Introduction
Gastric adenocarcinoma is one of the most frequent
malignant tumors, with a high worldwide mortality. The clinical
outcomes remain unsatisfactory, which is mainly due to a poor
understanding of the mechanism of gastric cancer development and a
lack of specific target gene therapy. To improve the survival rate,
the development of novel treatments against molecular targets is
crucial. Multiple genetic and epigenetic alterations are known to
be involved in the tumorigenesis and progression of gastric cancer.
Since Weinberg and Barbacid first cloned the C-Ha-Ras from human
urinary bladder cancer in 1982, investigations have been conducted
into the role of Ras in various malignant tumors (1–3). The
Ras protein activator like-1 (RASAL1) gene was previously
identified (4). The protein,
encoded by the RASAL1 gene, is a member of the GTPase activating
proteins (GAPs) family. It enhances the intrinsic GTPase activity
of Ras proteins, resulting in the inactive GDP-bound form of Ras,
thereby allowing control of cell proliferation and differentiation.
The RASAL1 gene is reportedly correlated with the formation and
development of colon cancer (5–6). In
this study, we examined RASAL1 expression in vitro and in
vivo and its clinicopathological significance in gastric
adenocarcinoma.
Materials and methods
Clinical cases
Patients and clinical tissue
specimens
A total of 50 patients diagnosed with primary
gastric adenocarcinoma, who underwent surgically partial or total
gastrectomy between August 2009 and March 2010 in the Affiliated
Zhongda Hospital of the Southeast University (Nanjing, China), with
available clinical information, were included in the study. No
patients received chemotherapy or radiotherapy prior to surgery.
The clinical stages and pathological features were defined
according to the TNM Cancer Staging System of the American Joint
Committee on Cancer. Paired primary gastric cancer and adjacent
normal tissues were collected. The specimens were formalin-fixed,
paraffin-embedded and cut into 4-μm sections, which were stained
with hematoxylin and eosin for histopathological type,
differentiation stage and immunohistochemical evaluation. Written
informed consent was obtained from all patients. The study was
approved by the ethics committee of Zhongda Hospital, Southeast
University.
Immunohistochemical analysis
Immunohistochemistry was used to detect the
expression of RASAL1 in the specimens using a SP kit (Beijing
Zhongshan Goldenbridge Biotechnology Company, China) according to
the manufacturer’s instructions. The working anti-human rabbit
RASAL1 polyclonal antibody (Abcam, Cambridge, UK) was diluted at
1:200. The results were judged by two observers independently.
RASAL1 expression was determined by assessing the percentage and
intensity of stained tumor cells. The percentages of positive cells
(percentage scores) were recorded as: <5% (score 0), 6–25%
(score 1), 26–50% (score 2) and >51% (score 3). The staining
intensities (intensity scores) were classified as: no staining
(score 0), light brown staining (score 1), brown staining (score 2)
and dark brown staining (score 3). RASAL1 staining positivity was
calculated using the formula: overall score = percentage score ×
intensity score. An overall score of <1, 2–3, 4–6 and >6 was
defined as negative (−), weak positive (+), moderate positive (++)
and strong positive (+++), respectively. For negative controls,
sections were processed as above but treated with 0.01 mol/l
phosphate-buffered saline instead of primary antibodies.
Experimental studies
Cell lines
The well-differentiated gastric adenocarcinoma cell
MKN-28, the moderately differentiated gastric adenocarcinoma cell
SGC-7901 and the poorly differentiated gastric adenocarcinoma cell
BGC-823 were obtained from the Shanghai Institute of Biochemistry
and Cell Biology, China. The immortalized normal gastric epithelial
cell line GES-l was obtained from the Shanghai Institute of
Digestive Disease, China. The cell lines were cultured and
maintained in RPMI-1640 media and supplemented with 10% fetal
bovine serum, penicillin and streptomycin in a humidified cell
incubator with an atmosphere of 5% CO2 at 37°C.
Evaluation of RASAL1 mRNA in human
gastric cancer cell lines
Total RNA was extracted from 2×105 cells
(MKN-28, SGC-7901, BGC-823 and GES-l) by TRIzol reagent
(Invitrogen, CA, USA). RNA (1 μg) was converted into cDNA using the
reverse transcription system with oligo-dT (Promega, Madison, WI,
USA). PCR was carried out for RASAL1 using the primers:
5′-TGGATTTCTCTTCTTGCGATTCT-3′ (forward) and
5′-TGTTGTCCCGAAGGTCAA-3′ (reverse) (5). With β-actin acting as an internal
control, the primers used were: 5′-TGCTATCCCTGTACGCCTCT-3′
(forward) and 5′-AGTACTTGCGCTCAGGAGGA-3′ (reverse) (7). The PCR conditions were 94°C for 2 min,
94°C for 30 sec, 55°C for 30 sec and 72°C for 1 min for 32 cycles.
PCR products were separated by 2% agarose gel electrophoresis,
stained with ethidium bromide (EB) and visualized using the
ImageMaster VDS system (GE Healthcare, UK). Electrophoresis strips
were analyzed by TotalLab 2.0 software (Nonlinear Dynamics Ltd).
The ratio of the integrated density values (IDVs) for the RASAL1
transcripts to those for the β-actin transcripts was calculated.
Subsequently, a comparison was made of the RASAL1 mRNA levels for
the gastric cancer and the normal gastric epithelial cell lines.
The experiments were repeated three times to verify the results and
the mean value for the RASAL1 mRNA expression was used for
subsequent analysis.
Western blotting analysis
Western blotting analysis was performed to detect
RASAL1 protein expression in the cell lines (MKN-28, SGC-7901,
BGC-823 and GES-l) according to standard protocol. Cells were lysed
in Mammalian Protein Extraction Reagent (M-PER) (Pierce, Rockford,
IL, USA) containing a cocktail of proteinase inhibitors (Bio-Rad,
Hercules, CA, USA). The lysed proteins were quantified by a
bicinchoninic acid protein assay kit. Subsequently, equal amounts
of proteins were electrophoresed on 10% SDS-polyacrylamide gels
(SDS-PAGE) and then electroblotted onto nitrocellulose (Bio-Rad).
The anti-human goat RASAL1 polyclonal antibody (Abcam) was used at
a 1:200 dilution. The signal was visualized with an alkaline
horseradish peroxidase-conjugated rabbit anti-goat antibody
(1:5000, Jingmei, Biotech, Beijing, China) and an enhanced
chemiluminescence detection system (Amersham Pharmacia Biotech,
Freiburg, Germany). The experiments were repeated three times to
verify the results and the mean value was used for subsequent
analysis.
Statistical analysis
Results were shown as the mean ± standard deviation
(SD). Statistical analysis was performed using SPSS 16.0 software.
A Chi-square test, t-test and rank sum test were used. P<0.05
was considered to indicate a statistically significant
difference.
Results
Clinical cases
Expression patterns of RASAL1 by
immunohistochemistry in gastric cancer tissues
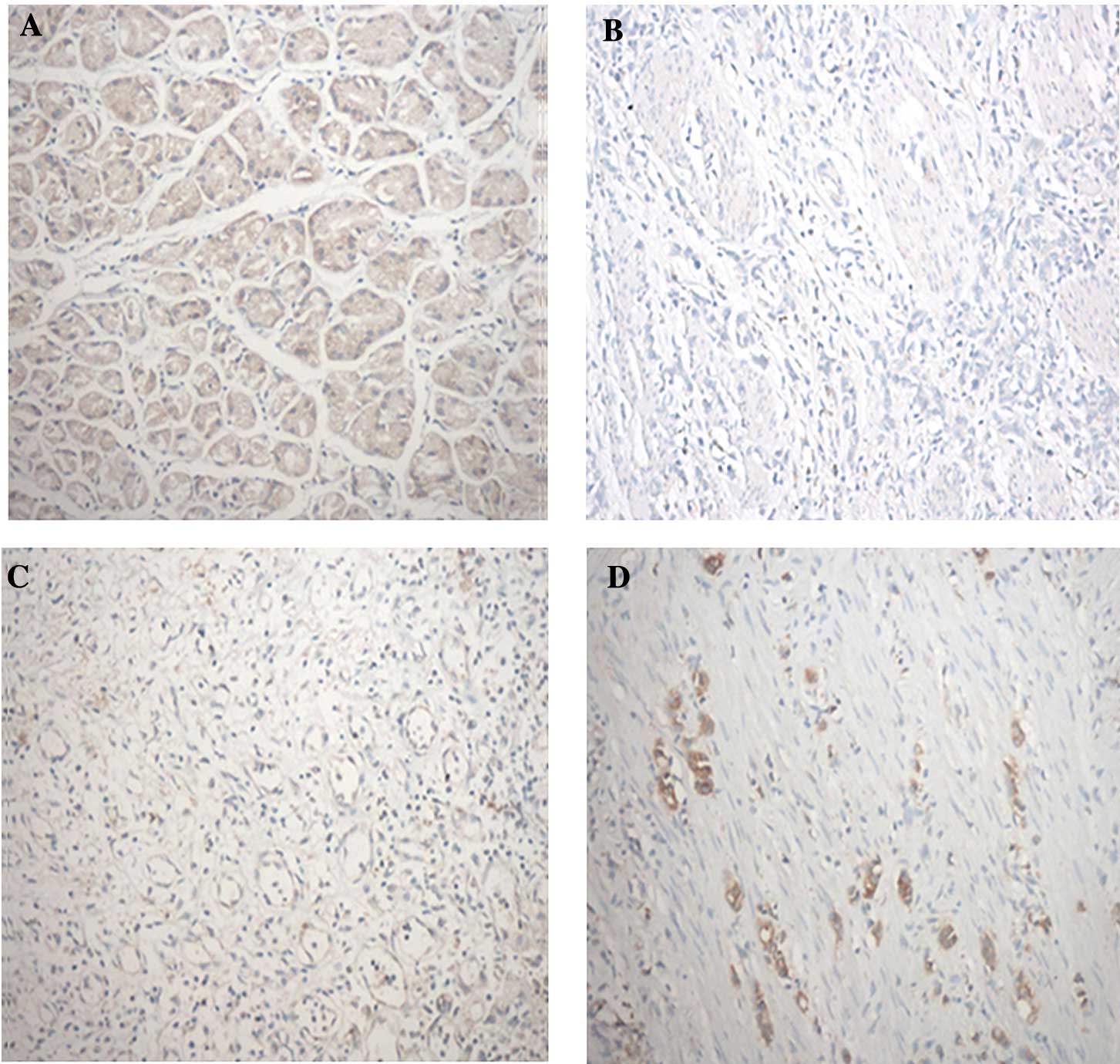
The RASAL1 protein status of 50 gastric carcinoma
and paired adjacent normal tissue samples were determined by
immunohistochemical staining (Fig.
1, Table I). The expression of
the RASAL1 protein was mainly observed in the cytoplasm. The
results showed that the RASAL1 protein expression was (−) in 12
cases, (+) in 23 cases, (++) in 13 cases and (+++) in 2 cases in
gastric carcinoma tissues and (++) in 16 cases and (+++) in 34
cases in adjacent normal tissues. The results indicated that RASAL1
protein expression is significantly reduced in gastric carcinoma
compared to adjacent normal tissues (p<0.05).
 | Table IRASAL1 protein in gastric carcinoma
and normal tissues. |
Table I
RASAL1 protein in gastric carcinoma
and normal tissues.
| | RASAL1 expression n
(%) |
|---|
| |
|
|---|
| Tissue | No. of cases (n) | − | + | ++ | +++ |
|---|
| Carcinoma | 50 | 12 (24.0) | 23 (46.0) | 13 (26.0) | 2 (4.0) |
| Normal | 50 | 0 (0.0) | 0 (0.0) | 16 (32.0) | 34 (68.0) |
Clinicopathological characteristics of
RASAL1 protein expression in gastric carcinoma
The correlation between clinicopathological
characteristics of gastric carcinoma tissue samples and the RASAL1
protein expression was examined (Table
II). This analysis revealed that the expression of the RASAL1
protein was decreased in gastric adenocarcinoma tissue compared to
normal gastric tissue (p<0.01). In gastric carcinoma tissues,
the reduced RASAL1 expression was associated with tumor size
(p=0.001), differentiation degree (p=0.001), invasion depth
(p=0.035) and lymph node metastasis (p=0.001), but not with age and
gender.
 | Table IICorrelation between RASAL1 protein and
clinicopathological characteristics in gastric carcinoma. |
Table II
Correlation between RASAL1 protein and
clinicopathological characteristics in gastric carcinoma.
| | RASAL1 expression
(n) | |
|---|
| |
| |
|---|
| Group | No. of cases (n) | − | + | ++ | +++ | p-value |
|---|
| Gender | | | | | | 0.370 |
| Male | 34 | 8 | 17 | 9 | 0 | |
| Female | 16 | 4 | 6 | 4 | 2 | |
| Age (yr) | | | | | | 0.554 |
| <60 | 19 | 5 | 7 | 6 | 1 | |
| ≥60 | 31 | 7 | 16 | 7 | 1 | |
| Tumor size (cm) | | | | | | 0.001 |
| <4 | 22 | 1 | 10 | 9 | 2 | |
| ≥4 | 28 | 11 | 13 | 4 | 0 | |
| Differentiation | | | | | | 0.001 |
| Well,
moderately | 21 | 0 | 8 | 11 | 2 | |
| Poorly | 29 | 12 | 15 | 2 | 0 | |
| Invasion
deptha | | | | | | 0.035 |
| m, sm | 10 | 1 | 4 | 3 | 2 | |
| mp or deeper | 40 | 11 | 19 | 10 | 0 | |
| Lymph node
metastasis | | | | | | 0.001 |
| Negative | 13 | 0 | 4 | 8 | 1 | |
| Positive | 37 | 12 | 19 | 5 | 1 | |
| TNM stage | | | | | | 0.034 |
| 1–2 | 16 | 1 | 5 | 10 | 0 | |
| 3–4 | 34 | 11 | 18 | 3 | 2 | |
Experimental studies
Expression of RASAL1 mRNA in human
gastric cancer cell lines
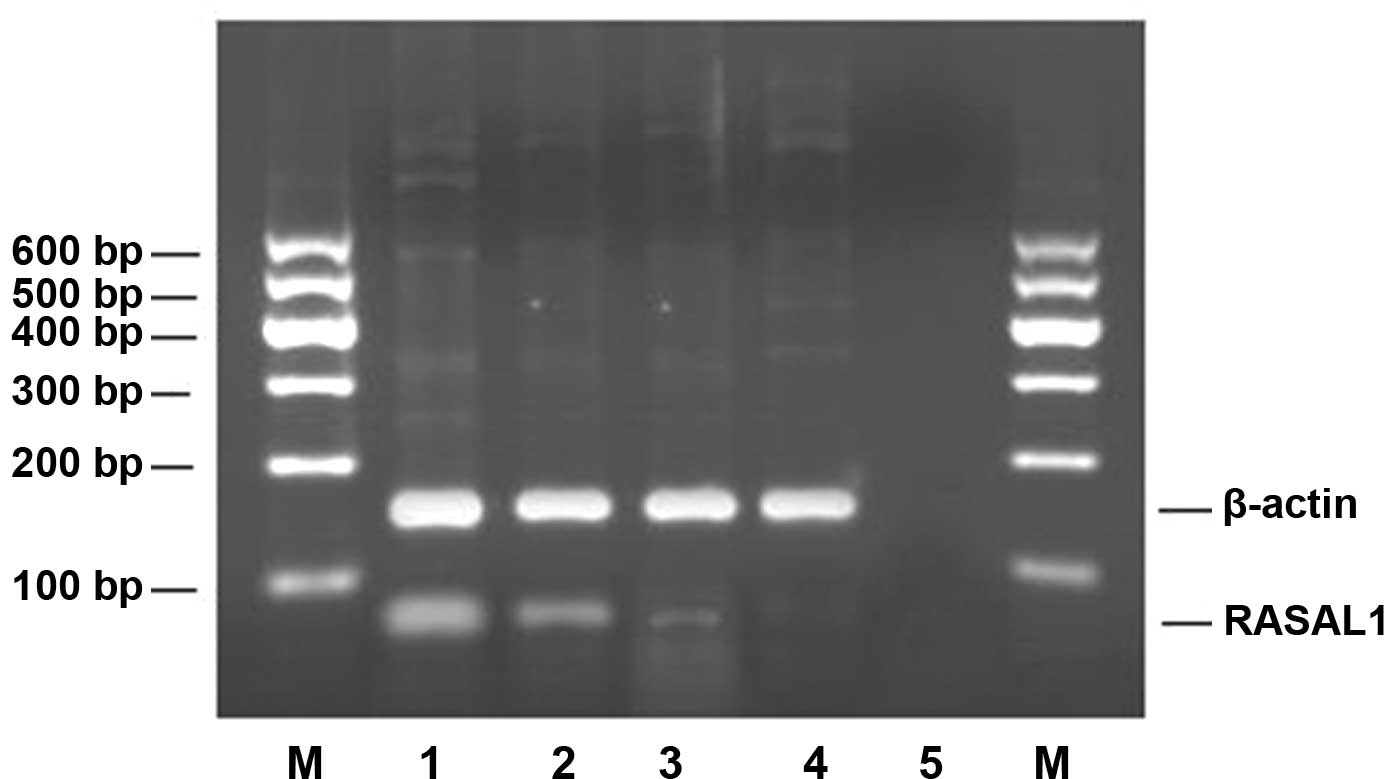
The results of RT-PCR revealed that the expression
of RASAL1 mRNA was decreased in the three tested gastric
adenocarcinoma cell lines compared to the normal gastric epithelial
cell line GES-l. The rates of down-regulated expression were 47.53%
for MKN-28, 85.80% for SGC-7901 and 95.86% for BGC-823 (Fig. 2, Table
III). The expression level of RASAL1 mRNA correlated with the
differentiation of the cells: cells with the poorest
differentiation had the lowest expression. Among the three types of
differentiated gastric cancer cells, the expression of RASAL1 mRNA
was different (p<0.01).
 | Table IIIExpression of RASAL1 mRNA in human
gastric adenocarcinoma cell lines. |
Table III
Expression of RASAL1 mRNA in human
gastric adenocarcinoma cell lines.
| Group | RASAL1/β-actin
(mean ± SD) | Rate of
downregulated expression (%) |
|---|
| GES-l | 0.507±0.005 | - |
| MKN-28 | 0.266±0.009 | 47.53a |
| SGC-7901 | 0.072±0.004 | 85.80a |
| BGC-823 | 0.021±0.003 | 95.86a |
Expression of RASAL1 protein in human
gastric cancer cell lines
The results of western blotting revealed that the
expression of RASAL1 protein was decreased in the three gastric
cancer cell lines compared to the normal gastric epithelial cell
line GES-l. The rates of downregulated expression were 40.12% for
MKN-28, 54.94% for SGC-7901 and 63.33% for BGC-823 (p<0.01)
(Fig. 3, Table IV). The expression levels of the
RASAL1 protein was also correlated with differentiation: cells with
the poorest differentiation had the lowest expression. Among the
three types of differentiated gastric cancer cells, the expression
of RASAL1 protein was different (F=3 059.420, p<0.01).
 | Table IVExpression of RASAL1 protein in human
gastric cancer cell lines. |
Table IV
Expression of RASAL1 protein in human
gastric cancer cell lines.
| Group | RASAL1/β-actin
(mean ± SD) | Rate of
downregulated expression (%) |
|---|
| GES-l | 0.810±0.008 | - |
| MKN-28 | 0.485±0.009 | 40.12a |
| SGC-7901 | 0.365±0.013 | 54.94a |
| BGC-823 | 0.297±0.005 | 63.33a |
Discussion
The Ras family comprises three members including
K-Ras, H-Ras and N-Ras that encode proteins known as p21, which
play a key role in transducing growth signals from the cell surface
to the cell nucleus. The activation of Ras signaling, such as the
‘Ras-RAF-MEK-ERK’ signaling cascade, causes cell proliferation,
differentiation and survival. The role of the Ras gene in the
development of malignant tumors has always been a focus of
research. Currently, it is known that the aberrant activation of
Ras signaling causes persistent cell proliferation and leads to
tumorigenesis (8).
Ras genes are well investigated due to their
frequent activation and mutation in human cancers. Abnormal Ras
signaling activation is mainly caused by mutations. Ras mutations
lead to constantly activated Ras signaling in the absence of
extracellular signals. Activating mutations of Ras are found in
20–25% of all human tumors and in up to 90% of certain tumor types.
While mutations of H-Ras and N-Ras do not frequently occur, high
levels of K-Ras mutations are found in leukemias, colon, pancreatic
and lung cancer (9). However, in
the absence of a mutation, it is difficult to explain the observed
high Ras signaling. For example, in some tumor entities, including
in gastric cancer, although Ras mutations are seldom detected, Ras
activity is still unusually high (10–11).
The underlying mechanisms regarding this remain largely
unknown.
Findings of other studies have shown a novel
mechanism for Ras activity regulation besides point mutation
(4–6). Ras has two structural conformations,
‘on’ and ‘off’. When Ras is bound to the nucleotide guanosine
diphosphate (GDP), it is in an ‘off’ state, whereas Ras bound to
guanosine triphosphate (GTP) is in an ‘on’ state. The activation
and deactivation of Ras is controlled by GAPs and guanine
nucleotide exchange factors (GEFs) (12). The GAPs have GTP enzyme
activity, which facilitate converting Ras from an active
GTP-bound state to an inactive GDP-bound state. Inversely, the GEFs
change Ras from an inactive GDP-bound state to an active GTP-bound
state. The balance between GEF and GAP activity determines the
guanine nucleotide status of Ras, thereby regulating Ras activity.
Since Ras GAPs switch off Ras signaling transduction pathways, the
genes encoding these RAS GAPs are considered to be potential tumor
suppressor genes. A number of studies have indicated that certain
members of the Ras GAP family are indeed tumor suppressors,
including neurofibromin (NF1) and DAB2IP (13–17).
The RASAL1 gene, which is located on chromosome 12
(12q23-q24), has an overall length of 1100 bp and a structure
mainly consisting of exon 1 and 2 and Ca2+-binding
sites. As a newly discovered gene, it was proved to regulate the
activity of the Ras signal transduction pathway (4). The RASAL1 gene possesses the
characteristics of Ras GAPs, which enhance the intrinsic Ras-GTPase
activity through hydrolyzing GTP into GDP, and thus is involved in
cell differentiation, generation and apoptosis. Jin et al
found that RASAL1 is expressed in almost all normal tissues,
including in the heart, kidney, gastrointestinal tract, pancreas,
lung, prostate and bone marrow, suggesting that RASAL1 genes
possess a significant role in maintaining various normal human
physiological functions (4). In
recent studies, RASAL1 was found to be downregulated in a variety
of tumor tissues, including naso-pharyngeal, breast, lung, liver
and esophageal cancer and lymphoma. By studying 152 patient
colorectal cancer tissues and 18 types of colon cancer cells, Ohta
et al found that the expression of RASAL1 decreased markedly
in colorectal cancer cells that contained the wild-type
K-Ras gene, but did not decrease in those colorectal cancer
cells with a mutant K-Ras gene (5). RASAL1 expression was detected in 46.9%
(30/64) of adenocarcinoma, 17.4% (8/46) of large adenoma and none
of the small adenoma samples (0/42). Based on the above study,
RASAL1 was found to inhibit tumor progression by downregulating the
Ras signal activity. It was also found that the ectopic expression
of RASAL1 in transfected colorectal cancer cells in culture
promoted Ras inactivation, which was confirmed by the depression of
ERK, the downstream effector in the Ras signaling transduction
pathway, as well as the suppression of the malignant phenotype in
colorectal cancer cells. In their study, Calvisi et al also
found that in the absence of Ras mutations, the downregulation of
RASAL1, as well as other Ras GAPs (DAB2IP and NF1), resulted in the
unrestrained activation of Ras signaling in the presence of
wild-type Ras in human hepatocarcinogenesis (12), but it is unknown whether or not this
also occurs in gastric cancer.
In our study, using immunohistochemistry, it was
found that of 50 cases of gastric cancer tissues 12 cases were (−),
23 cases (+), 13 cases (++) and 2 cases (+++); whereas in 50 cases
of normal gastric tissues 16 cases were (++) and 34 cases were
(+++). This finding indicates that the RASAL1 expression is
significantly reduced in gastric cancer tissues as well.
Furthermore, we found that a reduced RASAL1 expression was
associated with tumor size, differentiation degree, invasion depth
and lymph node metastasis, but not with age and gender. RT-PCR and
western blotting were used to detect the expression of RASAL1 in
three types of gastric adenocarcinoma cell lines, including well-,
moderately and poorly differentiated adenocarcinoma cells. The
results confirmed that RASAL1 expression decreased in the three
types of gastric adenocarcinoma cells compared with the normal
gastric epithelial cell line, and the expression level was
correlated with the differentiation: the poorest differentiation
had the lowest expression. In a recent study, Seto et al
investigated 10 gastric cancer cell lines by immunoblotting, and
found that RASAL1 expression was reduced in 6 out of the 10 cell
lines (18). These authors also
reported that the immunohistochemical analyses in primary gastric
tumors revealed that the RASAL1 expression was reduced in 23 out of
48 (48%) of the gastric cancers cell lines, but in none of the
adenomas (0/10). These results suggest that RASAL1 is important in
the tumorigenesis and development of gastric carcinoma. The results
of our study have shed light on the pathogenesis of gastric
carcinoma, and are a new therapeutic target for gastric carcinoma
treatment.
Epigenetic silencing has been found to be the key
mechanism responsible for the downregulated expression of the
RASAL1 gene in tumors. Evidence indicates that CpG island
methylation in the promoter of the RASAL1 gene may induce silencing
in multiple tumors, including in esophageal cancer, nasopharyngeal
carcinoma and colorectal cancer cells (4). Simultaneously, other studies have
demonstrated that in colon cancer cells, the gene methylation of
RASAL1 was reversed by using the methylation inhibitor
5-acetazolamide-2-cytosine deoxyriboside (5-Aza-CdR) (19). Kolfschoten et al, however,
found another important mechanism affecting RASAL1 expression.
Since RASAL1 is a transcription target of the anti-oncogene
pituitary homeobox 1 (PITX1), RASAL1 expression may be
downregulated by a decrease in PITX1 gene expression in human
gastric carcinogenesis (20–21).
In conclusion, our findings demonstrate that the
expression of RASAL1 is markedly decreased in gastric carcinoma
tissues and cell lines, and is associated with gastric carcinoma
differentiation degree and progression, suggesting that it is
important in the development of gastric carcinoma. The mechanisms
that contribute to the downregulated expression of RASAL1 in
gastric cancer require further investigation.
Acknowledgements
This study was supported by the Natural Science
Foundation of Jiangsu Province of China (No. BK2008301).
References
|
1
|
Parada LF, Tabin CJ, Shis C and Weinberg
RA: Human EJ bladder carcinoma oncogene is homologue of Harvey
sarcoma virus ras gene. Nature. 297:474–478. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fernández-Medarde A and Santos E: Ras in
cancer and developmental diseases. Genes Cancer. 2:344–358.
2011.
|
|
3
|
Cox AD and Der CJ: Ras history: the saga
continues. Small Gtpases. 1:2–27. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jin H, Wang X, Ying J, Wong AH, Cui Y,
Srivastava G, Shen ZY, Li EM, Zhang Q, Jin J, Kupzig S, Chan AT,
Cullen PJ and Tao Q: Epigenetic silencing of
Ca2+-regulated Ras GTPase-activating protein RASAL
defines a new mechanism of Ras activation in human cancers. Proc
Natl Acad Sci USA. 104:12353–12358. 2007.
|
|
5
|
Ohta M, Seto M, Ijichi H, Miyabayashi K,
Kudo Y, Mohri D, Asaoka Y, Tada M, Tanaka Y, Ikenoue T, Kanai F,
Kawabe T and Omata M: Decreased expression of the Ras
GTPase-activating protein RASAL1 is associated with colorectal
tumor progression. Gastroenterology. 136:206–216. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bernards A and Settleman J: Loss of the
Ras regulator RASAL1: another route to Ras activation in colorectal
cancer. Gastroenterology. 136:46–48. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang YW, Qu Y, Li JF, Chen XH, Liu BY, Gu
QL and Zhu ZG: In vitro and in vivo evidence of
metallopanstimulin-1 in gastric cancer progression and
tumorigenicity. Clin Cancer Res. 12:4965–4973. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Malumbres M and Barbacid M: RAS oncogenes:
the first 30 years. Nat Rev Cancer. 3:459–465. 2003.PubMed/NCBI
|
|
9
|
Karnoub AE and Weinberg RA: Ras oncogenes:
split personalities. Nat Rev Mol Cell Biol. 9:517–531. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kimura K, Nagasaka T, Hoshizima N,
Sasamoto H, Notohara K, Takeda M, Kominami K, Iishii T, Tanaka N
and Matsubara N: No duplicate KRAS mutation is identified on the
same allele in gastric or colorectal cancer cells with multiple
KRAS mutations. J Int Med Res. 35:450–457. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu ZM, Liu LN, Li M, Zhang QP, Cheng SH
and Lu S: Mutation detection of KRAS by high-resolution melting
analysis in Chinese with gastric cancer. Oncol Rep. 22:515–520.
2009.PubMed/NCBI
|
|
12
|
Calvisi DF, Ladu S, Conner EA, Seo D,
Hsieh JT, Factor VM and Thorgeirsson SS: Inactivation of Ras
GTPase-activating proteins promotes unrestrained activity of
wild-type Ras in human liver cancer. J Hepatol. 54:311–319. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kong Z, Xie D, Boike T, Raghavan P, Burma
S, Chen DJ, Habib AA, Chakraborty A, Hsieh JT and Saha D:
Downregulation of human DAB2IP gene expression in prostate cancer
cells results in resistance to ionizing radiation. Cancer Res.
70:2829–2839. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vigil D, Cherfils J, Rossman KL and Der
CJ: Ras superfamily GEFs and GAPs: validated and tractable targets
for cancer therapy? Nat Rev Cancer. 10:842–857. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bos JL, Rehmann H and Wittinghofer A: GEFs
and GAPs: critical elements in the control of small G proteins.
Cell. 129:865–877. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Iwashita S and Song SY: Ras GAPs: a
crucial regulator of extracellular stimuli for homeostasis of
cellular functions. Mol Biosyst. 4:213–222. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hölzel M, Huang S, Koster J, Ora I,
Lakeman A, Caron H, Nijkamp W, Xie J, Callens T, Asgharzadeh S,
Seeger RC, Messiaen L, Versteeg R and Bernards R: NF1 is a tumor
suppressor in neuroblastoma that determines retinoic acid response
and disease outcome. Cell. 142:218–229. 2010.PubMed/NCBI
|
|
18
|
Seto M, Ohta M, Ikenoue T, Sugimoto T,
Asaoka Y, Tada M, Mohri D, Kudo Y, Ijichi H, Tateishi K, Otsuka M,
Hirata Y, Maeda S, Koike K and Omata M: Reduced expression of RAS
protein activator like-1 in gastric cancer. Int J Cancer.
128:1293–1302. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu Q, Walker SA, Gao D, Taylor JA, Dai
YF, Arkell RS, Bootman MD, Roderick HL, Cullen PJ and Lockyer PJ:
CAPRI and RASAL impose different modes of information processing on
RAS due to contrasting temporal filtering of Ca2+. J
Cell Biol. 170:183–190. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kolfschoten IG, van Leeuwen B, Berns K,
Mullenders J, Beijersbergen RL, Bernards R, Voorhoeve PM and Agami
R: A genetic screen identifies PITX1 as a suppressor of RAS
activity and tumorigenicity. Cell. 121:849–858. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen YN, Chen H, Xu Y, Zhang X and Luo Y:
Expression of pituitary homeobox 1 gene in human gastric
carcinogenesis and its clinicopathological significance. World J
Gastroenterol. 14:292–297. 2008. View Article : Google Scholar : PubMed/NCBI
|