Introduction
Osteosarcoma (OS) in which the neoplastic osteoid is
produced by the proliferating spindle cell stroma of mesenchymal
derivation is the most common primary malignant bone tumor in
children and adolescents. A total of 10–15% of OS patients present
distant metastases at diagnosis (90% lung and 10% other locations)
(1). Despite significant clinical
improvements over the past few decades through the use of
combination chemotherapy and surgery, patients with metastasis or
who respond poorly to adjuvant chemotherapy continue to have an
extremely poor prognosis (2).
Therefore, the identification of specific molecular targets is
crucial in devising more targeted and appropriate therapeutic
strategies to improve patient survival.
Tumors have unlimited proliferative capacity, which
is closely linked to the maintenance of telomeres (3). The tumor cell prevents critical
telomere dysfunction by specific telomere maintenance mechanisms
(TMMs) referred to as telomerase activity (TA), alternative
lengthening of telomeres (ALT), and other unknown mechanisms used
to evade senescence and cell death (4). Similarly, OS cells need to maintain
telomeres stable by specific mechanisms. In this study, we
investigated the expression of two TMM-related components in human
OS cell lines, and showed the level of expression of mRNA and
proteins. Therefore, we may treat OS with molecular immunology
methods targeting ALT.
Materials and methods
Cell culture and antibodies
Human OS cell lines (MG-63, U2-OS and SAOS-2) and an
osteoblast cell line (hFOB 1.19) were obtained from the Type
Culture Collection of the Chinese Academy of Sciences (Shanghai,
China). U2-OS, MG-63 and SAOS-2 were cultured in RPMI-1640
(HyClone, USA) containing 10% heat-inactivated fetal bovine serum
(FBS; HyClone). hFOB 1.19 was cultured in DMEM/F12 (v/v) containing
10% heat-inactivated FBS and 0.3 mg/ml G418 (Sigma, St. Louis, MO,
USA). The OS cell lines were cultured at 37°C in a 5%
CO2 water-saturated atmosphere and hFOB 1.19 was
cultured at 34°C.
The primary antibodies used were: mouse monoclonal
anti-hTERT (ab5181, Abcam, Boston, MA, USA); mouse monoclonal
anti-PML (sc-966), anti-FEN1 (sc-28355), anti-MRE11 (sc-135992) and
anti-Rad52 (sc-365341) (all three purchased from Santa Cruz
Biotechnology, Santa Cruz, CA, USA); and mouse monoclonal
anti-β-actin (Fermentas, Burlington, Canada).
Whole-genome expression arrays
We used commercially available Illumina HumanWG-6
Expression BeadChips for whole genome expression analysis. Purified
cells were lysed in a TRIzol reagent (Invitrogen, Carlsbad, CA,
USA) following the manufacturer’s instructions using 1 ml TRIzol
reagent per 106 cells. Isolated total RNA was then
purified further using an Illumina Total Prep RNA amplification kit
(Ambion, Austin, TX, USA). Each purified RNA sample was assessed
for RNA concentration and purity (A260:280) using the Nano Drop
2000 Spectrophotometer. All information on RNA processing and
quality assessment is available. Total RNA (500 ng) was amplified
using the Illumina Total Prep RNA amplification kit according to
the manufacturer’s instructions (7). The biotinylated cRNA (1500 ng per
sample) was applied to Illumina HumanWG-6 v3 Expression BeadChips,
which provided whole genome transcriptomic coverage, and was
hybridized overnight at 58°C. Chips were washed and detected
according to the manufacturer’s instructions. The HumanWG-6 v3
Expression BeadChips were scanned on an Illumina BeadArray™ reader.
The BeadStudio software package included with the
Illumina® BeadStation 500GX system extracted gene
expression data from the images collected from the Illumina
BeadArray Reader.
Western blotting assays
Cell lines were washed with phosphate-buffered
saline (PBS) and lysed in lysis buffer (20 mM Tris-HCl, pH 7.4, 150
mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA, 50 μg/ml leupeptin, 30 μg/ml
aprotinin and 1 mM PMSF). Following centrifugation at 15,000 × g at
4°C for 10 min, the supernatants were separated on sodium dodecyl
sulfate polyacrylamide (SDS-PAGE) gel and transferred onto a
nitrocellulose membrane (Millipore, Billerica, MA, USA) in the
standard transfer buffer. After being blocked with 5% skim milk in
blocking solution (Tris-buffered saline containing 0.1% Tween-20,
TBST) at room temperature, the membrane was incubated with primary
antibodies diluted in blocking solution overnight at 4°C. The
antibodies and dilutions used were: anti-hTERT (1:1500), anti-PML
(1:200), anti-FEN-1 (1:200), anti-MRE11 (1:100), anti-Rad52 (1:200)
and anti-β-actin (1:1000). Following washing in TBST 3 times, the
secondary antibodies (1:3000–5000, Santa Cruz Biotechnology),
conjugated with HRP, were applied for 2 h at room temperature.
Following extensive washing in TBST, specific immuno-reactivity was
visualized using enhanced chemiluminescence (ECL) Western blotting
substrate (Pierce, Rockford, IL, USA) and the ECL system (Fusion
FX7, French). The relative levels were normalized to β-actin
expression.
Complementary DNA (cDNA) synthesis
TRIzol reagent (Invitrogen) was used to extract
total RNA from tissue samples and cell lines in accordance with the
manufacturer’s instructions. cDNA was synthesized using 0.5 μg of
total RNA and the FirstStrand synthesis kit (Fermentas). cDNA was
incubated for 60 min at 42°C, and the reaction was terminated by
heating at 70°C for 5 min.
Quantitative real-time PCR (qRT-PCR)
analysis
Relative mRNA expression was evaluated by qRT-PCR
performed with the Mx-3000P Real-time PCR system (Stratagene, La
Jolla, CA, USA) and real-time PCR kit (SYBR® Premix Ex
Taq™, Takara, Japan) according to the manufacturer’s instructions.
The primers (β-actin as the control gene) in qRT-PCR were
synthesized by Invitrogen. The reaction conditions were initial
denaturation at 95°C for 1 min followed by 40 cycles of
denaturation at 95°C for 15 sec, annealing at 56°C for 15 sec and
extension at 72°C for 45 sec. The final extension was performed at
72°C for 10 min. The total procedure was repeated nine times for
each reaction and the average value was calculated. In this
experiment, β-actin was used as the control gene. Levels of mRNA
expression were calculated based on the method of 2−ΔΔCT
(5).
Statistical analysis
Quantitative data were expressed as the mean ±
standard deviation (SD). P<0.05 was considered to be
statistically significant. Analysis was carried out using SPSS
13.0.
Results
Whole-genome expression arrays
The mRNA expression of the proteins related to the
maintenance of telomere length were listed (Table II) as reported in the literature.
Possible different genes including TRF1, TRF2, TERC, PML, RAD50,
MRE11A, FEN1 and FANCA were primarily identified, since there is a
2-fold difference in signal intensities (ratio >2 or <0.5 in
the Tables) between hFOB 1.19, which served as a control, and OS
cell lines such as MG-63, SAOS-2 and U2-OS.
 | Table IIExpression of different
telomere-associated proteins/mRNAs. |
Table II
Expression of different
telomere-associated proteins/mRNAs.
| mRNAs | hFOB 1.19 | MG-63 | U2-OS | SAOS-2 |
|---|
| Shelterin protein
complex |
| TRF1 | 6.4 | 15.8 | 7.4 | −2.3 |
| TRF2 | −16.8 | −3.2 | −10.1 | −6.1 |
| TPP1 | 15.4 | 94.3 | 174.1 | 138.5 |
| TINF2 | 2074.3 | 1963.7 | 2204.3 | 1826.5 |
| TERF2IP | 1485.7 | 1806 | 1974.9 | 2069 |
| POT1 | −10.1 | 21.2 | 43.7 | 34 |
|
Telomerase-associated proteins |
| TERC | 40.9 | 0.025 | 152.6 | 0.0013 |
| TERT | 11.4 | 0.9 | −0.8 | −0.1 |
| TEP1 | −7.6 | 5.8 | 11.6 | 13.7 |
| ALT-associated
proteins |
| PML | 18.8 | 23.7 | 45.6 | 30.7 |
| RAD50 | 74.6 | 143.5 | 192.7 | 203.1 |
| RAD51 | 200.4 | 193.6 | 151.3 | 125.1 |
| RAD52 | 20 | 37.7 | 14.5 | 33.6 |
| SMC5 | −14.8 | −12.7 | −17.5 | −23.8 |
| SMC6 | 117 | 74.5 | 35.5 | 58.5 |
| BLM | 459.7 | 396.1 | 388.6 | 532.2 |
| TOP3A | 630.3 | 730.2 | 846.4 | 783.9 |
| BLAP75 | 344.3 | 653.1 | 587.1 | 492.5 |
| MMS21 | 1788.4 | 3720.1 | 2370.4 | 1450.6 |
| MRE11A | 101 | 198.7 | 226.4 | 153.1 |
| FEN1 | 5856.5 | 8893 | 7145.3 | 6782.1 |
| MUS81 | 454.2 | 582.3 | 712.8 | 821.3 |
| FANCD2 | 436 | 433.7 | 324.6 | 398.2 |
| FANCA | 18.9 | 174.5 | 111.6 | 133.5 |
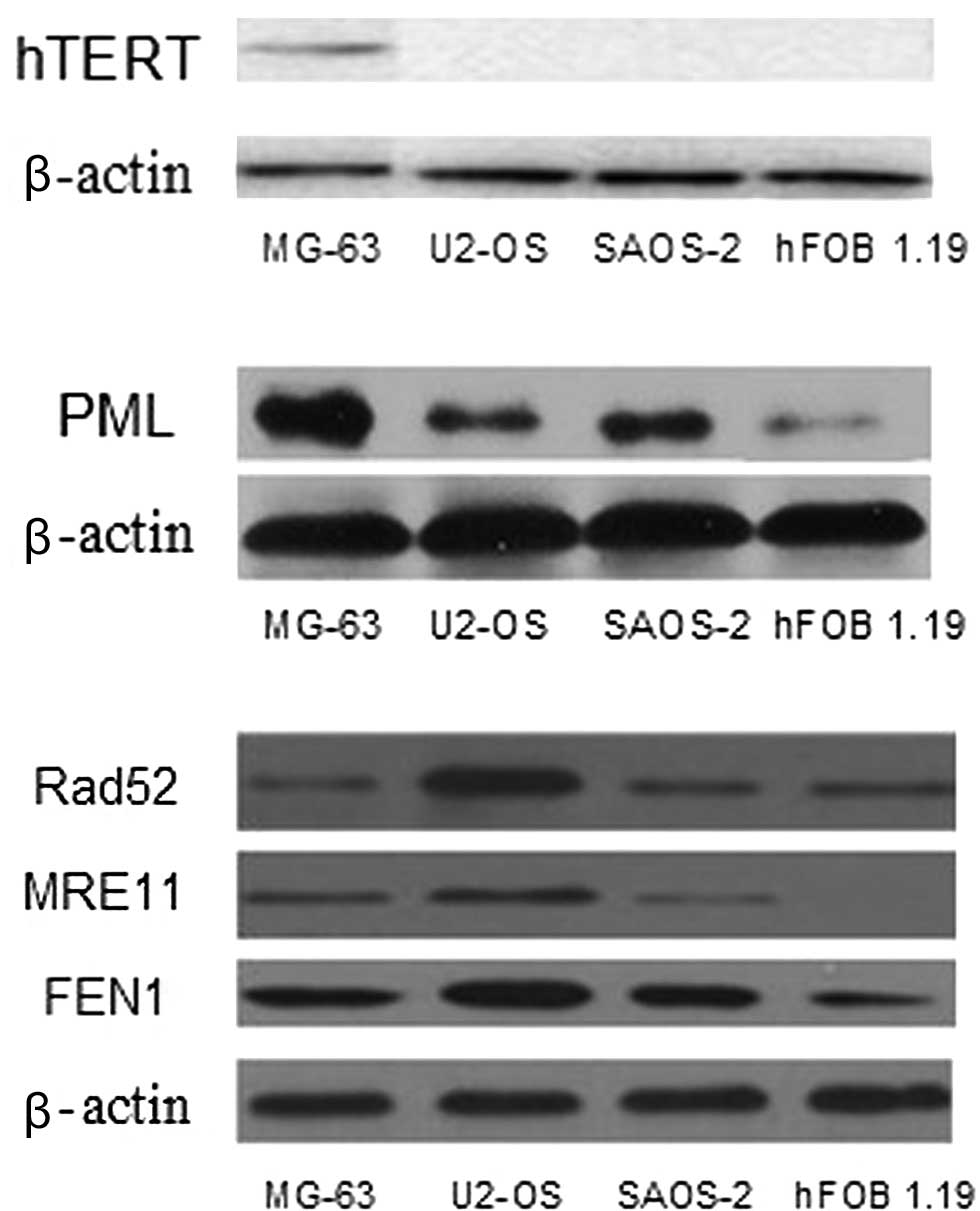
Western blot analysis
Western blot analysis shows that hTERT is expressed
in the MG-63 cell line, whereas it is negatively expressed in the
SAOS-2, U2-OS and hFOB 1.19 cell lines (Fig. 1A). Fig.
1B shows the varying levels of PML expression in all of the
cell lines, particularly in the MG-63 cell line, whereas PML is
less expressed in hFOB 1.19. Fig.
1C shows the different expression levels of Rad52, MRE11 and
FEN1 between the OS cell lines (MG-63, U2-OS and SAOS-2) and the
osteoblast cell line (hFOB 1.19).
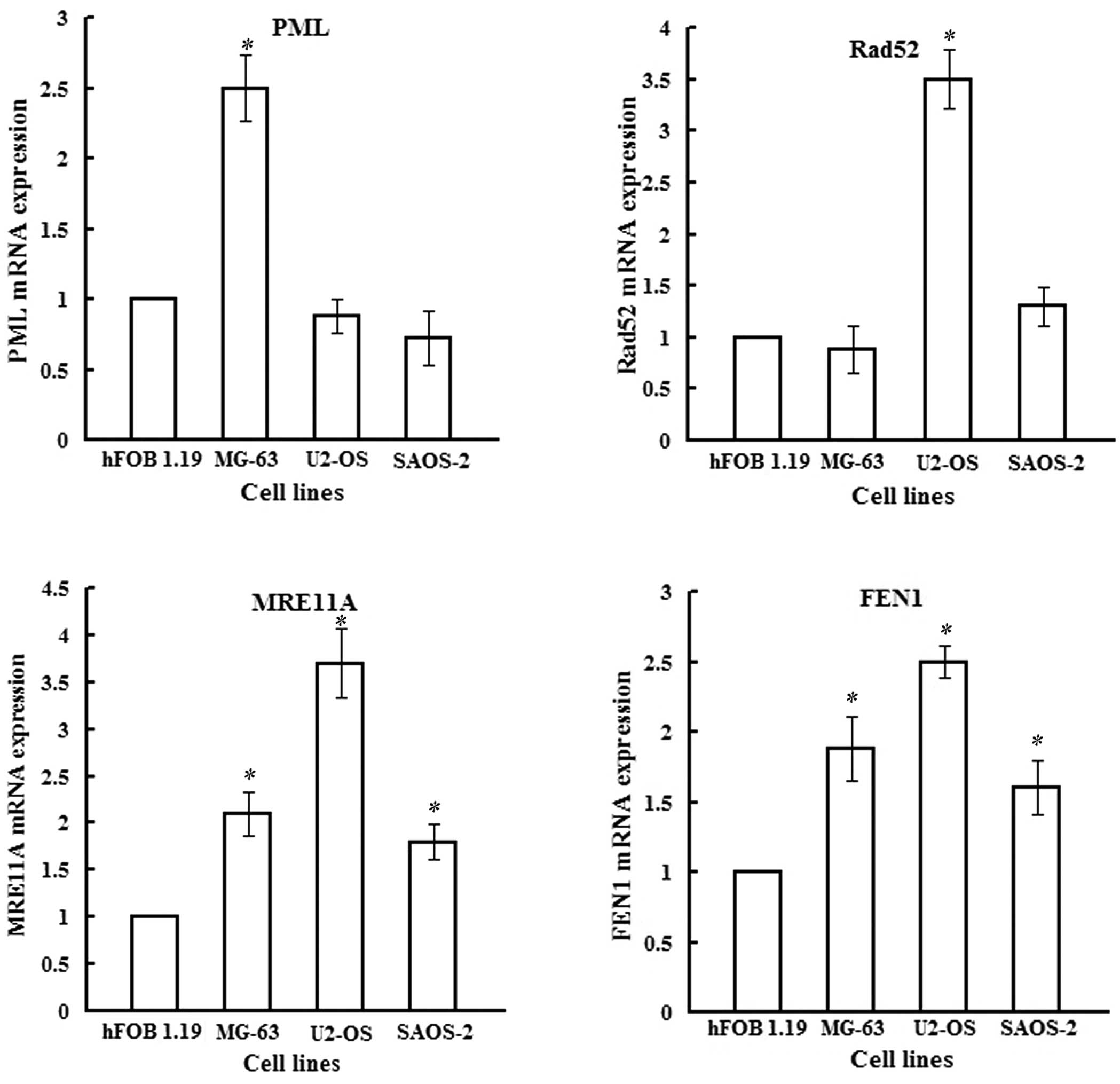
qRT-PCR analysis
The mRNA expression of hTERT was detected in the
MG-63 cell line, but was almost negative in U2-OS, SAOS-2 and hFOB
1.19. As for the mRNA level of PML, Fig. 2A shows varying levels of PML mRNA in
all four cell lines, particularly MG-63. The differences of PML,
MRE11A and FEN-1 between MG-63 and hFOB 1.19 achieved statistical
significance (P<0.05), as did Rad52, MRE11A and FEN-1 in U2-OS,
and MRE11A and FEN-1 in SAOS-2, compared to hFOB 1.19.
Discussion
Telomeres prevent chromosome ends from inducing a
DNA damage response and end-to-end fusions, which result in
chromosome breakage and recombination (7–9).
Telomeres shorten by 50–200 bp with each cell division, leading to
cell senescence and eventually death, as their ends cannot be
completely replicated by the DNA-replication machinery (the
end-replication problem) (10).
There are a number of adjuvant structures that
protect the telomere DNA, which is mostly double-stranded and has a
single-stranded terminus with 130–210 bases on average in human
cells (11). The telomeric DNA is
bound by the shelterin protein complex, which includes six subunit
proteins as follows: telomeric repeat-binding factor 1 (TRF1 or
TERF1), telomeric repeat-binding factor 2 (TRF2 or TERF2),
protection of telomeres 1 (POT1), tripeptidyl peptidase I (TPP1 or
PIP1, TINT1), TRF1-interacting nuclear protein 2 (TIN2 or TINF2)
and TRF2-interacting telomeric protein (RAP1 or TERF2IP). This
nucleoprotein complex prevents the chromosome end from being folded
as a DNA double-strand break (DSB) (12). In the present study, TRF1, TRF2,
TPP1 and POT1 were differentially expressed between OS and
osteoblast cell lines (Table II),
indicating that they may play a significant role in OS;
particularly TPP1 and POT1, which revealed insignificant
differences in the relative level of mRNAs compared to hFOB
1.19.
In humans, almost 85% of carcinomas maintain their
telomeres with telomerase, which synthesizes new telomeric DNA to
repair the ends of the chromosomes, whereas in normal tissues and
cells expression is almost negative. Thus, telomerase is a classic
target for developing an anti-tumor therapy (13). However, in 10–15% of tumors, DNA
replication is achieved through a mechanism known as ALT, which is
dependent on homologous recombination or other molecular
mechanisms. Therefore, ALT may be a significant new target for
tumor therapy. Tumors maintain the extreme heterogeneity of
telomere lengths using distinct nuclear structures called
ALT-associated promyelocytic leukemia (PML) bodies (APBs) (14). The PML protein is a zinc finger
transcription factor expressed as three major transcription
products due to alternative splicing. PML was first found in acute
promyelocytic leukemia (APL) and plays a significant role in genome
stability (15).
Besides the hTERT and PML, there are a number of
other related protein components of these two TMMs, which are
essential for telomere elongation or telomere loss prevention. As
is well known, human telomerase is composed of an RNA component
(hTR), a catalytic protein subunit (hTERT) and the
telomerase-associated protein (TEP1) (16,17).
However, hTR has a lack of specificity as it is widely expressed in
a number of tissues, even in those tissues without telomerase
activity (TA) (18,19). Few studies exist regarding TEP1
compared with the other two components of telomerase. Certain
findings indicate that the reverse transcriptase domain of hTERT
interacts with TEP1 (20). TEP1
also binds a small RNA (vault RNA, vRNA) within the cytoplasmic
vault complex (21). Inhibition of
TEP1 leads to greatly reduced vRNA levels, as well as a loss of
TEP1 and vRNA from the vault caps, but no reduction in TA or
telomere length (22,23). These results suggest that TEP1 is a
multifunctional RNA-binding protein and may not be essential for TA
and telomere length. Results of the present study have shown that
hTR (also known as TERC) and TEP1 were differentially expressed
between the two cell lines, whereas hTERT was negative in the
normal and OS cell lines. Therefore, to identify whether TEP1 plays
a physiological role in telomerase mechanisms, further experiments
should be conducted.
Findings of previous studies have shown that 31–87%
of OS tissues use the TA mechanism to maintain their telomere
length (24–27). On the other hand, numerous reports
in the available literature describe the absence of telomerase
activity or hTERT mRNA/protein expression in a number of OS cell
lines. The different extracellular matrix and detection methods
used may result in this diversity (28,29).
In OS, hTERT is a predictive indicator of worse prognosis, with a
trend in favor of shorter progression-free survival in patients
whose tumors expressed telomerase, and promotes the invasion
ability of telomerase-negative tumor cells in vitro
(28,30). Due to the different expression
between normal and malignant tissue, hTERT has been considered as a
significant potential target for tumor therapeutics. Therefore,
various telomerase inhibition therapies are currently in progress
including a lipid-modified thio-phosphoramidate oligonucleotide
(GRN163), which is the furthest along in clinical development, a
derivative of benzoic acid (BIBR1532) and a bisphosphonate
(31,32). However, there may be a number of
adverse effects of telomerase inhibition therapy on patients,
particularly in growing children, as certain normal cells express
telomerase, including hematopoietic stem, germ, immune and other
progenitor cells. Therefore, more investigations regarding the role
of telomerase are required.
Findings of certain studies have shown that TA and
ALT may elongate the telomere by various mechanisms, and even
coexist in the same immortal cells by transfection (33). Such findings suggest a significant
anti-tumor target, particularly for tumors that maintain the
telomere stability by ALT. Moreover, one hallmark of the ALT
mechanism is the presence of APBs, which contain PML bodies and
some telomere-related components.
The ALT mechanism is reportedly rare in epithelial
tumors but more common in tumors of neuroepithelial (e.g.,
astrocytoma) or mesenchymal origin (e.g. OS and liposarcoma).
However, although certain hypotheses have been put forward to
explain it, the basis of this tissue specificity has yet to be
elucidated (34).
Numerous proteins have been identified in APBs that
may be involved in ALT mechanisms, such as POT1, TRF1, TRF2, as
well as other proteins involved in homologous recombination repair,
including DNA repair protein RAD50, RAD51, RAD52, the structural
maintenance of chromosomes SMC5-SMC6 complex and the MRN complex
(35), and protein complexes that
include the BLM helicase, topoisomerase 3α (TOP3A) and BLAP75
(BLM-associated polypeptide 75) (36,37).
The (SMC5)-SMC6 complex, which is involved in telomere elongation,
is composed of SMC5, SMC6, and methyl methanesulfonate-sensitivity
21 (MMS21). MMS21 may sumoylate TRF1, TRF2, TIN2 and RAP1 to
elongate telomeres based on SMC5 and SMC6. The MRN complex, which
is composed of meiotic recombination 11 (MRe11), Rad50 and Nijmegen
breakage syndrome protein (NBS1), is the first protein complex to
be identified as necessary for ALT-mediated telomere maintenance
(38).
MRe11 is a nuclear 3′–5′ exonuclease/endonuclease
that associates with Rad50 and affects homologous recombination,
telomere maintenance and DNA double-strand break repair. MRe11 is
not detected in osteoblast cell lines by Western blot analysis.
Rad52, which interacts with Rad51, forms a heptameric ring that
binds single-stranded DNA ends and catalyzes the DNA-DNA
interaction necessary for the annealing of complementary strands
(39,40). Recent studies have demonstrated that
other proteins, including flap endonuclease 1 (FEN1), MUS81, the
Fanconi anaemia group D2 (FANCD2) and Fanconi anaemia group A
(FANCA) are also significant for ALT mechanisms (41–43).
These proteins are significant in telomere maintenance, as shown by
previous studies (44,45). This difference in Rad52, MRe11 and
FEN1 protein levels indicates that these cell lines are dependent
on ALT to different extents. MRN, MUS81, FEN1 and TOP3A all bind
TRF2, suggesting that reducing relative TRF2 saturation limits
control over these proteins at chromosomal telomeres (36).
In this study, the expression of hTERT, PML, Rad52,
MRe11 and FEN1 at mRNA and protein levels show the difference
between the OS cell lines and the osteoblast cell line. On the
other hand, other related proteins including RAD50, BLAP75, MRE11A,
FEN1, MUS81 and FANCA are expressed more in OS cell lines than in
hFOB 1.19 in whole-genome expression arrays. RAD50 and MRe11A have
been shown to play a role in telomere elongation, whereas BLAP75,
FEN1, MUS81 and FANCA are considered to be significant proteins
that prevent telomere loss in an ALT mechanism. Therefore, we
suggest that there are more proteins in OS cell lines that prevent
telomere loss rather than telomere elongation, and these may become
significant therapeutic targets in the treatment of OS in the
future.
In conclusion, OS cell lines maintain their telomere
length primarily through the ALT mechanism. A number of other
proteins regulate this process in OS. Therefore, anti-ALT therapy
may be a significant method used treat OS, which requires in-depth
study of the ALT mechanism.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (no. 30772185), the Fundamental
Research Funds for the Central Universities (no. 303275884,
201130302020010), Hubei Provincial Natural Science Foundation (no.
2009CDB288) and the funds of Zhongnan Hospital, Wuhan Universities
(no. 2009–22).
References
|
1
|
Arndt CA and Crist WM: Common
musculoskeletal tumors of childhood and adolescence. N Engl J Med.
341:342–352. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Endo-Munoz L, Cumming A, Sommerville S,
Dickinson I and Saunders NA: Osteosarcoma is characterised by
reduced expression of markers of osteoclastogenesis and antigen
presentation compared with normal bone. Br J Cancer. 103:73–81.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tabori U and Dome JS: Telomere biology of
pediatric cancer. Cancer Invest. 25:197–208. 2007. View Article : Google Scholar
|
|
4
|
Brachner A, Sasgary S, Pirker C, et al:
Telomerase- and alternative telomere lengthening-independent
telomere stabilization in a metastasis-derived human non-small cell
lung cancer cell line: effect of ectopic hTERT. Cancer Res.
66:3584–3592. 2006. View Article : Google Scholar
|
|
5
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Smith CC, Aylott MC, Fisher KJ, Lynch AM
and Gooderham NJ: DNA damage responses after exposure to DNA-based
products. J Gene Med. 8:175–85. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Smogorzewska A, Karlseder J,
Holtgreve-Grez H, Jauch A and de Lange T: DNA ligase IV-dependent
NHEJ of deprotected mammalian telomeres in G1 and G2. Curr Biol.
12:1635–1644. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
D’Adda di Fagagna F, Reaper PM,
Clay-Farrace L, et al: A DNA damage checkpoint response in
telomere-initiated senescence. Nature. 426:194–198. 2003.PubMed/NCBI
|
|
9
|
Bakkenist CJ, Drissi R, Wu J, Kastan MB
and Dome JS: Disappearance of the telomere dysfunction-induced
stress response in fully senescent cells. Cancer Res. 64:3748–3752.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shay JW and Wright WE: Hayflick, his
limit, and cellular ageing. Nat Rev Mol Cell Biol. 1:72–76. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Makarov VL, Hirose Y and Langmore JP: Long
G tails at both ends of human chromosomes suggest a C strand
degradation mechanism for telomere shortening. Cell. 88:657–666.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Palm W and de Lange T: How shelterin
protects mammalian telomeres. Annu Rev Genet. 42:301–334. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Artandi SE and DePinho RA: Telomeres and
telomerase in cancer. Carcinogenesis. 31:9–18. 2010. View Article : Google Scholar
|
|
14
|
Weinrich SL, Pruzan R, Ma L, et al:
Reconstitution of human telomerase with the template RNA component
hTR and the catalytic protein subunit hTRT. Nat Genet. 17:498–502.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Henson JD, Neumann AA, Yeager TR and
Reddel RR: Alternative lengthening of telomeres in mammalian cells.
Oncogene. 21:598–610. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu ZX, Zou WX, Lin P and Chang KS: A role
for PML3 in centrosome duplication and genome stability. Mol Cell.
17:721–732. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nakayama J, Tahara H, Tahara E, et al:
Telomerase activation by hTRT in human normal fibroblasts and
hepatocellular carcinomas. Nat Genet. 18:65–68. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Blasco MA, Funk W, Villeponteau B and
Greider CW: Functional characterization and developmental
regulation of mouse telomerase RNA. Science. 269:1267–1270. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Avilion AA, Piatyszek MA, Gupta J, Shay
JW, Bacchetti S and Greider CW: Human telomerase RNA and telomerase
activity in immortal cell lines and tumor tissues. Cancer Res.
56:645–650. 1996.PubMed/NCBI
|
|
20
|
Beattie TL, Zhou W, Robinson MO and
Harrington L: Polymerization defects within human telomerase are
distinct from telomerase RNA and TEP1 binding. Mol Biol Cell.
11:3329–3340. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kickhoefer VA, Stephen AG, Harrington L,
Robinson MO and Rome LH: Vaults and telomerase share a common
subunit, TEP1. J Biol Chem. 274:32712–32717. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kickhoefer VA, Liu Y, Kong LB, et al: The
telomerase/vault-associated protein TEP1 is required for vault RNA
stability and its association with the vault particle. J Cell Biol.
152:157–164. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu Y, Snow BE, Hande MP, et al:
Telomerase-associated protein TEP1 is not essential for telomerase
activity or telomere length maintenance in vivo. Mol Cell Biol.
20:8178–8184. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ulaner GA, Huang HY, Otero J, et al:
Absence of a telomere maintenance mechanism as a favorable
prognostic factor in patients with osteosarcoma. Cancer Res.
63:1759–1763. 2003.PubMed/NCBI
|
|
25
|
Terasaki T, Kyo S and Takakura M: Analysis
of telomerase activity and telomere length in bone and soft tissue
tumors. Oncol Rep. 11:1307–1311. 2004.PubMed/NCBI
|
|
26
|
Fujiwara-Akita H, Maesawa C, Honda T,
Kobayashi S and Masuda T: Expression of human telomerase reverse
transcriptase splice variants is well correlated with low
telomerase activity in osteosarcoma cell lines. Int J Oncol.
26:1009–1016. 2005.PubMed/NCBI
|
|
27
|
Sotillo-Piñeiro E, Sierrasesúmaga L and
Patiñno-García A: Telomerase activity and telomere length in
primary and metastatic tumors from pediatric bone cancer patients.
Pediatr Res. 55:231–235. 2004.PubMed/NCBI
|
|
28
|
Yu ST, Chen L, Wang HJ, Tang XD, Fang DC
and Yang SM: hTERT promotes the invasion of telomerase-negative
tumor cells in vitro. Int J Oncol. 35:329–336.
2009.PubMed/NCBI
|
|
29
|
Jegou T, Chung I, Heuvelman G, et al:
Dynamics of telomeres and promyelocytic leukemia nuclear bodies in
a telomerase-negative human cell line. Mol Biol Cell. 20:2070–2082.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sanders RP, Drissi R, Billups CA, Daw NC,
Valentine MB and Dome JS: Telomerase expression predicts
unfavorable outcome in osteosarcoma. J Clin Oncol. 22:3790–3797.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Herbert BS, Gellert GC, Hochreiter A, et
al: Lipid modification of GRN163, an N3′→P5′ thio-phosphoramidate
oligonucleotide, enhances the potency of telomerase inhibition.
Oncogene. 24:5262–5268. 2005.PubMed/NCBI
|
|
32
|
Pascolo E, Wenz C, Lingner J, et al:
Mechanism of human telomerase inhibition by BIBR1532, a synthetic,
non-nucleosidic drug candidate. J Biol Chem. 277:15566–15572. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Perrem K, Colgin LM, Neumann AA, Yeager TR
and Reddel RR: Coexistence of alternative lengthening of telomeres
and telomerase in hTERT-transfected GM847 cells. Mol Cell Biol.
21:3862–3875. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Costa A, Daidone MG, Daprai L, et al:
Telomere maintenance mechanisms in liposarcomas: association with
histologic subtypes and disease progression. Cancer Res.
66:8918–8924. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bhattacharyya S, Sandy A and Groden J:
Unwinding protein complexes in ALTernative telomere maintenance. J
Cell Biochem. 109:7–15. 2010.PubMed/NCBI
|
|
36
|
Mankouri HW and Hickson ID: The RecQ
helicase-topoisomerase III-Rmi1 complex: a DNA structure-specific
‘dissolvasome’? Trends Biochem Sci. 32:538–546. 2007.PubMed/NCBI
|
|
37
|
Raynard S, Zhao W, Bussen W, et al:
Functional role of BLAP75 in BLM-topoisomerase IIIalpha-dependent
holliday junction processing. J Biol Chem. 283:15701–15708. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jiang WQ, Zhong ZH, Henson JD, Neumann AA,
Chang AC and Reddel RR: Suppression of alternative lengthening of
telomeres by Sp100-mediated sequestration of the MRE11/RAD50/NBS1
complex. Mol Cell Biol. 25:2708–2721. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sugawara N, Wang X and Haber JE: In vivo
roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated
recombination. Mol Cell. 2:209–219. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Miyazaki T, Bressan DA, Shinohara M, Haber
JE and Shinohara A: In vivo assembly and disassembly of Rad51 and
Rad52 complexes during double-strand break repair. EMBO J.
3:939–949. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Saharia A and Stewart SA: FEN1 contributes
to telomere stability in ALT-positive tumor cells. Oncogene.
28:1162–1167. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zeng S, Xiang T, Pandita TK, et al:
Telomere recombination requires the MUS81 endonuclease. Nat Cell
Biol. 11:616–623. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fan Q, Zhang F, Barrett B, Ren K and
Andreassen PR: A role for monoubiquitinated FANCD2 at telomeres in
ALT cells. Nucleic Acids Res. 37:1740–1754. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wu G, Jiang X, Lee WH and Chen PL:
Assembly of functional ALT-associated promyelocytic leukemia bodies
requires Nijmegen Breakage Syndrome 1. Cancer Res. 63:2589–2595.
2003.PubMed/NCBI
|
|
45
|
Henson JD, Cao Y, Huschtscha LI, et al:
DNA C-circles are specific and quantifiable markers of
alternative-lengthening-of-telomeres activity. Nat Biotechnol.
27:1181–1185. 2009. View
Article : Google Scholar : PubMed/NCBI
|