Introduction
Renal cell carcinomas (RCCs) constitute 2–3% of
adult cancers and include clear cell RCC (ccRCC) (75%), papillary
RCC (PRCC) (10%) and chromophobe RCC (ChRCC) (5%) subtypes
(1,2). Accurate classification of RCCs is
crucial, since different histopathologic subtypes require specific
therapeutic management strategies due to their markedly different
prognoses and responses to therapy (3). The von Hippel-Lindau (VHL) tumor
suppressor gene at chromosome 3p25 is inactivated in over 70% of
sporadic ccRCCs, and constitutional VHL mutation carriers have a
high lifetime risk of developing ccRCC (4). Molecular-targeting drugs for advanced
RCC were designed based on the VHL-hypoxia inducible factor (HIF),
vascular endothelial growth factor receptor (VEGFR),
platelet-derived growth factor (PDGF) and transforming growth
factor transduction pathways (5),
as well as the mammalian target of the rapamycin pathway (6). However, the therapeutic effect of
molecular-targeting drugs in PRCC and ChRCC is less clear, since
fewer studies are available on the genetic alterations in these
subtypes than those in ccRCC. Therefore, newly developed
high-density single nucleotide polymorphism (SNP) array was applied
to investigate novel and common genetic change in the three RCC
subtypes.
A number of cytogenetic studies have identified the
chromosomal region responsible for RCC. Recent advances in
array-based comparative genomic hybridization (CGH) technology have
allowed chromosomal regions to be examined in much more detail,
thus revolutionizing understanding of gene-copy abnormalities
(7). The introduction of
high-density SNP genotyping technology to genomic profiling, termed
SNP-CGH, is a further advance, since the simultaneous measurement
of signal intensity variations and changes in allelic composition
allows for the detection of copy number changes and copy-neutral
loss-of-heterozygosity (LOH) events (8). Furthermore, SNP-CGH has the advantage
that candidate genes are readily accessed via the SNP tag number. A
few SNP-CGH studies have been performed in RCC, using the 10K array
(2) and the 307K array in ccRCC
(9). In this study, SNP-CGH was
used to detect chromosomal aberrations in each of the three types
of RCC using whole-genome genotyping with Human CNV370-Duo DNA
Analysis BeadChip, which covers the 370K SNPs over a median spacing
of 4.9 kb in the human genome.
Materials and methods
Tumor samples, normal control and DNA
extraction
Primary RCC tumors were surgically resected at
Kyushu University Hospital. Samples of carcinoma tissue were frozen
in liquid nitrogen immediately after surgery and stored in deep
freeze at −80°C until use. The RCC specimens were
histopathologically diagnosed as pure ccRCC, PRCC or ChRCC by a
single pathologist. Mixed histopathologic types of RCC were
excluded from this study. The proportion of tumor cells in a tissue
section was confirmed to be >70% in all of the tumor tissues.
The frozen blocks were then subjected to DNA extraction. DNA from
peripheral blood was used as the normal control DNA for each
patient. DNA extraction was performed according to standard
protocols (10). Written informed
consent was obtained from the patients. The study was approved by
the Institutional Review Board.
Whole-genome SNP array analysis
The Human CNV370-Duo DNA Analysis BeadChip system
(Illumina, San Diego, CA, USA) was used, as previously described
(11). The allele balance and
log2 ratio (tumor/normal) were visualized using the
Genome Browser on BeadStudio ver3 (Illumina).
Statistical analysis
Two-tailed Student's t-tests were used to evaluate
the allelic changes and clinical factors, such as stage, tumor
grade and prognosis (data not shown).
Results
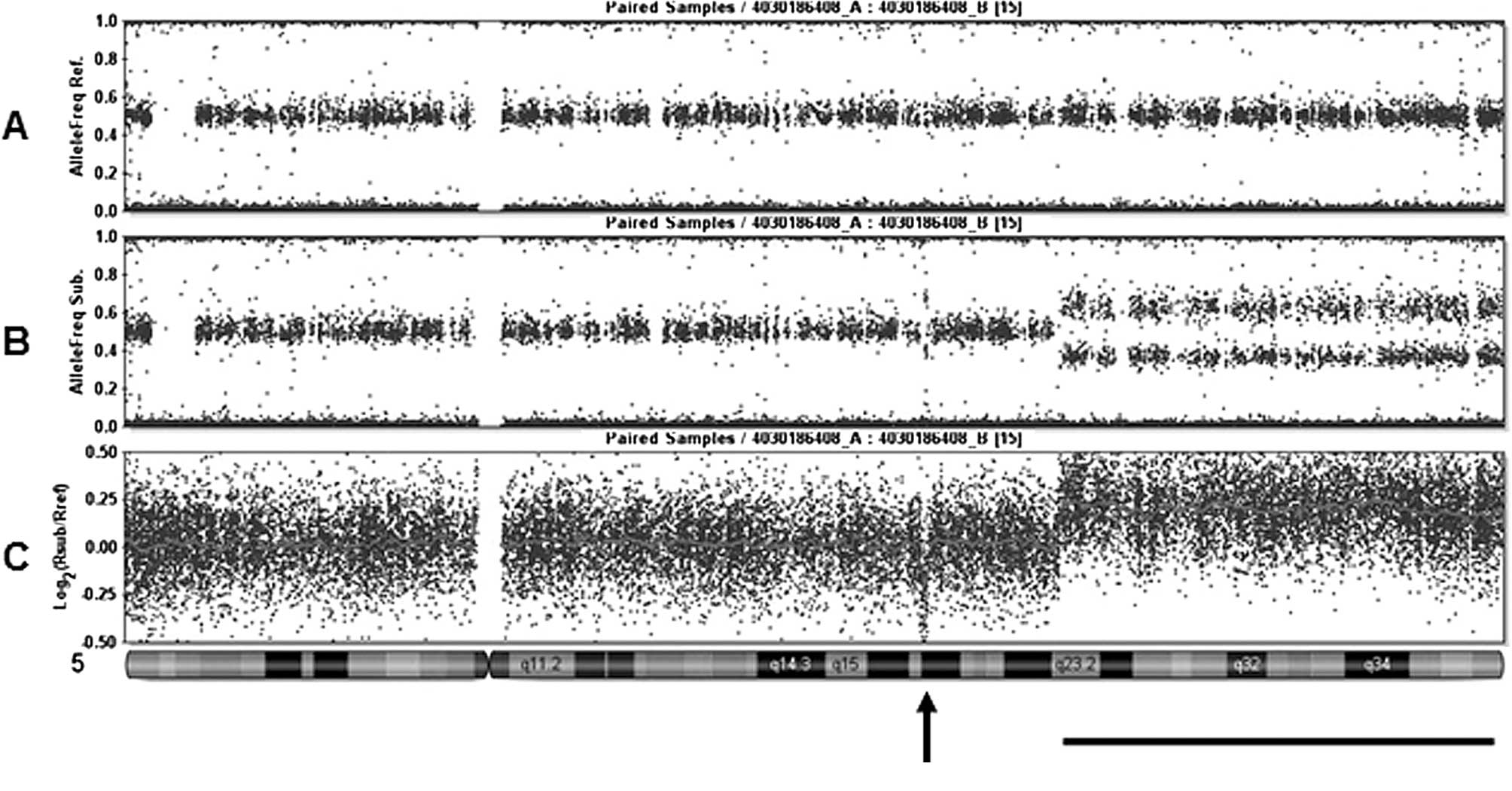
Fig. 1 shows a
representative result using the Human CNV370-Duo DNA Analysis
BeadChip system and the allele frequency of each SNP typing from
(A) normal DNA, (B) tumor DNA and (C) log2 ratio
(tumor/normal). A comparison of the three lines showed that a focal
deletion and partial chromosomal amplification of 5q were
identical. Since the chromosomal position of analyzed SNP is
available on the public database (http://www.ncbi.nlm.nih.gov/SNP/), genes involved in
observed chromosomal aberrations are precisely identified. For
example, the 1.45 Mb focal deletion shown in Fig. 1 is flanked by rs104114206 and
rs105568833. The deletion contains only one protein-coding gene as
a candidate tumor suppressor.
The most common genetic loss in ccRCC was identified
in 3p, which was found in 95% (21/22 samples) of the carcinomas
(Table I), suggesting that 3p loss
is early stage in clear cell carcinogenesis. Other frequent changes
were losses of 1p (23%), 3q (46%), 8p (32%), 8q (22%), 9p (27%), 9q
(27%) and 18q (23%), and gains of 5q (32%), 7p (27%), 7q (27%) and
1q (23%) (Table I). Furthermore,
microamplifications and microdeletions of <1 Mb were detected in
4 and 18 regions, respectively, in ccRCC (Table II). Among these, the microdeletion
of 10q23.31 was identified in two tumors. Moreover, five genes,
including PTEN, were identified as candidate tumor
suppressor genes in the SNP-based genomic database. The other
microamplifications and microdeletions observed are shown in
Table II.
 | Table IA summary of chromosomal gains and
losses in renal cell carcinomas. |
Table I
A summary of chromosomal gains and
losses in renal cell carcinomas.
|
Loss | Gain |
|---|
|
|
|
|---|
| Complete | Partial | Complete | Partial |
|---|
|
|
| | |
|---|
| Hemi | Neut | Mix | Hemi | Neut | Mix | | |
|---|
|
|
|
|
|
|
|
|
|
|---|
| cc | P | Ch | cc | P | Ch | cc | P | Ch | cc | P | Ch | cc | P | Ch | cc | P | Ch | cc | P | Ch | cc | P | Ch |
|---|
| 1p | 5 | 14 | 100 | | | | 5 | | | 14 | 14 | | | | | | | | | | | 9 | | |
| 1q | | 14 | 100 | | | | | | | | 14 | | | | | | | | 23 | | | | | |
| 2p | | | 100 | 5 | | | | | | | | | | | | | | | | | | | | |
| 2q | | | 100 | | | | | | | 14 | | | | | | | | | | | | 9 | 14 | |
| 3p | 55 | 29 | | | 14 | | | | | 36 | | 13 | 5 | | | | | | | | | | | |
| 3q | 14 | 14 | | | 29 | | | | | 27 | | | 5 | | | | | | | | | 5 | | |
| 4p | 14 | 14 | | | | | | | | | | | | | | | | | | | | | | |
| 4q | 14 | 14 | | | | | | | | 5 | | | | | | | | | | | | | | |
| 5p | | | 12.5 | | | | | | | | | | | | | | | | 9 | | | | | |
| 5q | | | 12.5 | | | | | | | 9 | | | | | | | | | 9 | | | 23 | 14 | |
| 6p | 9 | | 100 | | | | | | | | 14 | | | | | | | | | | | | 14 | |
| 6q | 9 | | 100 | | 14 | | | | | 9 | | | | | | | | | | | | | | |
| 7p | | | | 9 | | | | | | | | | | | | | | | 27 | 14 | | | | |
| 7q | | | | 9 | | | | | | | | | | | | | | | 27 | 14 | | | | 13 |
| 8p | | | 50 | 5 | | | | | | 9 | | | | | | | | | | 29 | | | | |
| 8q | 5 | | 50 | 9 | | | | | | 5 | | | | | | | | | | 29 | | | 14 | |
| 9p | 18 | | | 5 | | | | | | 5 | 14 | | | | | | | | | | | | | |
| 9q | 23 | 14 | | 5 | | | | | | | | | | | | | | | | | | | | |
| 10p | | | 100 | | | | | | | 5 | 14 | | | | | | | | | | | | 14 | |
| 10q | | | 100 | | | | | | | 5 | 14 | | 5 | | | | | | | 14 | | | | |
| 11p | 5 | | | | | 13 | | | | | | | 5 | | | | | | | | | 5 | 14 | |
| 11q | 5 | | | | | 13 | | | | 5 | 29 | | | | | | | | | | | 5 | 14 | |
| 12p | | | | 5 | | | | | | | | | | | | | | | 5 | 29 | | | | |
| 12q | | | | 5 | | | | | | | | | | | | | | | 5 | 29 | | | 14 | |
| 13p | | | 75 | 5 | | | | | | | | | | | | | | | | | | | | |
| 13q | | | 75 | 5 | | | 5 | | | | | | | | | | | | | | | | | |
| 14p | 27 | | | | | | | | | | | | | | | | | | | | | | | |
| 14q | 27 | | | | | | | | | | 14 | | | | | | | | | | | | | |
| 15p | | | | 5 | | | | | | | | | | | | | | | | | | | | |
| 15q | | | | 5 | | | | | | | | | | | | | | | | | | | | |
| 16p | | | | 5 | | | | | | | 14 | | | | | | | | | 29 | | | | |
| 16q | 5 | | | 5 | | | | | | | | | | | | | | | | 29 | | | | |
| 17p | | | 100 | | | | | | | 9 | | | | | | | | | | 14 | | | | |
| 17q | | | 100 | | | | | | | | | | | | | | | | | 14 | | 9 | 14 | |
| 18p | 9 | 14 | 25 | 9 | | | | | | | | | | | | | | | | | | | | |
| 18q | 14 | 14 | 25 | 9 | | | | | | | 14 | | | | | | | | | | | | | |
| 19p | | | | 5 | | | | | | | | | | | | | | | | | | | | |
| 19q | | | | 5 | | | | | | | | | | | | | | | | | | | | |
| 20p | | 14 | | | | | | | | | | | | | | | | | 5 | 43 | | | | |
| 20q | | | | | | | | | | | | | | | | | | | 5 | 43 | | 5 | 14 | |
| 21p | | | 50 | | | | | | | | | | | | | | | | | 14 | | | | |
| 21q | | | 50 | | | | | | | | | | | | 13 | | | | | 14 | | | | |
| 22p | | 29 | | 5 | | | | | | | | | | | | | | | | | | | | |
| 22q | | 29 | | 5 | | | | | | | | | | | | | | | | | | | | |
 | Table IIThe chromosomal locations of
microamplifications (G) and microdeletions (L). |
Table II
The chromosomal locations of
microamplifications (G) and microdeletions (L).
| ccRCC |
| G | 1p34.2 |
| G | 13q14.2-14.3 |
| G | 13q22.1-22.2 |
| G | 5q21.2 |
| L | 1p36.12-35.3 |
| L | 2q33.3 |
| L | 2q14.3 |
| L | 2q14.3-2q21.1 |
| L | 2q21.2 |
| L | 2q32.3 |
| L | 4p15.33 |
| L | 4p14 |
| L | 5q21.3 |
| L | 6q12 |
| L | 6q12 |
| L | 6q12-13 |
| L | 6q25.1 |
| L | 8q21.11 |
| L | 10q23.31 |
| L | 10q23.31 |
| L | 11p15.1 |
| L | 18q12.1 |
| PRCC |
| G | 2q21.1 |
| G | 10p12.1 |
| G | 10p12.31 |
| G | 10p11.22 |
| G | 10q22.1 |
| G | 10q22.2 |
| L | 1q24.2-25.1 |
| L | 8q24.23 |
| L | 10p15.2-15.1 |
| L | 10q21.1 |
| L | 10q21.3 |
| L | 10q21.3 |
| L | 10q22.2 |
| L | 14q21.2 |
| L | 16p13.2 |
| ChRCC |
| L | 3p22.1 |
| L | 14q13.1 |
| L | 21q11.2-q21.1 |
| L | 21q11.12-22.2 |
| L | 20p12.1 |
The most frequent chromosomal losses (43%) in PRCC
were observed in 3p and 3q, followed by 29% of losses in 1p, 1q,
11q, 18q, 22p and 22q (Table I).
The highest frequencies of chromosomal gains in PRCC were noted in
20q (57%), 20p (43%), 8q (43%) and 12q (43%). Microamplifications
and microdeletions of <1 Mb were detected in six and nine
regions, respectively, in PRCC (Table
II). A total of four microdeletions in 10q12–22 were identified
(Table II), but the lesions did
not overlap each other, suggesting that common tumor suppressor
genes are not located in this lesion. A database search for SNPs in
microdeleted areas of 8q24.23 and 10q21.3 identified only one
candidate tumor suppressor gene (data not shown).
By contrast, loss of entire chromosomes was a
specific characteristic of the genetic alterations in ChRCC
(Tables I and III). The complete allelic loss of the
homologous chromosome 1, 2, 6, 10 or 17 was common in the 8
patients with ChRCC (Table III).
Furthermore, only one partial chromosomal loss and gain was
detected (Table I) and only five
microdeletions and no microamplifications were observed in ChRCC
(Table II) using high-density SNP
array. This result strongly suggests that loss of entire
chromosomes is the main event in ChRCC and partial and microgenetic
changes rarely contribute to ChRCC carcinogenesis.
 | Table IIIThe common hemi-chromosomal losses
specifically observed in ChRCC. |
Table III
The common hemi-chromosomal losses
specifically observed in ChRCC.
| Chromosome | Pt.1 | Pt.2 | Pt.3 | Pt.4 | Pt.5 | Pt.6 | Pt.7 | Pt.8 |
|---|
| 1 | | | | | | | | |
| 2 | | | | | | | | |
| 3 | | | | | | | | |
| 4 | | | | | | | | |
| 5 | | | | | | | | |
| 6 | | | | | | | | |
| 7 | | | | | | | | |
| 8 | | | | | | | | |
| 9 | | | | | | | | |
| 10 | | | | | | | | |
| 11 | | | | | | | | |
| 12 | | | | | | | | |
| 13 | | | | | | | | |
| 14 | | | | | | | | |
| 15 | | | | | | | | |
| 16 | | | | | | | | |
| 17 | | | | | | | | |
| 18 | | | | | | | | |
| 19 | | | | | | | | |
| 20 | | | | | | | | |
| 21 | | | | | | | | |
| 22 | | | | | | | | |
| X | | | | | | | | |
| Y | | | | | | | | |
Copy-neutral LOH, which was undetectable in previous
LOH studies, was identified in a total of 37 chromosomal lesions
(32 complete and 5 partial losses) (Table I), which is a strong advantage in
this SNP array analysis. However, no significant correlations were
noted between the allelic changes and clinical factors, such as
stage, tumor grade and prognosis (data not shown).
Discussion
The identification of effective multiple tyrosine
kinase inhibitors, including sunitinib and sorafenib, is a crucial
development in the treatment of metastatic RCC (5). These drugs inactivate the VHL gene,
leading to an accumulation of HIF-1α, followed by the activation of
VEGF, PDGF and epidermal growth factors (5). However, the effects of these drugs on
PRCC and ChRCC are not well known, since the molecular differences
among the different histopathological subtypes of RCC have not been
intensively studied and recent trials have mostly been restricted
to ccRCC patients (12).
In this study, high-resolution SNP array analysis
was used to identify the detailed genomic alterations in the three
different histopathologic subtypes of RCC. The results demonstrated
that the genetic profiles of the three subtypes were inherently
different from each other, while 3p loss was the most frequent
change in ccRCC, as well as PRCC. This change suggests that the
effectiveness of multiple tyrosine kinase inhibitors should be
limited to ccRCC. Notably, the treatment effects of sunitinib and
sorafenib in a limited number of patients with PRCC and ChRCC
showed poorer clinical responses compared to ccRCC (12). The results of this clinical trial
were consistent with the genetic backgrounds identified in the
present study.
ccRCCs are characterized by the deletion of the
short arm of chromosome 3 (4,13),
loss of 6p, 8p, 9pq and 14q (14).
Yoshimoto et al previously analyzed chromosomal copy number
aberrations in RCC using array-based CGH, using a genome-wide
scanning array with 2304 BAC and PAC clones covering the entire
human genome at a resolution of approximately 1.3 Mb (7). In their analysis of 30 ccRCC samples,
these authors found losses of 3p25.1-p25.3 (77%), 3p21.31-p22.3
(81%), 3p14.1-p14.2 (77%), 8p23.3 (31%), 9q21.13-qter (19%) and
14q32.32-qter (38%), and gains of chromosomes 5q33.1-qter (58%),
7q11.22-q35 (35%) and 16p12.3-p13.12 (19%) (7). Recently, Chen et al (9) reported the SNP profiles of 80 patients
with ccRCC determined using Illumina's 307K SNP array. These
investigators reported that the most common LOH was 3p (69 cases),
followed by chromosome losses at 8p, 6q and 14q, while the most
frequent chromosome gains were at 5q (32 cases), including 10
entire 5q amplifications and 21 large amplifications. The results
were similar to those of the present study. However, Chen et
al only analyzed ccRCCs, and not PRCCs nor ChRCCs.
Few studies have used high-density whole-genomic
analysis in PRCC. In a study by Klatte et al (15) a cytogenetic analysis was performed
to distinguish between the tumor profiles of type I and type II
PRCCs. The authors found that loss of chromosome 1p and 3p, and
gain of 5q were exclusively observed in type II PRCC, whereas
trisomy 17 was more frequent in type I (15). Analysis of the genetic profiles of
the PRCC cases in the present study, which comprised six cases of
type II and one case of type I, were compatible with this previous
report, and the single case of type I PCRR showing trisomy 17
(Table I).
We analyzed the patterns of regional gain and loss
in the eight samples of ChRCC. The patterns of genomic alterations
noted in these ChRCCs differed from those in the ccRCCs and PRCCs.
Recurrent genomic losses were detected on chromosomes 1, 2, 6, 8,
10, 13, 17 and 21. Previous studies reported a loss of the entire
chromosome arm of chromosomes 1, 2, 6 or 10; and 1, 2, 6, 8, 10,
13, 17 or 21 in ChRCC, using conventional CGH (16) and array-based CGH (7), respectively. Our results confirmed
these findings for ChRCC using high-density analysis. We also
observed few partial chromosomal changes (Table I) or micro-genetic alterations
(<1 Mb) (Table III),
suggesting that genetic alterations in small regions, rather than
entire chromosomal changes, rarely occur in ChRCC. This entire
chromosomal loss may be an initial event in the carcinogenesis of
ChRCC.
Contamination with normal cells is occasionally a
source of error in the genetic analysis of tumor samples. Peiffer
et al addressed the effects of tumor heterogeneity and
mosaicism on the detection limits and showed that Illumina's SNP
array assay was able to detect LOH in tumor samples combined with
67% normal stroma (8). We confirmed
that the proportion of tumor cells in a tissue section was over
70%, and therefore postulate that this system provided a reliable
means of determining the detailed genetic profile of RCCs.
Acknowledgements
We acknowledge support from the following grants:
Health Sciences Research Grants for Clinical Research for Evidenced
Based Medicine and Grants-in-Aid for Cancer Research (016), from
the Ministry of Health, Labour and Welfare, Japan; Research
Promotion Grant from The Tokyo Biochemical Research Foundation,
Japan; Grant-in-Aid of Cancer Research from the Fukuoka Cancer
Society, Japan. We thank Noriko Hakoda and Seiko Kamori for their
technical assistance.
References
|
1
|
Thoenes W, Storkel S and Rumpelt HJ:
Histopathology and classification of renal cell tumors (adenomas,
oncocytomas and carcinomas). The basic cytological and
histopathological elements and their use for diagnostics. Pathol
Res Pract. 181:125–143. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Monzon FA, Hagenkord JM, Lyons-Weiler MA,
et al: Whole genome SNP arrays as a potential diagnostic tool for
the detection of characteristic chromosomal aberrations in renal
epithelial tumors. Mod Pathol. 21:599–608. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Herrmann E, Brinkmann OA, Bode ME, et al:
Histologic subtype of metastatic renal cell carcinoma predicts
response to combined immunochemotherapy with interleukin 2,
interferon alpha and 5-fluorouracil. Eur Urol. 51:1625–1631. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Clifford SC, Prowse AH, Affara NA, Buys CH
and Maher ER: Inactivation of the von Hippel-Lindau (VHL) tumour
suppressor gene and allelic losses at chromosome arm 3p in primary
renal cell carcinoma: evidence for a VHL-independent pathway in
clear cell renal tumourigenesis. Genes Chromosomes Cancer.
22:200–209. 1998. View Article : Google Scholar
|
|
5
|
Eto M and Naito S: Molecular targeting
therapy for renal cell carcinoma. Int J Clin Oncol. 11:209–213.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mulders P: Vascular endothelial growth
factor and mTOR pathways in renal cell carcinoma: differences and
synergies of two targeted mechanisms. BJU Int. 104:1585–1589. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yoshimoto T, Matsuura K, Karnan S, et al:
High-resolution analysis of DNA copy number alterations and gene
expression in renal clear cell carcinoma. J Pathol. 213:392–401.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Peiffer DA, Le JM, Steemers FJ, et al:
High-resolution genomic profiling of chromosomal aberrations using
Infinium whole-genome genotyping. Genome Res. 16:1136–1148. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen M, Ye Y, Yang H, et al: Genome-wide
profiling of chromosomal alterations in renal cell carcinoma using
high-density single nucleotide polymorphism arrays. Int J Cancer.
125:2342–2348. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yokomizo A, Mai M, Tindall DJ, et al:
Overexpression of the wild type p73 gene in human bladder cancer.
Oncogene. 18:1629–1633. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pfeifer D, Pantic M, Skatulla I, et al:
Genome-wide analysis of DNA copy number changes and LOH in CLL
using high-density SNP arrays. Blood. 109:1202–1210. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Choueiri TK, Plantade A, Elson P, et al:
Efficacy of sunitinib and sorafenib in metastatic papillary and
chromophobe renal cell carcinoma. J Clin Oncol. 26:127–131. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Higgins JP: Gene array studies in renal
neoplasia. Scientific World Journal. 6:502–511. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Thrash-Bingham CA, Salazar H, Freed JJ,
Greenberg RE and Tartof KD: Genomic alterations and instabilities
in renal cell carcinomas and their relationship to tumor pathology.
Cancer Res. 55:6189–6195. 1995.PubMed/NCBI
|
|
15
|
Klatte T, Pantuck AJ, Said JW, et al:
Cytogenetic and molecular tumor profiling for type 1 and type 2
papillary renal cell carcinoma. Clin Cancer Res. 15:1162–1169.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Speicher MR, Schoell B, du Manoir S, et
al: Specific loss of chromosomes 1, 2, 6, 10, 13, 17, and 21 in
chromophobe renal cell carcinomas revealed by comparative genomic
hybridization. Am J Pathol. 145:356–364. 1994.PubMed/NCBI
|