Introduction
Studies have been conducted on various factors
involved in defining the prognosis or manipulating the effects of
chemotherapy in order to improve the selection of therapeutic
approaches and their outcomes in various types of tumors (1). The prognostic factors commonly
assessed are clinicopathological, including tumor diameter, status
of lymphatic metastasis and nuclear atypia. In cases of prostate
cancer, tumor stage, the prognostic factors include Gleason scores
and nuclear grades. For breast cancer, therapeutic modalities are
selected by employing the factors that predict the efficacy of
treatment. Such factors include immunohistochemical analysis of the
expression of hormone receptors (ER and PgR) that define the
indication for endocrine therapy and testing for excessive
expression of HER2 protein to determine the indication for the use
of trastuzumab, a molecular-targeted therapeutic agent. For
prostate cancers, however, no useful molecular markers, such as
those cited above, exist to predict the outcome and efficacy of the
drug therapy. DNA/RNA synthesis is essential for the proliferation
of cancer cells, and for the enzymes that control the synthetic
process, thymidylate synthase (TS), orotate phosphoribosyl
transferase (OPRT) and thymidine phosphorylase (TP) in the de
novo pathway are cited. Dihydropyrimidine dehydrogenase (DPD)
has been identified for the enzymes involved in the breakdown of
these nucleic acids. These enzymes play key roles in the metabolism
of pyrimidine base, a component of DNA/RNA, while they are equally
important in the metabolism of 5-FU anticancer agents (2–8).
In cancers involving the stomach, colon and breast,
where 5-FU anticancer agents are considered to be the key drugs,
the levels of expression of enzymes involved in nucleic acid
metabolism, such as TS, DPD, TP and OPRT, have been investigated as
markers for determining prognosis. Subsequently, numerous studies
have been conducted on possible prognosis-deciding factors, as well
as factors that predict the efficacy of 5-FU anticancer agents
(13). On the other hand, it has
been reported that the levels of expression of mRNA for these
enzymes differ in each tumor (9),
indicating a need to evaluate each tumor individually.
Concerning prostate cancers, the expression levels
of these enzymes may be related to prognosis. The expression of TS
was evaluated in prostate cancers using a staining method to
observe its relationship with the prognosis (10). This study suggested a potential role
of TS as a prognostic factor.
The level of gene expression within tumor tissues
may be elucidated when the mRNA of the target gene is extracted and
its expression quantified using the real-time (RT)-PCR method.
However, in prostate cancers, the prostate is characteristically
dotted by tumor tissues and it is difficult to quantify only the
mRNA within tumor cells. To solve this problem, the Danenberg tumor
profile (DTP) which employs the microdissection technique was used.
In this method, the tumor tissue is isolated from the excised
prostatic sample for subsequent analysis, enabling a more accurate
evaluation of the mRNA expression level.
If the gene expression of nucleic acid-metabolizing
enzymes (represented by TS) is found to be a possible factor in
determining prognosis, patients who are likely to exhibit a poor
response would be separated from those undergoing the standard
therapy in order to formulate the subsequent therapeutic
approaches. Therefore, a retrospective study was conducted to
discover whether the expression levels of TS, DPD, TP and OPRT
(enzymes related to the metabolism of 5-FU drugs) can be used as
prognostic markers.
Patients and methods
Case series
A total of 172 patients who were diagnosed with
prostate cancer and underwent radical prostatectomies between 1996
and 2006 were recruited. Prostate tissues were collected from the
surgical specimens and the relationship between the mRNA expression
of nucleic acid-metabolizing enzymes and clinicopathological
factors was retrospectively examined. Table I lists the backgrounds of the
patients. Maximum androgen blockade was conducted in a neoadjuvant
setting for 77 patients. The pathological specimens of the study
were reviewed by a single pathologist. The median for the
prostate-specific antigen (PSA) value at the initial examination
was 7.2 ng/ml (range 2.8–111.2), while the median observation
period was 4.05 years (range 0.6–16.3). Approval was obtained at
commencement of the study from the ethics committee of our
institution, and the determination and analysis of mRNAs were
conducted on each of the 172 patients from whom informed consent
was obtained.
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| No. of patients | 172 |
| Median age
(years) | 68 |
| Range | 49–77 |
| Clinical stage |
| B0 | 91 |
| B1 | 39 |
| B2 | 28 |
| C | 5 |
| Unknown | 9 |
| Histological
type |
|
Well-differentiated | 65 |
| Moderately
differentiated | 66 |
| Poorly
differentiated | 17 |
| Gleason score |
| 6 | 17 |
| 7 | 104 |
| 8–10 | 41 |
| Median PSA
(ng/ml) | 7.2 |
| Range | 2.8–111.2 |
Determination of the gene expression
level of nucleic acid-metabolizing enzymes by RT-PCR
Following an examination of the hematoxylin and
eosin (H&E)-stained slides by the pathologist, the
formalin-fixed, paraffin-embedded (FFPE) specimens were subjected
to microtomy in order to prepare the slides. The mRNA (the level of
gene expression) of each of the nucleic acid-metabolizing enzymes
(TS, DPD, OPRT and TP) contained in the tumor tissues on the slides
was analyzed using the DTP method (Response Genetics, Inc., Los
Angeles, CA, United States Patent No. 6,248,535). This method
allowed the laser capture microdissection technique (11), by which tumor cells are selectively
extracted from the FFPE, and the quantitative real-time RT-PCR
method to be combined for relative quantitative determination of
the mRNA of the target gene. This combination was carried out to
prepare the cDNA following RNA extraction, and a RT-PCR was
conducted by the Taq Man method to amplify the target cDNA
sequence. The expression level of each mRNA (for TS, DPD, TP or
OPRT) was computed from the ratio against the expression of
β-actin, a housekeeping gene.
Statistical analysis
The relationship between the expression level of
mRNA for TS, DPD, TP or OPRT and the clinicopathological factors
was evaluated: to analyze the differences in the mean of mRNA
expression among the subgroups, log 10 (mRNA) was used for a normal
distribution; to compare two groups, a t-test was conducted in a
state in which a parametric method was possible; and to compare
three or more groups, tests were conducted by employing an analysis
of variance. To examine whether the expression of each mRNA can be
a prognostic factor, a receiver operating characteristic (ROC)
analysis was conducted to obtain an appropriate cut-off value for
each mRNA that was to be used for prognosis prediction.
Additionally, the cases in this analysis were stratified according
to the PSA relapse and the expression level of each mRNA (low or
high): true positive (TP); false negative (FN); false positive (FP)
and true negative (TN). Then, ‘sensitivity [= TP/(TP + FN)]’ and
‘specificity [= TN/(FP + TN)]’ were computed and the level of mRNA
expression at which ‘sensitivity - (1 - specificity)’ is maximized
was used as the cut-off value. For the analysis of survival time,
PSA relapse-free survival (PSA-RFS) was computed by applying the
Kaplan-Meier method and using the day of surgery as the starting
point and the first day of the confirmed relapse of PSA as the day
when the event occurred. To find the differences among subgroups, a
log-rank test was used. The level of significance was set at
p<0.05. Statistical software, JMP ver7.0.1 (SAS Institute, Inc.)
was used for the statistical analysis.
Results
Evaluation of the level of enzyme
expression (for TS, DPD, TP and OPRT) related to 5-FU
metabolism
The expression level for TS, DPD, TP and OPRT in the
excised prostatic tissue was determined. The analysis was possible
in 94, 83, 97 and 92 patients, respectively, and the mean level of
expression for each mRNA was 1.805, 0.69, 3.52 and 0.85,
respectively.
A comparative evaluation was performed in order to
investigate possible correlations between the mRNA expression of
each enzyme (TS, DPD, TP or OPRT) and various clinicopathological
factors related to prostate cancers. Table II shows the mean of mRNA
expression. No significant differences were found in patient age,
tumor stage and differentiation or level of mRNA expression.
However, the expression of TS and OPRT was significantly higher in
those cases exhibiting poorly differentiated carcinoma in their
tumor tissues. When contrasted to the total Gleason scores, the
increase from 6 to 7, then to 8–10 was positively correlated with
increases in TS, DPD, TP and OPRT expression; however, only TP
showed a significant difference. Regarding the correlation between
the levels of PSA at the initial examination and the mRNA
expression, the OPRT expression was significantly higher in the
‘PSA-high’ group (≥5.6 ng/ml) in contrast to the ‘PSA-low’ group
(<5.6 ng/ml). Among the patients in whom PSA relapse was
confirmed, the expression of TS and OPRT was significantly higher.
When the groups with and without pre-surgical treatment were
compared, the expression of DPD and TP was significantly higher and
that of OPRT significantly lower in the former group.
 | Table IIRelationship between
clinicopathological factors and mRNA expression. |
Table II
Relationship between
clinicopathological factors and mRNA expression.
| Mean of mRNA
expression (n) |
|---|
|
|
|---|
| TS | P-value | DPD | P-value | TP | P-value | OPRT | P-value |
|---|
| Age (years) |
| <65 | 1.75 (29) | NS | 0.81 (24) | NS | 6.31 (29) | NS | 0.94 (28) | NS |
| ≥65 | 2.12 (65) | | 0.82 (59) | | 5.17 (68) | | 0.96 (64) | |
| Stage |
| B0 | 1.83 (59) | NS | 0.74 (46) | NS | 4.22 (52) | NS | 0.87 (52) | NS |
| B1 | 2.14 (19) | | 0.98 (14) | | 7.07 (20) | | 1.06 (19) | |
| B2 | 1.94 (12) | | 0.96 (13) | | 5.22 (14) | | 0.79 (12) | |
| C | 2.90 (4) | | 0.87 (4) | | 18.1 (4) | | 1.53 (4) | |
| Histological
type |
|
Well-differentiated | 1.99 (39) | NS | 0.81 (32) | NS | 5.67 (40) | NS | 0.90 (40) | NS |
| Moderately
differentiated | 1.80 (40) | | 0.80 (39) | | 5.64 (42) | | 0.92 (38) | |
| Poorly
differentiated | 2.32 (9) | | 0.94 (6) | | 4.51 (9) | | 1.17 (9) | |
| Including poorly
differentiated type |
| − | 1.78 (69) | <0.05 | 0.84 (61) | NS | 5.93 (72) | NS | 0.87 (67) | <0.05 |
| + | 2.51 (19) | | 0.73 (16) | | 4.08 (19) | | 1.15 (20) | |
| Total Gleason
score |
| 6 | 1.34 (6) | NS | 0.68 (5) | NS | 3.29 (7) | <0.05 | 0.70 (6) | NS |
| 7 | 1.97 (68) | | 0.80 (59) | | 4.74 (69) | | 0.94 (64) | |
| 8–10 | 2.34 (19) | | 0.90 (18) | | 8.53 (20) | | 1.08 (21) | |
| PSA (ng/ml) |
| <5.6 | 1.80 (30) | NS | 0.83 (27) | NS | 4.70 (33) | NS | 0.81 (30) | <0.05 |
| ≥5.6 | 2.04 (58) | | 0.82 (51) | | 6.09 (58) | | 0.99 (57) | |
| PSA relapse |
| − | 1.81 (68) | <0.01 | 0.87 (62) | NS | 5.28 (72) | NS | 0.86 (68) | <0.05 |
| + | 2.55 (20) | | 0.66 (15) | | 4.06 (19) | | 1.19 (19) | |
| Neoadjuvant
therapy |
| − | 2.00 (74) | NS | 0.72 (64) | <0.01 | 4.13 (75) | <0.01 | 0.98 (73) | <0.01 |
| + | 1.71 (17) | | 1.21 (17) | | 11.0 (19) | | 0.65 (16) | |
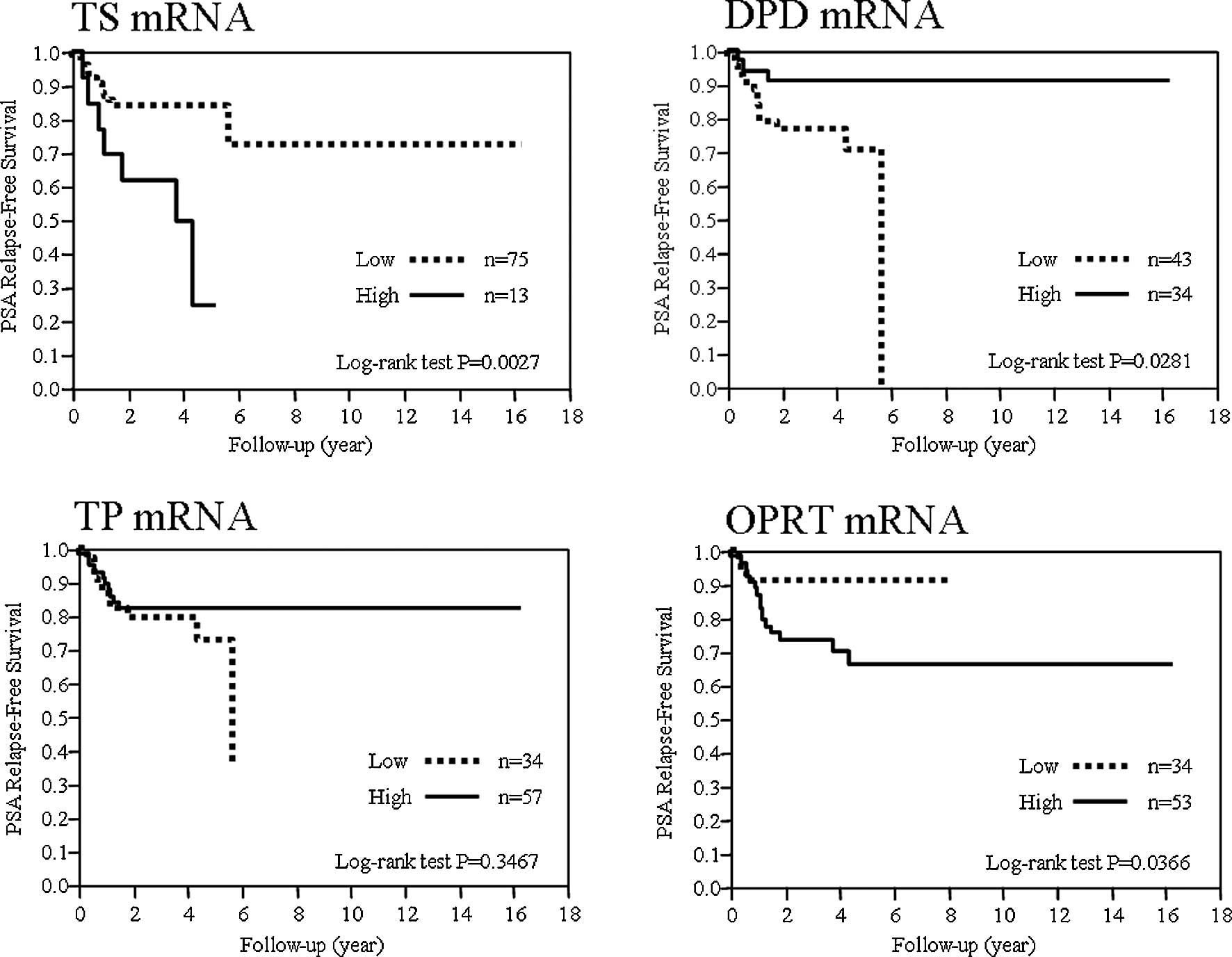
The expression of TS, DPD, TP and OPRT
and its potential role as a prognostic factor
To evaluate the potential use of each mRNA
expression as a prognostic factor, the PSA-RFS was examined for the
expression of each mRNA. The cut-off values for mRNAs for TS, DPD,
TP and OPRT determined by the ROC analysis were 2.98, 0.84, 3.09
and 0.68, respectively. mRNAs exceeding the cut-off values were
defined as the high groups and those below the cut-off values, the
low groups, and their correlation with prognosis was examined. The
prognosis was significantly poorer in the TS-high, DPD-low and
OPRT-high groups (Fig. 1).
The 5-year PSA-RFS was 84, 24.62, 70.78, 91.08,
72.59, 82.25, 91.18 and 66.2% in TS-low and -high, DPD-low and
-high, TP-low and -high and OPRT-low and -high groups,
respectively.
Discussion
In the present study, the results of the analysis of
relapse-free survival time stratified by the mRNA expression
indicated that the prognosis is significantly poorer in cases when
the TS mRNA level was over 2.98, OPRT mRNA exceeded 0.68 or the DPD
expression level was below 0.84. It was suggested that these cases
require additional treatment or that the planning of a new
therapeutic modality was necessary to improve prognosis. Studies on
the relationship between mRNA expression and prognosis have been
conducted on various types of tumor, but only a few focused on
prostate cancers (10). The data
presented in this study are beneficial to future studies.
TS is known to be a target enzyme for 5-FU, and its
potential role as a prognostic factor has been described for
various types of tumor species. Patients with positive or high
levels of TS exhibited a poor prognosis (12–15),
which was in agreement with our results. DPD is a 5-FU decomposing
enzyme and is crucial for determining sensitivity to this
anticancer agent. Previous studies found that cases that were
exposed to 5-FU preparations and showed a high DPD level responded
poorly to these therapeutic agents or the prognosis was found to be
poor (16–18). These cases had been treated with
5-FU preparations and it was considered that the DPD expression
levels affected the drug effects. In this study, few patients were
exposed to 5-FU preparations, but contrary to the results of the
earlier studies, the prognosis was poor in the DPD-low group. TP
did not affect prognosis in the present study, but it was reported
that in cases of metastatic colorectal cancer the effect of 5-FU is
limited when the TP mRNA expression is high in the tumor tissue
(19). In another study, a
satisfactory prognosis is described in those cases with high levels
of TP expression among patients who underwent post-operative
adjuvant chemotherapy (20). OPRT
is a 5-FU phosphorylating enzyme. In cancers of the digestive
system, OPRT defines the prognosis and 5-FU sensitivity, and
patients with a low expression of this enzyme are associated with a
poor prognosis (13,21,22).
Results of this study, however, showed that a poor prognosis in the
group with a high OPRT expression. Of note is that the relationship
between each mRNA expression and prognosis varies, depending on the
type of cancer.
Compared to tumors affecting other organs, prostate
cancers are characterized by tumors scattered throughout the
prostate gland. Therefore, macroscopic differentiation of the tumor
from normal tissues is difficult. In early tumors with a surgical
indication, in particular, the tumor volume is limited, making the
diagnosis even more difficult. In addition, tumor and interstitial
tissue are intermingled. Subsequently, when mRNA is extracted
directly from the tumor tissue of an excised specimen, the
expression level of mRNA in the tumor cells may not be evaluated
accurately. In the present study, on the other hand, the DTP method
was utilized and it was hypothesized that the volume of the mRNA
expressed was accurately determined. However, the DTP method also
has limitations, such as non-availability under certain conditions
for the preservation or fixation of the FFPE specimens and
limitations regarding facilities where the determination can be
made. When compared to immunostaining, the DTP method is more
beneficial due to accurate quantification of mRNA expression. It is
crucial to select a method of determination that is most suitable
for the research objective.
In examining the mRNA expression of these enzymes in
cancerous prostatic tissue, the group with high PSA levels at the
initial examination exhibited significantly higher levels of OPRT
mRNA expression, indicating the possibility that OPRT is a
prognostic factor for prostate cancers. In the cases exhibiting a
high expression of TS and OPRT mRNA, including certain cases with
poorly differentiated carcinoma, this suggests that a correlation
with malignant potential exists.
When a group of patients who had received
pre-operative treatment was compared to patients who had not such
treatment, it was noted that in the former group, the DPD and TP
mRNA expression was high, whereas the OPRT mRNA expression was low.
Few studies have reported that the pre-operative endocrine therapy
of prostate cancers affects the level of mRNA expression of nucleic
acid-metabolizing enzymes (23). It
is possible that the pre-operative treatment exerted a direct
effect on the mRNA expression patterns in the tumor tissue, or a
relative decrease of tumor cells that are considered to be
associated with high malignancy resulted in an apparent change in
the expression level. However, the mechanism by which endocrine
therapy affects nucleic acid-metabolizing enzymes is inadequately
elucidated in this study.
This study was conducted on patients with localized
cancers, but the determination of mRNA of these enzymes may also
contribute to the prognostic evaluation of patients with advanced
cancers. If individualized treatment becomes a reality by using
prognostic factors, selective treatment may become an option for
patients in whom 5-FU preparations show more promising results.
In conclusion, among patients with prostate cancers,
those with a poor prognosis can be identified by determining the
levels of TS, DPD and OPRT mRNA expression in the excised tumors.
Such an mRNA expression appears to be useful as a prognostic
factor.
References
|
1
|
Mok TS, Wu YL, Thongprasert S, et al:
Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N
Engl J Med. 361:947–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Heggie GD, Sommadossi JP, Cross DS, Huster
WJ and Diasio RB: Clinical pharmacokinetics of 5-fluorouracil and
its metabolites in plasma, urine, and bile. Cancer Res.
47:2203–2206. 1987.PubMed/NCBI
|
|
3
|
Van Kuilenburg AB: Dihydropyrimidine
dehydrogenase and the efficacy and toxicity of 5-fluorouracil. Eur
J Cancer. 40:939–950. 2004.PubMed/NCBI
|
|
4
|
Danenberg PV: Thymidylate synthetase – a
target enzyme in cancer chemotherapy. Biochim Biophys Acta.
473:73–92. 1977.
|
|
5
|
Pinedo HM and Peters GF: Fluorouracil:
biochemistry and pharmacology. J Clin Oncol. 6:1653–1664.
1988.PubMed/NCBI
|
|
6
|
Longley DB, Harkin DP and Johnston PG:
5-Fluorouracil: mechanisms of action and clinical strategies. Nat
Rev Cancer. 3:330–338. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peters GJ, Laurensse E, Leyva A, Lankelma
J and Pinedo HM: Sensitivity of human, murine, and rat cells to
5-fluorouracil and 5′-deoxy-5-fluorouridine in relation to
drug-metabolizing enzymes. Cancer Res. 46:20–28. 1986.
|
|
8
|
Peters GJ, van Groeningen CJ, Laurensse EJ
and Pinedo HM: A comparison of 5-fluorouracil metabolism in human
colorectal cancer and colon mucosa. Cancer. 68:1903–1909. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fukui Y, Oka T, Nagayama S, Danenberg PV,
Danenberg KD and Fukushima M: Thymidylate synthase,
dihydropyrimidine dehydrogenase, orotate phosphoribosyltransferase
mRNA and protein expression levels in solid tumors in large scale
population analysis. Int J Mol Med. 22:709–716. 2008.
|
|
10
|
Li Y, Mizutani Y, Shiraishi T, et al:
Prognostic significance of thymidylate synthase expression in
patients with prostate cancer undergoing radical prostatectomy.
Urology. 69:988–995. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Emmert-Buck MR, Bonner RF, Smith PD, et
al: Laser capture microdissection. Science. 274:998–1001. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ichikawa W, Takahashi T, Suto K, et al:
Thymidylate synthase predictive power is overcome by irinotecan
combination therapy with S-1 for gastric cancer. Br J Cancer.
91:1245–1250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ichikawa W, Takahashi T, Suto K, et al:
Simple combinations of 5-FU pathway genes predict the outcome of
metastatic gastric cancer patients treated by S-1. Int J Cancer.
119:1927–1933. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hakamada Y, Tsuchida A, Arima M, et al:
Prognostic predictors in breast cancer patients with postoperative
5-fluorouracil-based chemotherapy. Int J Mol Med. 16:309–314.
2005.PubMed/NCBI
|
|
15
|
Miyoshi T, Kondo K, Toba H, et al:
Predictive value of thymidylate synthase and dihydropyrimidine
dehydrogenase expression in tumor tissue, regarding the efficacy of
postoperatively administered UFT (tegafur+uracil) in patients with
non-small cell lung cancer. Anticancer Res. 27:2641–2648.
2007.PubMed/NCBI
|
|
16
|
Etienne MC, Chéradame S, Fischel JL, et
al: Response to fluorouracil therapy in cancer patients: the role
of tumoral dihydropyrimidine dehydrogenase activity. J Clin Oncol.
13:1663–1670. 1995.PubMed/NCBI
|
|
17
|
Horiguchi J, Yoshida T, Koibuchi Y, et al:
DPD activity and immunohistochemical DPD expression in human breast
cancer. Oncol Rep. 11:65–72. 2004.PubMed/NCBI
|
|
18
|
Salonga D, Danenberg KD, Johnson M, et al:
Colorectal tumors responding to 5-fluorouracil have low gene
expression levels of dihydropyrimidine dehydrogenase, thymidylate
synthase, and thymidine phosphorylase. Clin Cancer Res.
6:1322–1327. 2000.
|
|
19
|
Metzger R, Danenberg K, Leichman CG, et
al: High basal level gene expression of thymidine phosphorylase
(platelet-derived endothelial cell growth factor) in colorectal
tumors is associated with nonresponse to 5-fluorouracil. Clin
Cancer Res. 4:2371–2376. 1998.
|
|
20
|
Saito S, Tsuno N, Nagawa H, et al:
Expression of platelet-derived endothelial cell growth factor
correlates with good prognosis in patients with colorectal
carcinoma. Cancer. 88:42–49. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ochiai T, Nishimura K, Noguchi H, et al:
Prognostic impact of orotate phosphoribosyl transferase among
5-fluorouracil metabolic enzymes in resectable colorectal cancers
treated by oral 5-fluorouracil-based adjuvant chemotherapy. Int J
Cancer. 118:3084–3088. 2006. View Article : Google Scholar
|
|
22
|
Ichikawa W, Uetake H, Shirota Y, et al:
Both gene expression for orotate phosphoribosyltransferase and its
ratio to dihydropyrimidine dehydrogenase influence outcome
following fluoropyrimidine-based chemotherapy for metastatic
colorectal cancer. Br J Cancer. 89:1486–1492. 2003. View Article : Google Scholar
|
|
23
|
Inoue T, Segawa T, Shiraishi T, et al:
High-grade and hormone-treated prostate cancer express high levels
of thymidylate synthase. BJU Int. 98:197–200. 2006. View Article : Google Scholar : PubMed/NCBI
|