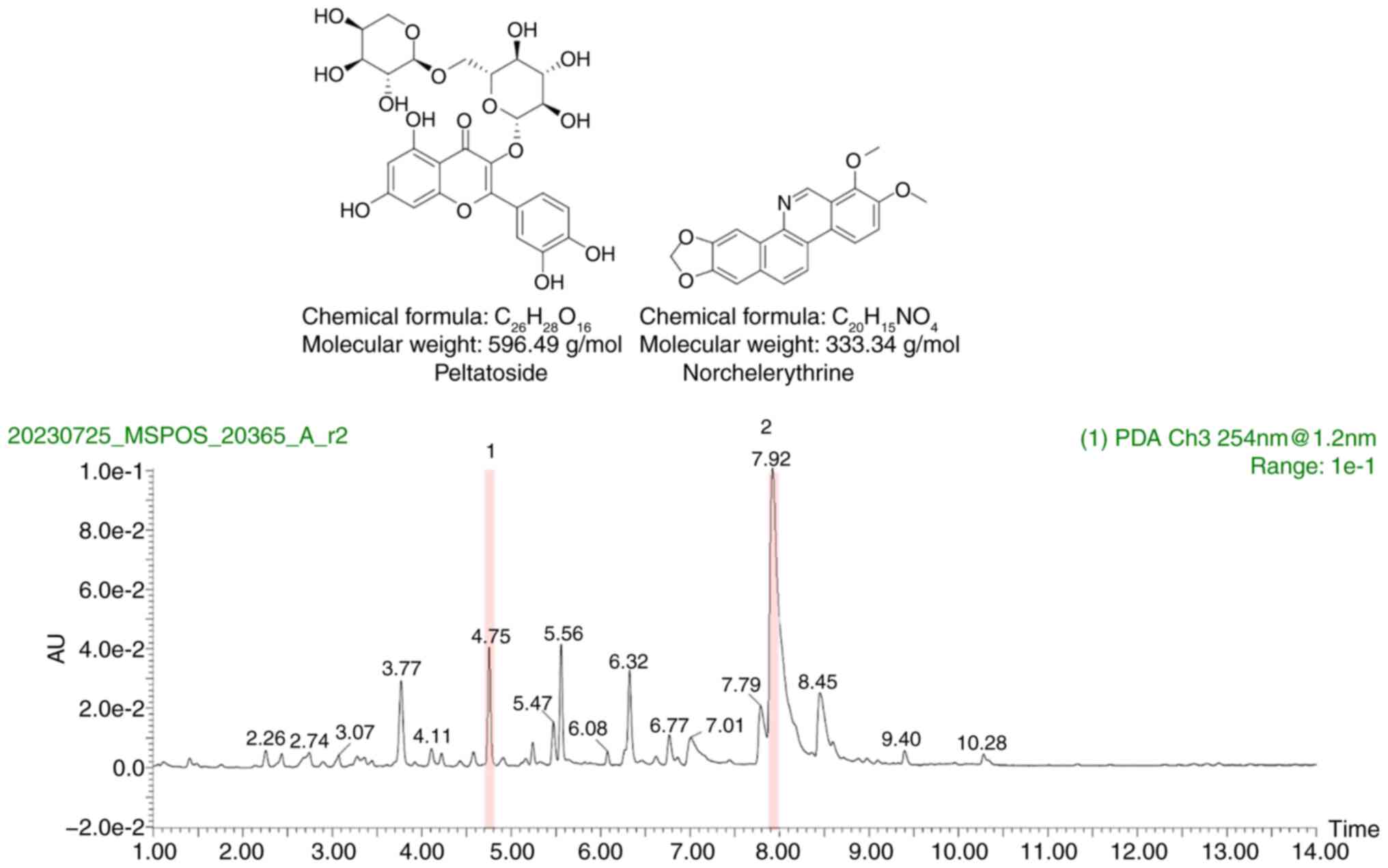
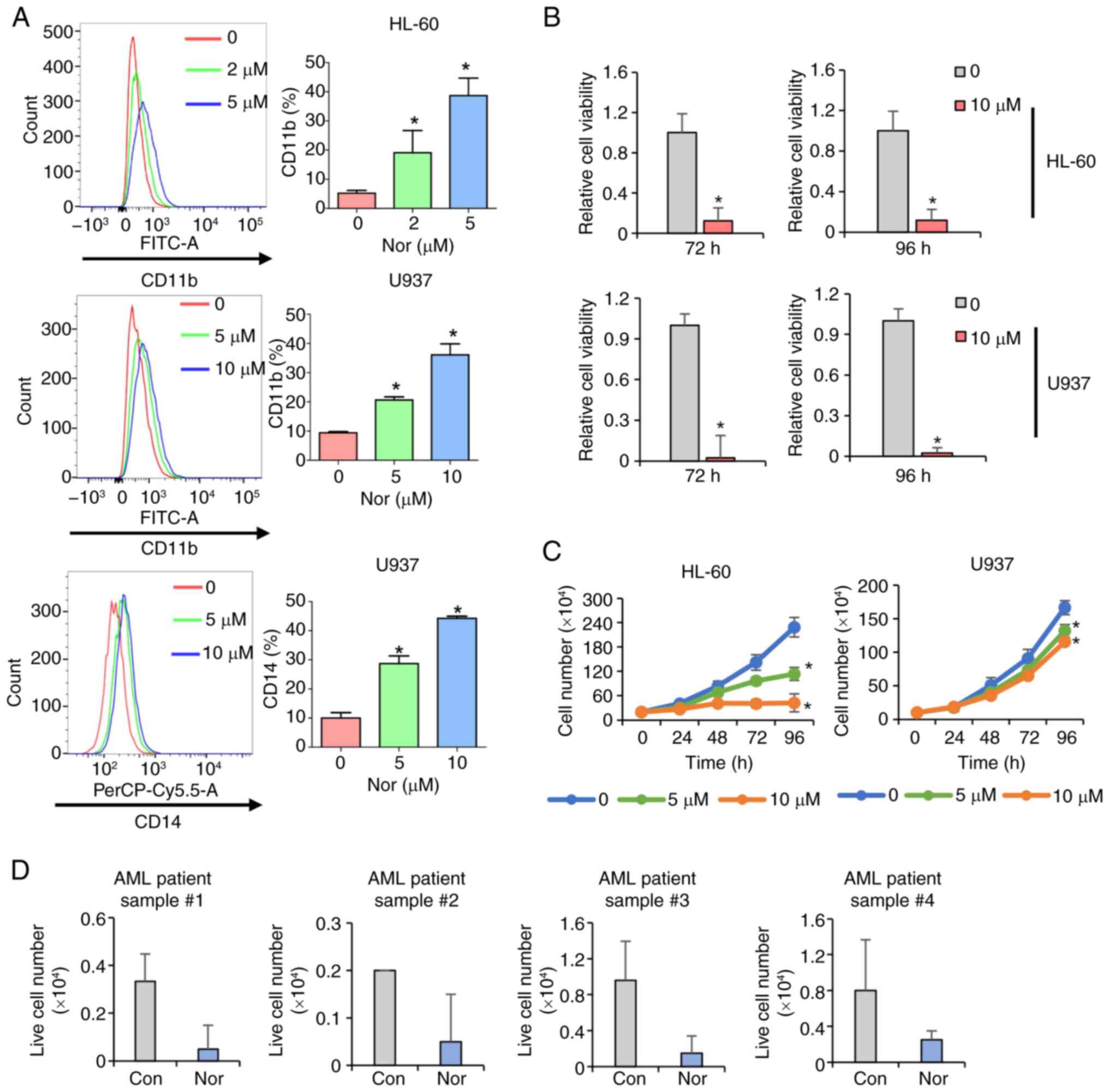
Norchelerythrine in CIP induces
differentiation
The compounds in CIP that induce differentiation in
AML cells were identified and quantified by UPLC-QTOF-MS on the CIP
extract. Two notably peaks were identified and labeled peltatoside
and norchelerythrine (Fig. 4).
Peltatoside did not have any significant effect on the viability or
proliferation of U937 and THP-1 cells (Fig. S3A and B). These findings
suggested that peltatoside is not the differentiation-inducing
compound present in CIP. Norchelerythrine significantly increased
the fluorescence intensity of CD11b and CD14 in HL-60 and U937
cells (Fig. 5A). Furthermore,
treatment of HL-60 and U937 with norchelerythrine resulted in
significantly decreased cell viability and proliferation in a
dose-dependent manner (Fig. 5B and
C). Moreover, norchelerythrine decreased the number of viable
cells in primary AML cells from patients harboring IDH1, ETS
variant transcription factor 6, Lysine methyltransferase 2A (KMT2A)
and CCAAT/enhancer binding protein α mutation (Figs. 5D and S4). Myeloid lineage cells distribution
was not altered by norchelerythrine, suggesting that
norchelerythrine did not have any effect on normal hematopoiesis in
mice (Fig. S5).
The effects of Corydalis genus plant extract
on AML cells were examined based on the hypothesis that plants of
the same genus have comparable compound compositions and similar
biological effects. C. speciosa Maxim. extract increased
expression of the CD11b surface marker (Fig. S6A). Similarly to CIP, the extract
of C. speciosa Maxim. decreased cell viability and increased
the rate of apoptosis (Fig. S6B and
C). Consistent with these results, norchelerythrine, but not
peltatoside, was also observed in the UPLC-QTOF-MS chromatogram of
the C. speciosa Maxim. extract (Fig. S7). These findings suggest that
norchelerythrine may be one of the major compounds in CIP
responsible for promoting cell differentiation and inhibiting cell
proliferation in AML.
ROS-mediated DNA damage response is
involved in norchelerythrine-induced differentiation
ROS influence cellular signaling processes key for
cell proliferation and differentiation. In particular, ROS
concentration plays a regulatory role in the differentiation of
cells during hematopoiesis (60,61). The mechanism of
norchelerythrine-induced differentiation was examined using a
fluorescent probe (MitoSOX Red) to detect ROS levels in the
presence or absence of norchelerythrine. The cells treated with
norchelerythrine had higher intracellular ROS levels than those
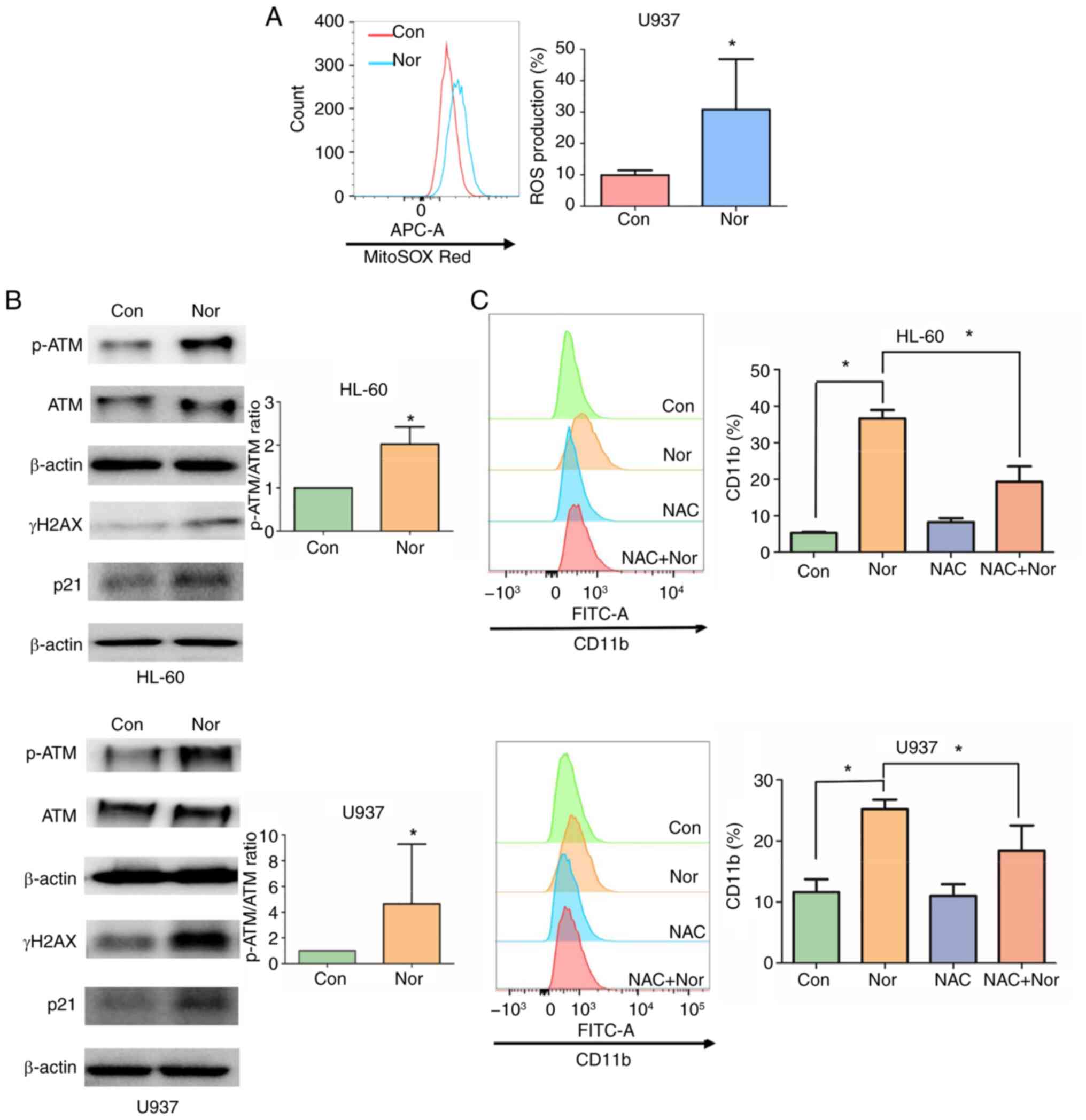
treated with DMSO (Fig. 6A).
Previous studies have suggested that alkaloids promote ROS
accumulation, leading to DNA damage (62-64). DNA damage in human and murine
myeloid leukemia cells is associated with myeloid differentiation
and cell cycle arrest (65,66). Therefore, the present study
examined whether norchelerythrine activates DNA damage response via
ROS production. Norchelerythrine treatment led to increased levels
of H2AX phosphorylation at Ser139 (γH2AX), which is a marker of DNA
double-strand breaks. In addition, levels of phosphorylated ATM and
p21, but not total ATM, increased following exposure to
norchelerythrine (Fig. 6B). The
ROS scavenger NAC was used to determine if ROS accumulation
mediated differentiation of AML cells. NAC treatment reduced the
differentiation induced by norchelerythrine significantly (Fig. 6C), suggesting that ROS generated
by norchelerythrine were involved in AML differentiation. These
findings suggest that ROS and DNA damage response are critical in
norchelerythrine-induced differentiation.
Discussion
The primary chemotherapy of AML, the 7 + 3 regimen,
has not changed for several decades. The 5-year survival rate is
poor, particularly for patients aged >65 years. Patients with
APL are successfully treated with differentiation therapy,
highlighting the need to develop novel agents for treating AML
(67,68). The present study showed that among
100 plant extracts, CIP extracts were the most effective in
overcoming differentiation arrest in AML. Furthermore, UPLC-QTOF-MS
analysis identified norchelerythrine as one of the key compounds in
CIP, which exerted anti-leukemic efficacy in vitro. The
present mechanistic study found that the ROS generated by
norchelerythrine were responsible for AML differentiation and the
inhibition of cell proliferation through DNA damage response (DDR)
activation.
Our previous study demonstrated that CIP exhibits
cytotoxicity in diffuse large B cell lymphoma cells (35). To the best of our knowledge, the
present study is the first to reveal that CIP has anti-leukemic
activity in AML. CIP contains several alkaloids, including
corynoline and acetylcorynoline (69-75). To the best of our knowledge, the
present study is the first to demonstrate that CIP contains
norchelerythrine, a phytochemical with diverse biological
activities, such as inhibitory effects against several
microorganisms, such as Staphylococcus aureus,
Pseudomonas aeruginosa, Enterococcus faecalis and
Escherichia coli (76). In
addition, norchelerythrine has antifeedant activity against
Tribolium castaneum, causing damage to stored grain products
(77). Studies have assessed its
cytotoxic effects on human hematoma, cervical carcinoma and gastric
cancer and murine lymphocytic leukemia cells in vitro
(78-80). However, the impact of
norchelerythrine on AML has been unexplored. To the best of our
knowledge, the present study is the first to reveal the anti-AML
activity of norchelerythrine and explore the underlying
mechanisms.
Cells respond differently to ROS and DDR; leukemic
cells differentiate but hematopoietic stem cells exit quiescence
and differentiate (81). The
hypothesis that ROS serves a pivotal role in
norchelerythrine-mediated AML differentiation is supported by
direct FACS measurements showing increased ROS levels and
inhibition of these effects by NAC, a ROS scavenger. These data
suggest that norchelerythrine engages the tumor-suppressive
signaling pathway shared by chemotherapeutic drugs, such as
cisplatin and doxorubicin.
Norchelerythrine effectively induces apoptosis in
samples from patients with AML, including samples with
myelodysplasia-related changes (MRCs). AML-MRCs include patients
with ≥20% of blasts, prior history of myelodysplastic syndrome
(MDS) or MDS/myeloproliferative neoplasm, a cytogenetic abnormality
related to MDS and multilineage dysplasia. AML-MRC account for up
to 48% of all adult AML cases and mainly affects elderly patients,
showing a poor prognosis with lower remission rates and shorter
overall survival time compared with other AML subtypes (82,83). Here, norchelerythrine decreased
the number of viable cells in two samples from patients with
AML-MRC, suggesting it may be an effective treatment for these
patients.
The present study did not identify the direct target
of norchelerythrine. Quantseq 3′ mRNA-seq was conducted in
CIP-resistant AML cells. Upregulated genes such as ANKRD18A, ITM2A,
and BLMH may be involved in inhibiting differentiation and could be
potential targets of CIP and norchelerythrine. The observation that
several genes upregulated in CIP-resistant cells have no reported
association with myeloid differentiation provides novel avenues for
research in AML. Further studies are needed to understand their
functions and potential impacts on AML.
The present results suggested that norchelerythrine
exhibits anti-leukemic effects by inducing myeloid differentiation,
decreasing cell viability and causing cell cycle arrest.
Norchelerythrine inhibits cell proliferation in samples from
patients with AML harboring various mutations. Norchelerythrine
mechanistically activated the DDR by generating ROS. However, the
present study was limited to in vitro experiments and lacks
supporting in vivo evidence, leaving the anti-tumor activity
and toxicity of norchelerythrine in vivo unclear. Further
in vivo studies in leukemic mouse models will be required to
evaluate the preclinical therapeutic potential of norchelerythrine
in AML.
In summary, the present study showed that CIP and
norchelerythrine exhibited anti-leukemic effects by generating ROS,
leading to activation of DDR and the subsequent induction of
terminal differentiation in AML cells. Based on these findings, CIP
and norchelerythrine hold promise as novel therapeutic candidates
for treating AML. In addition, considering that differentiation
therapy with ATRA and other agents is being evaluated in various
types of cancer, including hepatocellular carcinoma (25,84), further research is warranted to
explore their full therapeutic potential and mechanisms of action
in clinical settings.
Supplementary Data
Availability of data and materials
The data generated in the present study may be found
in the Gene Expression Omnibus under accession number GSE280425 or
at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE280425).
Authors' contributions
JL designed and performed the experiments and wrote
the manuscript. BJ and CK performed the experiments. JL and SWK
confirm the authenticity of all the raw data. HK, JL, BJ, CK, TJK,
YS, SHL, HJS and SWK analyzed the data. TJK, SHL, SWK and HJS
supervised the study. YS revised the manuscript. SWK designed and
conceived the study and wrote the manuscript. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was conducted according to the
Declaration of Helsinki and approved by the Institutional Review
Board of Pusan National University Hospital (approval no.
2403-010-137). Written informed consent was obtained from all
participants included in the study. The animal protocol was
reviewed and approved by the Pusan National
University-Institutional Animal Care and Use Committee (approval
no. PNU 2022-0239).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Use of artificial intelligence tools
During the preparation of this work, AI tools were
used to improve the readability and language of the manuscript.
Subsequently, the authors revised and edited the content produced
by the AI tools as necessary, taking full responsibility for the
ultimate content of the present manuscript.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Research
Foundation funded by the Ministry of Education (grant nos.
2020R1I1A2075060, 2022R1F1A1074989 and 2022R1A4A5031503), Republic
of Korea.
References
|
1
|
Estey E and Döhner H: Acute myeloid
leukaemia. Lancet. 368:1894–1907. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pollyea DA, Kohrt HE and Medeiros BC:
Acute myeloid leukaemia in the elderly: A review. Br J Haematol.
152:524–542. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Saultz JN and Garzon R: Acute myeloid
leukemia: A concise review. J Clin Med. 5:332016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Döhner H, Weisdorf DJ and Bloomfield CD:
Acute myeloid leukemia. N Engl J Med. 373:1136–1152. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Park HJ and Gregory MA: Acute myeloid
leukemia in elderly patients: New targets, new therapies. Aging
Cancer. 4:51–73. 2023. View Article : Google Scholar
|
|
6
|
Robak T and Wierzbowska A: Current and
emerging therapies for acute myeloid leukemia. Clin Ther.
31:2349–2370. 2009. View Article : Google Scholar
|
|
7
|
Newell LF and Cook RJ: Advances in acute
myeloid leukemia. BMJ. 375:n20262021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kantarjian H, Kadia T, DiNardo C, Daver N,
Borthakur G, Jabbour E, Garcia-Manero G, Konopleva M and Ravandi F:
Acute myeloid leukemia: current progress and future directions.
Blood Cancer J. 11:412021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Saygin C and Carraway HE: Emerging
therapies for acute myeloid leukemia. J Hematol Oncol. 10:932017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Parisi E, Draznin J, Stoopler E, Schuster
SJ, Porter D and Sollecito TP: Acute myelogenous leukemia: Advances
and limitations of treatment. Oral Surg Oral Med Oral Pathol Oral
Radiol Endod. 93:257–263. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bhansali RS, Pratz KW and Lai C: Recent
advances in targeted therapies in acute myeloid leukemia. J Hematol
Oncol. 16:292023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Short NJ, Konopleva M, Kadia TM, Borthakur
G, Ravandi F, DiNardo CD and Daver N: Advances in the treatment of
acute myeloid leukemia: New drugs and new challenges. Cancer
Discov. 10:506–525. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kadia T, Ravandi F, Cortes J and
Kantarjian H: New drugs in acute myeloid leukemia. Ann Oncol.
27:770–778. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
van Dijk AD, de Bont ESJ and Kornblau SM:
Targeted therapy in acute myeloid leukemia: Current status and new
insights from a proteomic perspective. Expert Rev Proteomics.
17:1–10. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Godwin C, Gale R and Walter R: Gemtuzumab
ozogamicin in acute myeloid leukemia. Leukemia. 31:1855–1868. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ehninger A, Kramer M, Röllig C, Thiede C,
Bornhäuser M, von Bonin M, Wermke M, Feldmann A, Bachmann M,
Ehninger G and Oelschlägel U: Distribution and levels of cell
surface expression of CD33 and CD123 in acute myeloid leukemia.
Blood Cancer J. 4:e2182014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tabata R, Chi S, Yuda J and Minami Y:
Emerging immunotherapy for acute myeloid leukemia. Int J Mol Sci.
22:19442021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Taussig DC, Pearce DJ, Simpson C,
Rohatiner AZ, Lister TA, Kelly G, Luongo JL, Danet-Desnoyers GA and
Bonnet D: Hematopoietic stem cells express multiple myeloid
markers: Implications for the origin and targeted therapy of acute
myeloid leukemia. Blood. 106:4086–4092. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Walter RB, Appelbaum FR, Estey EH and
Bernstein ID: Acute myeloid leukemia stem cells and CD33-targeted
immunotherapy. Blood. 119:6198–6208. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Damle NK and Frost P: Antibody-targeted
chemotherapy with immunoconjugates of calicheamicin. Curr Opin
Pharmacol. 3:386–390. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Castelli G, Pelosi E and Testa U: Targeted
therapies in the treatment of adult acute myeloid leukemias:
Current status and future perspectives. Int J Hematol Oncol.
5:143–164. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yan M and Liu Q: Differentiation therapy:
A promising strategy for cancer treatment. Chin J Cancer. 35:32016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Madan V and Koeffler HP: Differentiation
therapy of myeloid leukemia: Four decades of development.
Haematologica. 106:26–38. 2021.
|
|
24
|
Stubbins RJ and Karsan A: Differentiation
therapy for myeloid malignancies: Beyond cytotoxicity. Blood Cancer
J. 11:1932021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
de Thé H: Differentiation therapy
revisited. Nat Rev Cancer. 18:117–127. 2018. View Article : Google Scholar
|
|
26
|
Veiga M, Costa EM, Silva S and Pintado M:
Impact of plant extracts upon human health: A review. Crit Rev Food
Sci Nutr. 60:873–886. 2020. View Article : Google Scholar
|
|
27
|
Altemimi A, Lakhssassi N, Baharlouei A,
Watson DG and Lightfoot DA: Phytochemicals: Extraction, isolation,
and identification of bioactive compounds from plant extracts.
Plants (Basel). 6:422017.PubMed/NCBI
|
|
28
|
Dixit S and Ali H: Anticancer activity of
medicinal plant extract-A review. J Chem Cheml Sci. 1:79–85.
2010.
|
|
29
|
Li W, Huang H, Zhang Y, Fan T, Liu X, Xing
W and Niu X: Anti-inflammatory effect of tetrahydrocoptisine from
Corydalis impatiens is a function of possible inhibition of TNF-α,
IL-6 and NO production in lipopolysaccharide-stimulated peritoneal
macrophages through inhibiting NF-κB activation and MAPK pathway.
Eur J Pharmacol. 715:62–71. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang C, Zhang C, Wang Z, Tang Z, Kuang H
and Kong ANT: Corynoline isolated from Corydalis bungeana Turcz.
exhibits anti-inflammatory effects via modulation of Nfr2 and
MAPKs. Molecules. 21:9752016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zheng J, Zhao Y, Lun Q, Song Y, Shi S, Gu
X, Pan B, Qu C, Li J and Tu P: Corydalis edulis Maxim. Promotes
insulin secretion via the activation of protein kinase Cs (PKCs) in
mice and pancreatic β cells. Sci Rep. 7:404542017. View Article : Google Scholar
|
|
32
|
Xu Z, Chen X, Zhang Q, Chen L and Wang Y:
Corydalis yanhusuo W.T. Wang extract inhibits MCF-7 cell
proliferation by inducing cell cycle G2/M arrest. Am J Chin Med.
39:579–586. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Oh MT, Eom HS and Chi GY:
Antiproliferative effect and apoptotic mechanism of extract of
Corydalis yanhusuo on human hepatocarcinoma cells. J Physiol Pathol
Korean Med. 21:1437–1449. 2007.
|
|
34
|
Lu JJ, Bao JL, Chen XP, Huang M and Wang
YT: Alkaloids isolated from natural herbs as the anticancer agents.
Evid Based Complement Alternat Med. 2012:4850422012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Habli Z, Toumieh G, Fatfat M, Rahal ON and
Gali-Muhtasib H: Emerging cytotoxic alkaloids in the battle against
cancer: Overview of molecular mechanisms. Molecules. 22:2502017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Spirin P, Shyrokova E, Lebedev T, Vagapova
E, Smirnova P, Kantemirov A, Dyshlovoy SA, Amsberg GV, Zhidkov M
and Prassolov V: Cytotoxic marine alkaloid 3,10-dibromofascaplysin
induces apoptosis and synergizes with cytarabine resulting in
leukemia cell death. Mar Drugs. 19:4892021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang XD, Li CY, Jiang MM, Li D, Wen P,
Song X, Chen JD, Guo LX, Hu XP, Li GQ, et al: Induction of
apoptosis in human leukemia cells through an intrinsic pathway by
cathachunine, a unique alkaloid isolated from Catharanthus roseus.
Phytomedicine. 23:641–653. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Silva SLR, Dias IRSB, Rodrigues ACBDC,
Costa RGA, Oliveira MS, Barbosa GADC, Soares MBP, Dias RB, Valverde
LF, Rocha CAG, et al: Emetine induces oxidative stress, cell
differentiation and NF-κB inhibition, suppressing AML
stem/progenitor cells. Cell Death Discov. 10:2012024. View Article : Google Scholar
|
|
39
|
Gupta K, Chakrabarti A, Rana S, Ramdeo R,
Roth BL, Agarwal ML, Tse W, Agarwal MK and Wald DN: Securinine, a
myeloid differentiation agent with therapeutic potential for AML.
PLoS One. 6:e212032011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu G, Liu T, Li H, Li Y, Li D and Li W:
c-MYC and reactive oxygen species play roles in tetrandrine-induced
leukemia differentiation. Cell Death Dis. 9:4732018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
42
|
Woo YR, Kwon CS, Lee JE, Jeon BE, Kim TJ,
Choo J, Seo YS and Kim SW: Ajania pacifica (Nakai) K. bremer and
humphries extract limits MYC expression to induce apoptosis in
diffuse large B cell lymphoma. Curr Issues Mol Biol. 46:4580–4594.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kwon CS, Lee JE, Jeon BE, Woo YR, Kim YS,
Kim JW, Park CJ, Jang SY and Kim SW: Anti-leukemic effects of
Idesia polycarpa Maxim branch on human B-cell acute lymphoblastic
leukemia cells. Curr Issues Mol Biol. 45:4035–4049. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lee JE, Kwon CS, Jeon BE, Kim WR, Lee DH,
Koh S, Kim HS and Kim SW: Genome-wide gene expression profiling
defines the mechanism of anticancer effect of colorectal cancer
cell-derived conditioned medium on acute myeloid leukemia. Genes
(Basel). 13:8832022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jeon BE, Kwon CS, Lee JE, Moon K, Cha J,
Park I, Koh S, Yoon M, Kim SW and Kim JN: Anticancer activity of
continentalic acid in B-cell lymphoma. Molecules. 26:68452021.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Vidriales MB, Orfao A, López-Berges MC,
González M, López-Macedo A, García MA, Galende J and San Miguel JF:
Light scatter characteristics of blast cells in acute myeloid
leukaemia: Association with morphology and immunophenotype. J Clin
Pathol. 48:456–462. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Mol BA, Wasinda JJ, Xu YF, Gentle NL and
Meyer V: 1,25-Dihydroxyvitamin D3 augments low-dose
PMA-based monocyte-to-macrophage differentiation in THP-1 cells. J
Immunol Methods. 532:1137162024. View Article : Google Scholar
|
|
48
|
Kelly LM, Englmeier U, Lafon I, Sieweke MH
and Graf T: MafB is an inducer of monocytic differentiation. EMBO
J. 19:1987–1997. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sykes DB, Kfoury YS, Mercier FE, Wawer MJ,
Law JM, Haynes MK, Lewis TA, Schajnovitz A, Jain E, Lee D, et al:
Inhibition of dihydroorotate dehydrogenase overcomes
differentiation blockade in acute myeloid leukemia. Cell.
167:171–186.e15. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hosseini M, Rezvani HR, Aroua N, Bosc C,
Farge T, Saland E, Guyonnet-Dupérat V, Zaghdoudi S, Jarrou L,
Larrue C, et al: Targeting myeloperoxidase disrupts mitochondrial
redox balance and overcomes cytarabine resistance in human acute
myeloid leukemia. Cancer Res. 79:5191–5203. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liu Q and Dong F: Gfi-1 inhibits the
expression of eosinophil major basic protein (MBP) during
G-CSF-induced neutrophilic differentiation. Int J Hematol.
95:640–647. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhou J, Bi C, Ching YQ, Chooi JY, Lu X,
Quah JY, Toh SH, Chan ZL, Tan TZ, Chong PS and Chng WJ: Inhibition
of LIN28B impairs leukemia cell growth and metabolism in acute
myeloid leukemia. J Hematol Oncol. 10:1382017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yoshino S, Yokoyama T, Sunami Y, Takahara
T, Nakamura A, Yamazaki Y, Tsutsumi S, Aburatani H and Nakamura T:
Trib1 promotes acute myeloid leukemia progression by modulating the
transcriptional programs of Hoxa9. Blood. 137:75–88. 2021.
View Article : Google Scholar :
|
|
54
|
Lee JW, Kim HS, Kim S, Hwang J, Kim YH,
Lim GY, Sohn WJ, Yoon SR, Kim JY, Park TS, et al: DACH1 regulates
cell cycle progression of myeloid cells through the control of
cyclin D, Cdk 4/6 and p21Cip1. Biochem Biophys Res Commun.
420:91–95. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Park SM, Cho H, Thornton AM, Barlowe TS,
Chou T, Chhangawala S, Fairchild L, Taggart J, Chow A, Schurer A,
et al: IKZF2 drives leukemia stem cell self-renewal and inhibits
myeloid differentiation. Cell Stem Cell. 24:153–165.e7. 2019.
View Article : Google Scholar :
|
|
56
|
Kirkey DC, Loeb AM, Castro S, McKay CN,
Perkins L, Pardo L, Leonti AR, Tang TT, Loken MR, Brodersen LE, et
al: Therapeutic targeting of PRAME with mTCRCAR T cells in acute
myeloid leukemia. Blood Adv. 7:1178–1189. 2023. View Article : Google Scholar :
|
|
57
|
Boyer T, Guihard S, Roumier C, Peyrouze P,
Gonzales F, Berthon C, Quesnel B, Preudhomme C, Behal H, Duhamel A,
et al: Tetraspanin CD81 is an adverse prognostic marker in acute
myeloid leukemia. Oncotarget. 7:62377–62385. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Prajapati S, Meydan C, Dillon R, Dunham N,
Fan H, Gandara JA, Lee T, Neelamraju Y, Sheridan C, Wang Z, et al:
Loss of CCAAT-enhancer binding protein delta promotes acute myeloid
leukemia cell proliferation and survival by upregulating cyclin D1
expression. Blood. 142(Suppl 1): S13802023. View Article : Google Scholar
|
|
59
|
Zhu Y, Park M, Murtadha M, Caserta E,
Nguyen LXT, Singer M, Estepa MD, Nigam L, Dona' AA, Sanchez JF, et
al: CD84 is a therapeutically targetable driver of leukemogenesis
via disruption of energy supply in acute myeloid leukemia. Blood.
140(Suppl 1): S89–S90. 2022. View Article : Google Scholar
|
|
60
|
Sauer H, Wartenberg M and Hescheler J:
Reactive oxygen species as intracellular messengers during cell
growth and differentiation. Cell Physiol Biochem. 11:173–186. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Prieto-Bermejo R, Romo-González M,
Pérez-Fernández A, Ijurko C and Hernández-Hernández Á: Reactive
oxygen species in haematopoiesis: Leukaemic cells take a walk on
the wild side. J Exp Clin Cancer Res. 37:1252018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Murata T, Kohno S, Ogawa K, Ito C,
Itoigawa M, Ito M, Hikita K and Kaneda N: Cytotoxic activity of
dimeric acridone alkaloids derived from Citrus plants towards human
leukaemia HL-60 cells. J Pharm Pharmacol. 72:1445–1457. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Long Q, Xiao X, Yi P, Liu Y, Varier KM,
Rao Q, Song J, Qiu J, Wang C, Liu W, et al: L20, a Calothrixin B
analog, induces intrinsic apoptosis on HEL cells through
ROS/γ-H2AX/p38 MAPK pathway. Biomed Pharmacother. 137:1113362021.
View Article : Google Scholar
|
|
64
|
Alhuthali HM, Bradshaw TD, Lim KH, Kam TS
and Seedhouse CH: The natural alkaloid Jerantinine B has activity
in acute myeloid leukemia cells through a mechanism involving
c-Jun. BMC Cancer. 20:6292020. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Santos MA, Faryabi RB, Ergen AV, Day AM,
Malhowski A, Canela A, Onozawa M, Lee JE, Callen E,
Gutierrez-Martinez P, et al: DNA-damage-induced differentiation of
leukaemic cells as an anti-cancer barrier. Nature. 514:107–111.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Nicolae CM, O'Connor MJ, Constantin D and
Moldovan GL: NFκB regulates p21 expression and controls DNA
damage-induced leukemic differentiation. Oncogene. 37:3647–3656.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
De Kouchkovsky I and Abdul-Hay M: Acute
myeloid leukemia: A comprehensive review and 2016 update. Blood
Cancer J. 6:e4412016. View Article : Google Scholar
|
|
68
|
Johnson DE and Redner RL: An ATRActive
future for differentiation therapy in AML. Blood Rev. 29:263–268.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhang Ying ZY, Qin MinJian QM and Xie
GuoYong XG: Analysis of alkaloid compositions from Corydalis incise
2008. https://www.cabidigitallibrary.org/doi/full/10.5555/20093000856.
|
|
70
|
Manske RHF: The alkaloids of fumariaceous
plants. XLIV. Corydalis incisa (Thunb.) Pers. and the constitutions
of adlumidine and capnoidine. J Am Chem Soc. 72:3207–3208. 1950.
View Article : Google Scholar
|
|
71
|
Kametani T, Ihara M and Honda T:
Morphinandienone alkaloids from Corydalis incisa. Phytochemistry.
10:1881–1883. 1971. View Article : Google Scholar
|
|
72
|
Nonaka G and Nishioka I: Alkaloids of
Corydalis incisa PERS. V. The structures of corydalispirone and
corydalisol. Chem Pharm Bull. 23:294–298. 1975. View Article : Google Scholar
|
|
73
|
Nonaka G and Nishioka I: Alkaloids of
Corydalis incisa PERS. III. The structures of corydamine
hydrochloride and N-formyl corydamine. Chem Pharm Bull.
21:1410–1414. 1973. View Article : Google Scholar
|
|
74
|
Nonaka G and Nishioka I: Alkaloids of
Corydalis incisa PERS. VI. The structures of benzo [c]
phenanthridine-type alkaloids, 12-hydroxycorynoline and
11-epicorynoline. Chem Pharm Bull. 23:521–526. 1975. View Article : Google Scholar
|
|
75
|
Sulaiman M, Jannat K, Nissapatorn V,
Rahmatullah M, Paul AK, de Lourdes Pereira M, Rajagopal M, Suleiman
M, Butler MS, Break MKB, et al: Antibacterial and antifungal
alkaloids from Asian angiosperms: Distribution, mechanisms of
action, structure-activity, and clinical potentials. Antibiotics
(Basel). 11:11462022. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Luo X, Pedro L, Milic V, Mulhovo S, Duarte
A, Duarte N and Ferreira MJ: Antibacterial benzofuran neolignans
and benzophenanthridine alkaloids from the roots of Zanthoxylum
capense. Planta Med. 78:148–153. 2012. View Article : Google Scholar
|
|
77
|
Wang CF, You CX, Yang K, Guo SS, Geng ZF,
Fan L, Du SS, Deng ZW and Wang YY: Antifeedant activities of
methanol extracts of four Zanthoxylum species and
benzophenanthridines from stem bark of Zanthoxylum schinifolium
against Tribolium castaneum. Ind Crops Prod. 74:407–411. 2015.
View Article : Google Scholar
|
|
78
|
Pang SQ, Wang GQ, Lin JS, Diao Y and Xu
RA: Cytotoxic activity of the alkaloids from Broussonetia
papyrifera fruits. Pharm Biol. 52:1315–1319. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Chang YC, Chang FR, Khalil AT, Hsieh PW
and Wu YC: Cytotoxic benzophenanthridine and benzylisoquinoline
alkaloids from Argemone mexicana. Z Naturforsch C J Biosci.
58:521–526. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Chen JJ, Fang HY, Duh CY and Chen IS: New
indolopyridoquinazoline, benzo[c]phenanthridines and cytotoxic
constituents from Zanthoxylum integrifoliolum. Planta Med.
71:470–475. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Weiss CN and Ito K: DNA damage: A sensible
mediator of the differentiation decision in hematopoietic stem
cells and in leukemia. Int J Mol Sci. 16:6183–6201. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Baer C, Walter W, Stengel A, Hutter S,
Meggendorfer M, Kern W, Haferlach C and Haferlach T: Molecular
classification of AML-MRC reveals a distinct profile and identifies
MRC-like patients with poor overall survival. Blood. 134(Suppl 1):
S27352019. View Article : Google Scholar
|
|
83
|
Arber DA and Erba HP: Diagnosis and
treatment of patients with acute myeloid leukemia with
myelodysplasia-related changes (AML-MRC). Am J Clin Pathol.
154:731–741. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zheng S, Bian H, Li J, Shen Y, Yang Y and
Hu W: Differentiation therapy: Unlocking phenotypic plasticity of
hepatocellular carcinoma. Crit Rev Oncol Hematol. 180:1038542022.
View Article : Google Scholar : PubMed/NCBI
|