Introduction
Chronic myelogenous leukemia (CML) is a
myeloproliferative disease characterized by the Philadelphia (Ph)
chromosome. This chromosome is created by a reciprocal t(9:22)
translocation which transfers the Abelson (ABL) oncogene on
chromosome 9 to the breakpoint cluster region (BCR) of chromosome
22, resulting in the formation of a fused BCR/ABL gene (1). BCR/ABL produces an abnormal tyrosine
kinase that causes aberrant myelopoiesis identified in CML. Variant
Ph chromosome translocations involving one or more chromosome
regions in addition to chromosomes 9 and 22 have been identified in
5–10% of CML patients (2). In these
variants, four-way Ph chromosome translocations are rare (3). The prognostic significance of variant
Ph chromosome CML remains unclear.
The progression of CML from the chronic phase (CP)
to blast crisis (BC) is frequently associated with non-random
secondary chromosomal aberrations, including +8, i(17q), +19 and an
extra Ph chromosome (4).
Since tyrosine kinase activity is required for the
transforming function of the BCR/ABL fusion protein, imatinib, a
specific inhibitor of the kinase, is an effective treatment for CML
patients. The 5-year estimated overall survival rate for patients
who receive imatinib as initial therapy is 89%. This rate is higher
than those reported in previous studies of CML treatment and only
7% of all patients progressed to the accelerated phase (AP) or BC
(5). In a previous study, deletions
on the derivative chromosome 9 [der(9)] were identified at a higher
frequency in patients with variant Ph translocations than in those
with classic Ph (45 and 17%, respectively) (6).
In the present study, a novel case of a Ph
chromosome-positive CML in BC was identified, with a four-way
rearrangement involving five chromosomal regions, 9p21, 9q34,
12p13.3, 20q11.2 and 22q11.2, an unbalanced translocation
der(7)t(7;8) (p11.2;q11.2), deletions of ABL and argininosuccinate
synthetase (ASS) genes at 9q34 on der(9), partial monosomies 8, 12
and an additional Ph chromosome. In addition, immunopheno-type
analysis indicated biphenotypic leukemia.
Materials and methods
Case report
In April 2011, a 22-year-old female presented with a
white blood cell count (WBC) of 97×109 cells/l
(neutrophils, 21; lymphocytes, 73; eosinophiles, 4; monocytes, 1;
and basophiles, 1%). The platelet count was 268×109
cells/l and the hemoglobin level was 9.1 g/dl. Physical examination
revealed splenomegaly and weight loss was noted. Chromosome
analysis using banding cytogenetics demonstrated a karyotype
consistent with clinical diagnosis of a CML in CP. The patient was
treated daily with Zyloric (300 mg) and hydroxyurea (500 mg) for
four days. LDH was 1860 U/l (normal, <460 U/l) and serum
alkaline phosphase was 348 U/l (normal, <232 U/l). In September
2011, the patient presented for the second time with a WBC count of
132.4×109 cells/l (neutrophils, 1; lymphocytes, 40; and
immature cells, 52%). Platelet count was 22×109/l and the
hemoglobin level was 10 g/dl. Imatinib mesylate (400 mg/day) was
administered for five months and following this period the
described symptoms were not observed. In October 2011, the patient
died for unknown reasons under treatment.
Chromosome analysis
Chromosome analysis using GTG-banding was performed
according to standard procedures (7) prior to chemotherapeutic treatment. A
total of 20 metaphase cells derived from unstimulated bone marrow
culture were analyzed. Karyotypes are described according to the
International System for Human Cytogenetic Nomenclature (8).
Molecular cytogenetics
Fluorescence in situ hybridization (FISH)
using LSI BCR/ABL+9q34 three color dual fusion translocation probe
(Abbott Molecular/Vysis, Des Plaines, IL, USA) and chromosome
enumeration probe (CEP) for chromosome 9 (Abbott Molecular/Vysis)
were applied according to the manufacturer’s instructions together
with a whole chromosome painting (WCP) probe for chromosomes 7, 8,
9, 12, 20 and 22 (MetaSystems, Altlussheim, Germany) (7). FISH using the corresponding chromosome
specific array-proven multicolor banding (aMCB) probe sets based on
microdis-section derived region-specific libraries was performed as
previously described (7). A minimum
of 20 metaphase spreads were analyzed, using a fluorescence
microscope (AxioImager.Z1 mot, Carl Zeiss Ltd., Hertfordshire, UK)
equipped with appropriate filter sets to discriminate between a
maximum of five fluorochromes and the counterstain DAPI
(4′,6-diamino-2-phenylindole). Image capture and processing were
performed using the ISIS imaging system (MetaSystems).
Reverse transcription polymerase chain
reaction (RT-PCR) for BCR/ABL fusion transcripts
Total RNA was extracted from the diagnostic
peripheral blood sample using the InviTrap RNA kit (Invitek GmbH,
Berlin, Germany) according to the manufacturer’s instructions. cDNA
was prepared from 5μg total RNA with the Genequality BCR-ABL kit
(AB Analitica, Padova, Italy) according to the manufacturer’s
instructions.
Flow cytometry immunophenotyping
Immunophenotyping of leukemic blasts was performed
as previously described (7).
DNA sequencing
Detection of BCR/ABL mutation domain was performed
using previously described primers (9).
Results
Karyotyping was performed prior to and following
chemotherapy treatment. Prior to chemotherapy, the karytype was
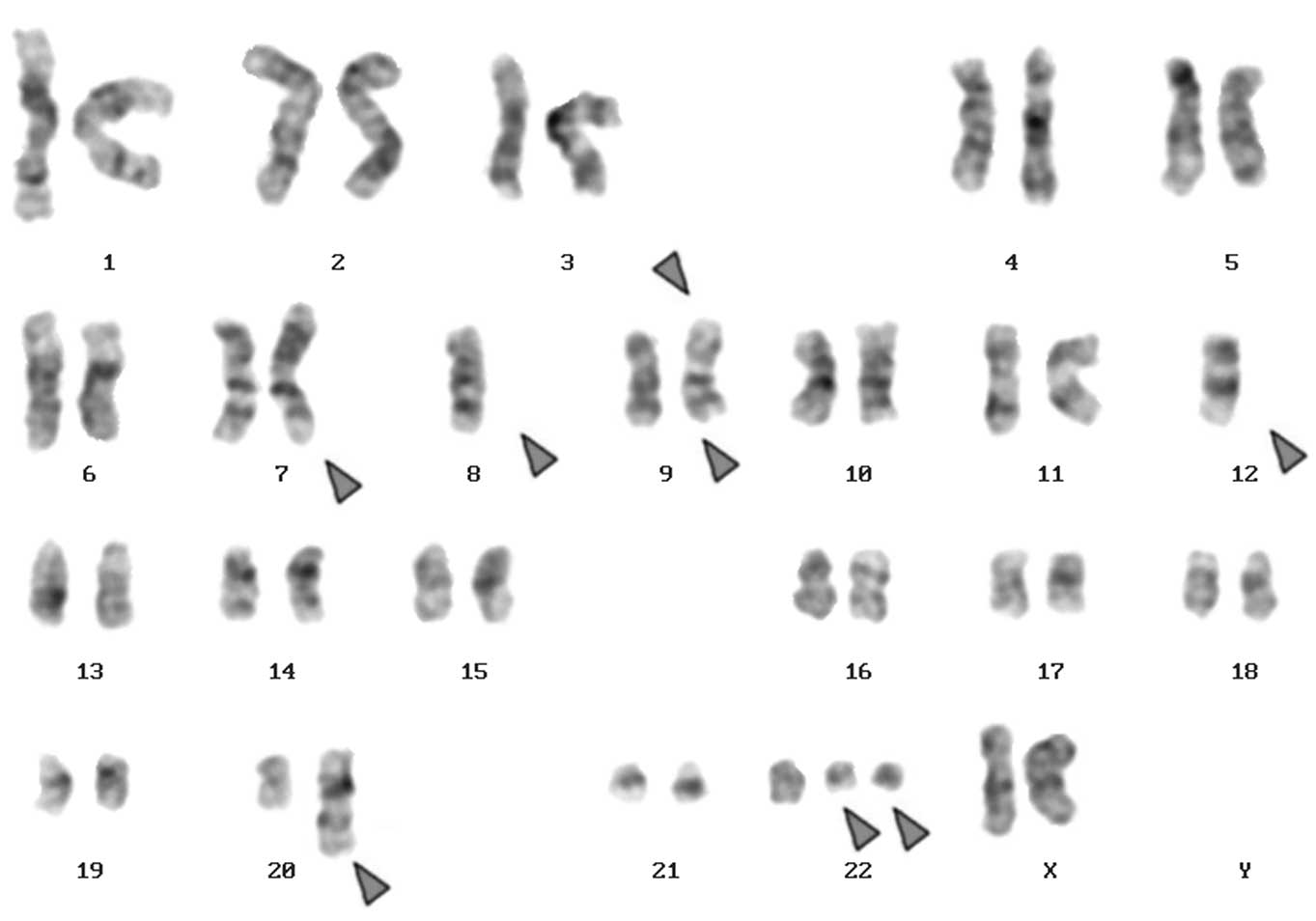
identified as 46,XX,t(9;22)[20] and following chemotherapy was
45,XX,der(7)t(7;8),-8,der(9)t(20;9;22),-12,der(12)(12;20),+der(22)t(9;22)×2[13]/45,XX,der(7)t(7;8),-8,t(9;22)
[7] (Fig. 1A). Number of cells is
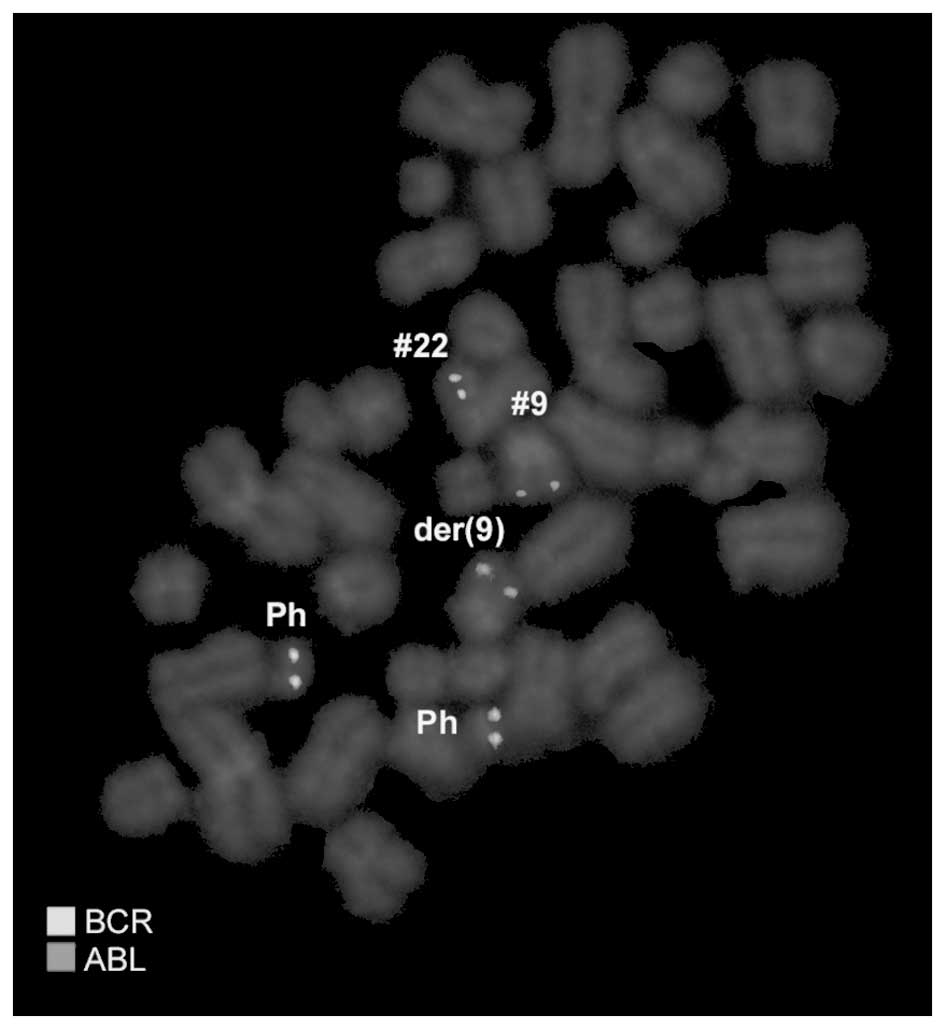
provided in square brackets. Dual-color FISH using a probe specific
for BCR, ABL and ASS genes revealed two typical Ph chromosomes with
the BCR/ABL fusion gene on the der(22). On the der(9), ABL and ASS
genes at 9q34 were deleted and the BCR gene was present (Fig. 2). Chromosomes 7, 8, 9, 12, 20 and 22
were observed using WCP and/or CEP probes (data not shown). RT-PCR
confirmed the presence of the BCR-ABL fusion (b3a2 transcript)
revealing a major M-BCR transcript, most often identified in CML
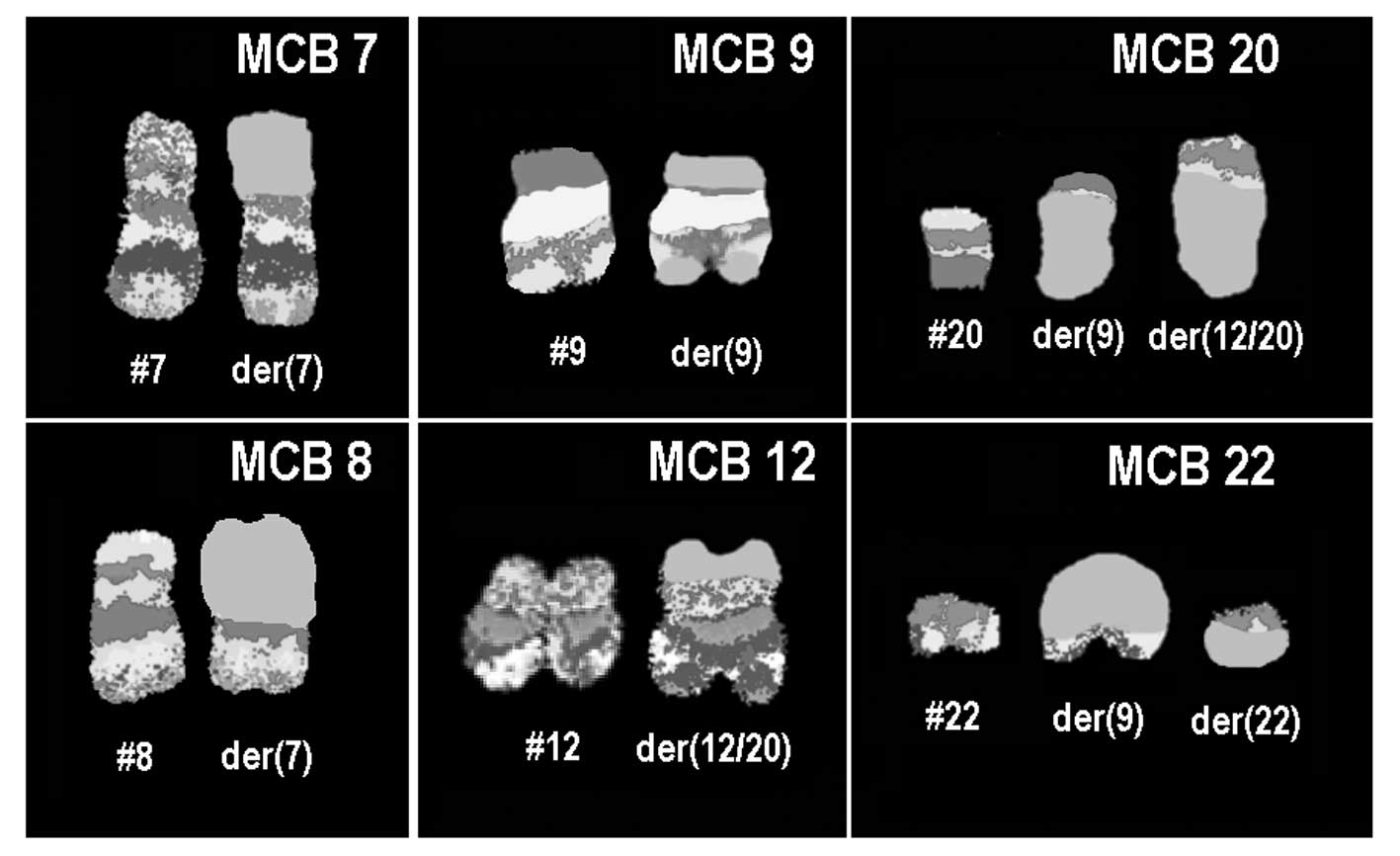
(data not shown). Finally, aMCB using probes for the corresponding
chromosomes was performed as previously reported (7) (Fig.
3). The following final karyotype was determined:
45,XX,der(7)t(7;8)(p11.2;q11.2),-8,der(9)
(20qter→20q11.2::9p21→9q34::22q11.2→22qter),-12,der(12)
(20pter→20q11.2::12p13.3→12q24.3::12q24.3→12q15∼21.1),
+der(22)t(9;22)(q34;q11.2)×2[13]/45,XX, der(7)t(7;8)
(p11.2;q11.2),-8,t(9;22)(q34;q11.2)[7]. Number of cells is provided
in square brackets.
Immunophenotypic analysis of peripheral blood
demonstrated that the abnormal cell population positivity reacted
with antibodies against CD45 (95%), HLADr (79%), CD19 (73%), CD34
(29%), CD10 (78%), CD33 (41%), CD18 (70%), CD32 (70%), CD22 (40%),
CD123 (61%), CD20 (40%), CD235a (63%), CD117 (30%), CD38 (59%) and
CD15 (60%). The cell population negativity reacted with additional
antibodies used. Expression profiles of multilineages indicated
that the patient had biphenotypic leukemia (10).
DNA sequencing of the BCR/ABL kinase domain
identified no mutations.
Discussion
In the present study, a novel case of Ph
chromosome-positive CML in BC with a four-way rearrangement was
observed, including five chromosomal regions, 9p21, 9q34, 12p13.3,
20q11.2, 22q11.2, an unbalanced translocation t(7;8) (p11.2;q11.2),
deletions of ABL and ASS genes on der(9), monosomies 8, 12 and an
additional Ph chromosome. To the best of our knowledge, these
chromosomal aberrations, particularly t(7;8)(p11.2;q11.2) have not
been previously observed in CML (11).
Four-way Ph translocation is extremely rare and only
anecdotal cases have been described in the imatinib era. In the
most recent study of CML, only 3/500 patients receiving imatinib
mesylate as a frontline therapy were observed to have a four-way
translocation (3).
The mechanism of development of this complex
rearrangement may include a primary standard t(9;22), followed by a
subsequent three-way translocation affecting chromosomes 12, 20 and
the der(9). The fusion BCR/ABL signal was identified on der(22) and
chromosome 22 had not rearranged with chromosome 20 or 12. These
observations are consistent with a common two-step rearrangement
process (12). Therefore, an
inherent implication of the two-step mechanism is that variant
translocations may be associated with a poorer prognosis (13).
Resistance to chemotherapy occurs as a result of
increased expression of the BCR-ABL kinase from genomic
amplification, clonal chromosomal evolution or mutations in the ABL
kinase of the BCR-ABL gene, affecting drug interaction or kinase
activity (14).
Submicroscopic ASS gene deletions in fused
chromosome 9 were previously reported to be important for
development of shortened CP and decreased overall survival,
associated with a poor prognosis and response to interferon and
imatinib mesylate (15,16).
Leukemias of ambiguous lineage are uncommon,
representing ∼4% of all acute leukemias, and frequently demonstrate
an aggressive disease course, with mean survival rates less than
those of leukemias derived from a single-cell lineage (17). No single chromosome abnormality is
unique to biphenotypic leukemia (18). In the present study, a complex
cytogenetic abnormality was identified using conventional and
molecular cytogenetics. Therefore, we hypothesize that leukemias of
ambiguous lineage associated with cytogenetic abnormalities
indicate a poorer prognosis than those without demonstrable
abnormalities.
Recurrent chromosomal deletions identified in
sporadic types of cancer often contain tumor suppressor genes
(TSGs). TSGs function in signaling networks that protect against
tumor initiation and progression and are inactivated by deletions,
point mutations or promoter hypermethylation (19). For example, TCR β (7p15) (20); DLC-1 (8p21.3–22), FEZ1 (8p22) and
LTPS (8p23) (21); p16INK4a, p14ARF
and p15INK4b (9p21) (22); and the
leukemogenesis-relevent ETV6 gene (12p)(23).
In conclusion, the present study reports a novel
case of a Ph chromosome-positive CML in BC with a four-way Ph
trans-location. The translocation is likely to result from a
two-step mechanism. In addition to an unbalanced translocation
der(7) t(7;8)(p11.2;q11.2), multiple partial chromosomal regions
were deleted, partial monosomies 8, 12 and an additional Ph
chromosome were identified. Immunophenotyping indicated that the
patient had biphenotypic leukemia. These observations represent an
adverse prognosis in CML. The patient died under treatment one
month after diagnosis.
Acknowledgements
The authors thank Professor I. Othman,
the Director General of the Atomic Energy Commission of SYRIA
(AECS) and Dr N. Mirali, Head of the Molecular Biology and
Biotechnology Department, for their support. The present study was
supported by the AECS, and in part by the DAAD,
Stefan-Morsch-Stiftung and the Monika-Kutzner-Stiftung.
References
|
1.
|
Sawyers CL: Chronic myeloid leukemia. N
Engl J Med. 340:1330–1340. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Johansson B, Fioretos T and Mitelman F:
Cytogenetic and molecular genetic evolution of chronic myeloid
leukemia. Acta Haematol. 107:76–94. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Adriana Z and Al Bahar S: Novel four-way
Ph translocation t(9;22;7;1)(q34;q11;q22;p13) in a chronic myeloid
leukemia patient receiving tyrosine kinase inhibitor therapy. Int J
Hematol. 95:315–319. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Sandberg AA: The Chromosomes in Human
Cancer and Leukemia. 2nd edition. Elsevier Science; New York, NY:
pp. 151–172. 1990
|
|
5.
|
Druker BJ, Guilhot F, O’Brien SG, Gathmann
I, Kantarjian H, Gattermann N, et al: Five-year follow-up of
patients receiving imatinib for chronic myeloid leukemia. N Engl J
Med. 355:2408–2417. 2006.PubMed/NCBI
|
|
6.
|
Reid A, Gribble SM, Huntly BJ, Andrews KM,
Campbell L, Grace CD, Wood ME, Green AR and Nacheva EP: Variant
Philadelphia translocations in chronic myeloid leukaemia can mimic
typical blast crisis chromosome abnormalities or classic t(9;22): a
report of two cases. Br J Haematol. 113:439–442. 2001. View Article : Google Scholar
|
|
7.
|
Al-Achkar W, Wafa A, Klein E and Aljapawe
A: Biclonal myelodysplastic syndrome involving six chromosomes and
monoallelic loss of RB1 - A rare case. Mol Cytogenet. 4:162011.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Shaffer L, Slovak M and Cambell L: ISCN
(2009): An International System for Human Cytogenetic Nomenclature.
1st edition. S. Karger AG; Basel: 2009
|
|
9.
|
Chien JH, Tang JL, Chen RL, Li CC and Lee
CP: Detection of BCR-ABL gene mutations in Philadelphia chromosome
positive leukemia patients resistant to STI-571 cancer therapy.
Leuk Res. 32:1724–1734. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Rothe G and Schmitz G: Consensus protocol
for the flow cyto-metric immunophenotyping of hematopoietic
malignancies. Working Group on Flow Cytometry and Image Analysis.
Leukemia. 10:877–895. 1996.PubMed/NCBI
|
|
11.
|
Mitelman F, Johansson B and Mertens F:
Mitelman Database of Chromosome Aberrations in Cancer. http://cgap.nci.nih.gov/Chromosomes/Mitelman.
Accessed October 3, 2012.
|
|
12.
|
Reid AG, Huntly BJP, Grace C, Green AR and
Nacheva EP: Survival implications of molecular heterogeneity in
variant Philadelphia-positive chronic myeloid leukaemia. Br J
Haematol. 121:419–427. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Bennour A, Sennana H, Laatiri MA, Khelif A
and Saad A: A masked BCR/ABL rearrangement in a case of chronic
myeloid leukemia with translocation t(3;9)(p14;q34). Cancer Genet
Cytogenet. 181:72–74. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Hochhaus A, Kreil S, Corbin AS, La Rosée
P, Müller MC, Lahaye T, Hanfstein B, Schoch C, Cross NCP, Berger U,
Gschaidmeier H, Druker BJ and Hehlmann R: Molecular and chromosomal
mechanisms of resistance to imatinib (STI571) therapy. Leukemia.
16:2190–2196. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Bacher U, Schnitter S, Kern W, Hiddemann
W, Haferlach T and Schoch C: The incidence of submicroscopic
deletions in reciprocal translocations is similar in acute myeloid
leukemia, BCR-ABL positive acute lymphoblastic leukemia and chronic
myeloid leukemia. Haematologica. 90:558–559. 2005.
|
|
16.
|
Bennour A, Sennana H, Laatiri MA, Elloumi
M, Khelif A and Saad A: Molecular cytogenetic characterization of
variant Philadelphia translocations in chronic myeloid leukemia:
genesis and deletion of derivative chromosome 9. Cancer Genet
Cytogenet. 194:30–37. 2009. View Article : Google Scholar
|
|
17.
|
Brunning RD, Matutes E, Borowitz M, et al:
Acute leukaemias of ambiguous lineage. World Health Organization
Classification of Tumours: Pathology and Genetics of Tumours of
Haemopoietic and Lymphoid Tissues. Jaffe ES, Harris NL, Stein H and
Vardiman JW: IARC Press; Lyon, France: pp. 106–107. 2001
|
|
18.
|
Carbonell F, Swansbury J, Min T, Matutes
E, Farahat N, Buccheri V, Morilla R, Secker-Walker L and Catovsky
D: Cytogenetic findings in acute biphenotypic leukaemia. Leukemia.
10:1283–1287. 1996.PubMed/NCBI
|
|
19.
|
Downward J: Targeting RAS signalling
pathways in cancer therapy. Nat Rev Cancer. 3:11–22. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Wlodarska I, Martin-Garcia N, Achten R, De
Wolf-Peeters C, Pauwels P, Tulliez M, de Mascarel A, Briere J,
Patey M, Hagelmeijer A and Gaulard P: Fluorescence in situ
hybridization study of chromosome 7 aberrations in hepatosplenic
T-cell lymphoma: isochromosome 7q as a common abnormality
accumulating in forms with features of cytologic progression. Genes
Chromosomes Cancer. 33:243–251. 2002. View Article : Google Scholar
|
|
21.
|
Qin LX: Chromosomal aberrations related to
metastasis of human solid tumors. World J Gastroenterol. 8:769–776.
2002.PubMed/NCBI
|
|
22.
|
Mancini M, Scappaticci D, Cimino G, Nanni
M, Derme V, Elia L, Tafuri A, Vignetti M, Vitale A, Cuneo A,
Castoldi G, Saglio G, Pane F, Mecucci C, Camera A, Specchia G,
Tedeschi A, Di Raimondo F, Fioritoni G, Fabbiano F, Marmont F,
Ferrara F, Cascavilla N, Todeschini G, Nobile F, Kropp MG, Leoni P,
Tabilio A, Luppi M, Annino L, Mandelli F and Foà R: A comprehensive
genetic classification of adult acute lymphoblastic leukemia (ALL):
analysis of the GIMEMA 0496 protocol. Blood. 105:3434–3441. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Andreasson P, Johansson B, Arheden K,
Billstrom R, Mitelman F and Hoglund M: Deletions of CDKN1B and ETV6
in acute myeloid leukemia and myelodysplastic syndromes without
cytogenetic evidence of 12p abnormalities. Genes Chromosomes
Cancer. 19:77–83. 1997. View Article : Google Scholar : PubMed/NCBI
|