Introduction
Chronic obstructive pulmonary disease (COPD) is a
complex disease characterized by irreversible airflow limitation,
abnormal permanent distal air-space enlargement and emphysema in
the lung. This is usually progressive and associated with an
abnormal inflammatory response of the lungs to noxious particles or
gases (1). COPD is a major and
increasing cause of morbidity and mortality worldwide (2). The World Health Organization has
predicted that it is likely to be the third leading cause of
mortality in the world by the year 2020. Although cigarette smoking
is widely accepted as the most significant risk factor for COPD,
only 10–15% of smokers develop the disease (3). This, together with the familial
clustering of early-onset COPD (4),
as well as a susceptibility to frequent exacerbation in these
individuals (5), strongly suggests
a genetic factor in the pathogenesis of COPD.
As the only well-defined genetic factor of COPD,
α1-antitrypsin deficiency, is rare in worldwide populations, other
genetic factors must be involved in the susceptibility and
development of COPD. Given that 95% of those who develop COPD are
smokers, oxidative stress caused by smoking is considered to be a
pathogenetic factor for COPD. Thus, the genes encoding enzymes that
protect the lung against smoke-induced oxidative stress have
received more attention and become the focus of genetic COPD
studies. Microsomal epoxide hydrolase (EPHX1) is an enzyme
essential for the metabolism of the highly reactive epoxide
intermediates produced by cigarette smoke and is expressed at
varying levels in the majority of tissues and cell types (6). Epidemiological studies have shown that
EPHX1 activity in the liver, lung and peripheral blood leucocytes
varies as much as 50-fold in Caucasian populations (7). The human EPHX1 gene is located on
chromosome 1q42.1 and consists of nine exons. Further studies have
reported that two common polymorphisms in the coding regions are
responsible for the variable enzyme activity, conferring decreased
activity by one mutation and increased activity by another
(8). The T to C transition in exon
3 changes tyrosine (Tyr) residue 113 to histidine (His), thus
reducing the enzyme activity by ∼50% (slow allele). The second, the
A to G transition in exon 4 changes histidine (His) residue 139 to
arginine (Arg) and produces an enzyme with an activity increased by
∼25% (fast allele) (8). Based on
these two single nucleotide polymorphisms (SNPs), the population
may be divided into four groups of putative EPHX1 phenotypes with
various activities (fast, normal, slow and extremely slow)
(8,9).
Since the initial report of an association between
increased susceptibility to COPD and EPHX1 113 His-His homozygosity
in Caucasian populations (9), a
number of studies have been performed to assess the association
between the EPHX1 polymorphisms and COPD in various populations
(10–32). However, the results of the studies
remain controversial and inconclusive. A number of studies
supported this association (12,13,18,19,22,24,26,27,29) or
reported an association with disease severity rather than
susceptibility (14,30), while others did not observe this
association (10,11,20,21,23,24,31).
The remaining studies reported a protective effect for this
polymorphism in the development of COPD (16,17).
The lack of replication and consistency in these studies occured,
most likely, due to the different ethnic populations used in the
various studies, the poor matching of cases and controls or the
small sample sizes lacking the statistical power to produce a
reliable conclusion in any individual study. Thus, a comprehensive
analysis is essential. A systematic review and meta-analysis of the
published data may be a powerful tool to aid the elucidation of
this association. Although three meta-analyses have been performed
to attempt to clarify this question, two completely different and
contradictory conclusions have been reported. Two studies reported
that the EPHX1 113 mutant homozygote was a risk factor for COPD
(26,33), while the other reported it to have a
protective effect (17). Since
then, additional studies of the association between EPHX1
polymorphisms and COPD have been reported.
In the present study, a new comprehensive
meta-analysis was performed to systematically investigate the
association between the EPHX1 polymorphisms and an individual’s
susceptibility to COPD. A total of 24 published case-control
studies were included in this meta-analysis. The putative EPHX1
enzyme activity and risk of COPD were predicted by single
polymorphism of T113C/A139G and combined double polymorphisms.
Analyses stratified by ethnicity, sample size, cigarette smoking
status of the controls and Hardy-Weinberg equilibrium (HWE)
violation of the controls were performed to explore the variations
which affect the final conclusion.
Materials and methods
Literature search
A literature search was performed in PubMed,
MEDLINE, Embase, Wanfang Database and Weipu Database for studies
published up to April 2012 to use in the present meta-analysis. The
key terms were ‘EPHX1 or microsomal epoxide hydrolase or EPHX or
EPOX or HYL1 or MEH’, ‘COPD or chronic obstructive pulmonary
disease’ and ‘polymorphism’ in various combinations. The search was
limited to articles published in English and Chinese. References
matching the above criteria were also searched manually to identify
additional studies.
Inclusion and exclusion criteria
The eligible investigations met the following
criteria: i) the studies were case-control studies designed to
explore the association between at least one of the two EPHX1 gene
polymorphisms (T113C and A139G) and COPD susceptibility; ii) the
studies provided data on the distribution of EPHX1 gene
polymorphisms in the case-control population; iii) the studies were
published in the English or Chinese language. Studies were excluded
if they did not contain enough data for meta-analysis, were
abstracts or reviews or were duplicated within other included
studies.
Data extraction
Two authors searched the studies and extracted the
data according to the above inclusion and exclusion criteria. The
following characteristics were collected from the eligible studies:
first author, year of publication, country of studied population,
ethnicity, sample size of the cohorts, number of COPD patients,
number of controls, source of the control group (population-based
or hospital-based) and the EPHX1 gene T113C and A139G allele and
genotype distributions among the case and control groups. Different
case-control groups within one study were considered to be
independent studies. The cigarette smoking status of the controls
was strategically classified as current smokers, ex-smokers and
non-smokers.
Statistical analyses
ORs with 95% CIs were calculated to assess the
association between EPHX1 gene polymorphisms and COPD risk. The
wild type homozygotes, TT of T113C and AA of A139G, were used as
reference genotypes. Thus the comparisons were mutant homozygote or
heterozygote vs. reference genotype (CC vs. TT, CT vs. TT, GG vs.
AA and AG vs. AA ). The ORs of COPD risk associated with EPHX1
enzymatic activity were estimated using the normal activity as the
reference group (extemely slow vs. normal, slow vs. normal and fast
vs. normal). Stratified analyses were performed using ethnicity,
sample size, cigarette smoking status of the controls and HWE
violation of the controls.
Heterogeneity across the publications was assessed
with the Cochran’s χ2 test (Q-test) (34) and P<0.10 was considered to
indicate statistically significant heterogeneity. The I2
test was also performed to evaluate heterogeneity between studies.
A high heterogeneity was considered to be present when
I2>50% and much higher when I2>75%
(35). A higher heterogeneity is a
common phenomenon in genetic association studies (36). The pooled OR was calculated by a
fixed-effect model (using the Mantel-Haenszel method) or a
random-effect model (using the DerSimonian-Laird method) according
to the heterogeneity among the studies (37,38).
The statistical significance of the ORs was analyzed by the Z test
and P<0.05 was considered to indicate statistically significant
differences. Random effect meta-regression models with restricted
maximum likelihood estimation were performed to evaluate the
variance among the individual ORs when heterogeneity was detected.
The pre-specified possible sources of inter-study heterogeneity
were: ethnicity of a population, source of the control group,
sample size and HWE violation. Publication bias was assessed using
a funnel plot and Egger’s and Begg’s tests (39,40).
The funnel plot was asymmetrical when there was evidence of
publication bias.
Statistical analyses were performed using the Revman
5.1 software and STATA 10.0 software. The P-value was two-tailed
and P<0.05 was considered to indicate a statistically
significant difference.
Results
Study characteristics
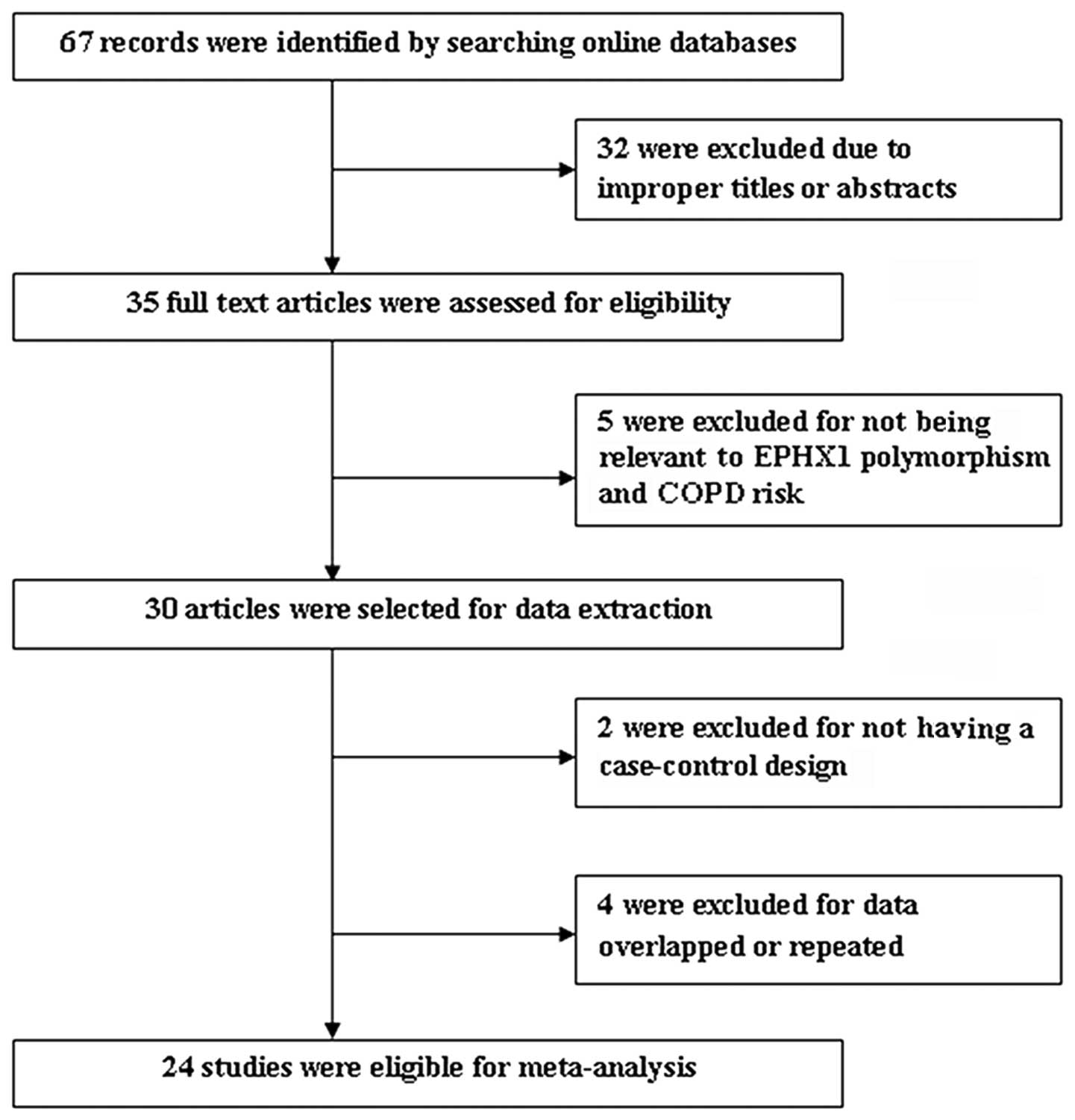
As shown in Fig. 1,
a total of 67 results were identified after an initial search of
the selected electronic databases. After screening the titles and
abstracts, 35 articles were selected for further review. Among
them, five studies were excluded for not referring to the
association between the EPHX1 gene polymorphism and COPD risk; two
were excluded since they were not case-control studies between the
EPHX1 gene polymorphism and COPD risk; and four were excluded due
to the data overlapping with that of another study. Thus, a total
of 24 studies were suitable for meta-analysis (9–32).
Among them, 11 studies were performed in Caucasian populations
(9,12,13,16,17,22,23,25,26,29,31),
12 studies were performed in Asian populations (10,11,14,15,18–21,27,28,30,32)
and one was performed in an African population (24). The article by Smith and Harrison had
two independent studies and thus these were considered separately
(9). The study by Chen et al
evaluated only the association between the EPHX1 113 T/C gene
polymorphism and COPD risk (27).
The total number of samples involved in the 24 eligible studies was
51,142, which included 8,259 COPD patients and 42,883 controls. The
HWE test had been calculated for the T113C and A139G polymorphisms
in the control groups for all the included 24 papers. For the T113C
locus, 15 studies (10,14,16,17,20,22–31)
obeyed the HWE and 10 (9,11–13,15,18,19,21,32)
deviated. For the A139G locus, with the exception of three studies
(18,21,28),
the remaining 21 studies (9–17,19,20,22–26,29–32,36)
obeyed the HWE. Among the overall studies, 17 (9,10,12,13,15,18–20,22–26,28,29,31)
further evaluated the putative EPHX1 enzyme activity and COPD risk
using the method described by Hassett et al(8). The characteristics of each study are
presented in Table I.
 | Table ICharacteristics of the studies
included in the meta-analysis. |
Table I
Characteristics of the studies
included in the meta-analysis.
| First author
(ref) | Year | Country | Ethnicity | Case | Control | Genotyping
Method |
|---|
| Smith CA (9) | 1997 | UK | Caucasian | 68 | 203 | PCR-RFLP |
| Smith CA (9) | 1997 | UK | Caucasian | 94 | 203 | PCR-RFLP |
| Takeyabu K
(10) | 2000 | Japan | Asian | 79 | 58 | Sequencing |
| Yim JJ (11) | 2000 | Korea | Asian | 83 | 76 | PCR-RFLP |
| Yoshikawa M
(30) | 2000 | Japan | Asian | 40 | 140 | PCR-RFLP |
| Rodriguez F
(12) | 2002 | Spain | Caucasian | 79 | 146 | PCR-RFLP and
SSCP |
| Zhang RB (20) | 2002 | China | Asian | 55 | 52 | PCR-RFLP |
| Budhi A (28) | 2003 | Japan | Asian | 63 | 172 | PCR-RFLP |
| Korytina GF
(13) | 2003 | Russia | Caucasian | 91 | 164 | PCR-RFLP |
| Park SS (32) | 2003 | Korea | Asian | 58 | 79 | PCR-RFLP |
| Cheng SL (14) | 2004 | China | Asian | 184 | 212 | PCR-RFLP |
| Xiao D (15) | 2004 | China | Asian | 100 | 100 | PCR-RFLP |
| Hersh CP (16) | 2005 | USA | Caucasian | 304 | 441 | Taqman |
| Park JY (29) | 2005 | USA | Caucasian | 131 | 262 | PCR-RFLP |
| Brøgger J (17) | 2006 | Norway | Caucasian | 248 | 244 | Taqman |
| Fu WP (18) | 2006 | China | Asian | 256 | 266 | Sequencing and
PCR-RFLP |
| Matheson MC
(31) | 2006 | Australia | Caucasian | 72 | 220 | ARMS |
| Vibhuti A (19) | 2007 | India | Asian | 202 | 136 | PCR-RFLP |
| Chappell S
(23) | 2008 | European
countries | Caucasian | 1,017 | 912 | Taqman |
| Zidzik J (22) | 2008 | Slovakia | Caucasian | 217 | 160 | PCR-RFLP |
| Lakhdar R (24) | 2009 | Tunisia | African | 234 | 182 | PCR-RFLP |
| Zheng Q (21) | 2009 | China | Asian | 80 | 87 | PCR-RFLP |
| Penyige A (25) | 2010 | Hungary | Caucasian | 272 | 301 | Taqman |
| Chen CZ (27) | 2011 | China | Asian | 105 | 103 | PCR-RFLP |
| Lee J (26) | 2011 | Denmark | Caucasian | 4,127 | 37,964 | Taqman |
Quantitative data synthesis
EPHX1 113 mutant homozygote and COPD
risk
After pooling the data from the 25 studies (with
Smith and Harrison’s considered as two separate studies) for
meta-analysis, the associations between the EPHX1 113 mutant
homozygote and heterozygote and the COPD risk were analyzed.
Summary ORs are shown in Table II.
The overall OR showed that the 113 mutant homozygote was
significantly associated with an increased risk of COPD (OR, 1.33;
95% CI, 1.06–1.69). Heterogeneity existed among the studies
(P<0.0001; I2, 73%), so a random-effect model was
used for the analysis (Fig. 2).
 | Table IISummary ORs for the association of
the EPHX1 113 mutant homozygote and heterozygote with COPD
risk. |
Table II
Summary ORs for the association of
the EPHX1 113 mutant homozygote and heterozygote with COPD
risk.
| Study group | N | 113CC vs. TT OR
(95% CI) | P-value | P-value for
heterogeneity | I2
(%) | 113CT vs. TT OR
(95% CI) | P-value | P-value for
heterogeneity | I2
(%) |
|---|
| Total | 25 | 1.33
(1.06–1.69) | 0.02 | <0.0001 | 73 | 1.12
(0.96–1.30) | 0.14 | <0.00001 | 69 |
| Ethnicity | | | | | | | | | |
| Asian | 12 | 1.07
(0.76–1.52) | 0.69 | 0.002 | 62 | 1.07
(0.69–1.65) | 0.77 | <0.00001 | 80 |
| Caucasian | 12 | 1.61
(1.12–2.31) | 0.01 | <0.0001 | 81 | 1.08
(0.96–1.22) | 0.20 | 0.10 | 37 |
| Sample size | | | | | | | | | |
| >200 | 19 | 1.54
(1.17–2.02) | 0.002 | <0.0001 | 78 | 1.16
(0.98–1.37) | 0.09 | <0.00001 | 75 |
| <200 | 6 | 0.80
(0.55–1.15) | 0.23 | 0.76 | 0 | 0.92
(0.64–1.33) | 0.67 | 0.35 | 11 |
| Smoking status | | | | | | | | | |
| Smokers | 14 | 1.11
(0.84–1.48) | 0.46 | 0.0005 | 64 | 1.06
(0.82–1.38) | 0.65 | <0.00001 | 77 |
| Non-smokers | 11 | 1.86
(1.14–3.04) | 0.01 | <0.00001 | 81 | 1.12
(0.93–1.35) | 0.23 | 0.06 | 44 |
| Controls in
HWE | 15 | 1.07
(0.86–1.34) | 0.52 | 0.004 | 56 | 0.98
(0.86–1.12) | 0.76 | 0.03 | 45 |
| Asian | 6 | 0.86
(0.52–1.43) | 0.57 | 0.04 | 56 | 0.75
(0.45–1.22) | 0.25 | 0.01 | 65 |
| Caucasian | 8 | 1.09
(0.85–1.39) | 0.51 | 0.03 | 55 | 1.02
(0.93–1.12) | 0.71 | 0.28 | 18 |
In the analysis stratified by ethnicity, the study
populations were divided into two subgroups, one comprising Asian
individuals (12 studies) and the comprising Caucasian individuals
(12 studies). The pooled ORs showed a significant association
between the 113 mutant homozygote and COPD risk in the Caucasian
subgroup (OR, 1.61; 95% CI, 1.12–2.31), but this was not
significant in Asian subgroup (OR, 1.07; 95% CI, 0.76–1.52). For
sample size, the studies were stratified into two subgroups, one
comprising studies with >200 subjects and one with <200
subjects. In these subgroups, a significant association between the
113 mutant homozygote and COPD risk was observed in the >200
subject subgroup (OR, 1.54, 95% CI, 1.17–2.02), but not in the
<200 subgroup (OR, 0.80, 95% CI, 0.55–1.15; Table II).
For case-control studies, a deviation from the HWE
of the control groups may lead to an erroneous result and imply
insufficient random sampling. Thus the 15 studies in which the
controls were in HWE were analyzed. Six studies were in Asian
populations, eight populations were Caucasian and one was African.
There were significant heterogeneities among the populations
(P=0.004; I2, 56%). The pooled OR by the random-effects
model showed no significant association between the 113 mutant
homozygote and COPD risk (OR, 1.07; 95% CI, 0.86–1.34). When these
15 studies were stratified by ethnicity, no statistically
significant associations were detected (Asian: OR, 0.86; 95% CI,
0.52–1.43; Caucasian: OR, 1.09; 95% CI, 0.85–1.39).
Analysis stratified by the cigarette smoking status
of the controls was performed. The controls were strategically
classified as current smokers, ex-smokers and non-smokers. A
moderate difference was observed between the cigarette smokers and
non-smokers (Table II). The OR for
the 113 mutant homozygote vs. wildtype homozygote in non-smokers
was 1.86 (95% CI, 1.14–3.04) and 1.11 (95% CI, 0.84–1.48) in
smokers.
EPHX1 113 mutant heterozygote and COPD
risk
In the genetic model of the 113 mutant heterozygote
vs. wildtype homozygote, 25 studies were pooled by the
random-effects model since heterogenetity existed among the studies
(P<0.0001; I2, 69%). Overall, the OR was 1.12 (95%
CI, 0.96–1.30) and was not significant (Table II), which suggested that the 113
mutant heterozygote was not associated with COPD risk. Analyses
stratified by ethnicity, sample size, smoking status of the
controls and HWE violation of the controls were performed to
further examine the association between the 113 mutant heterozygote
and COPD risk. The results are presented in Table III. All the results of the
stratified analyses were consistent with the main analysis.
 | Table IIISummary ORs for the association of
the putative EPHX1 enzyme activity (extremely slow and slow enzyme
activity) with COPD risk. |
Table III
Summary ORs for the association of
the putative EPHX1 enzyme activity (extremely slow and slow enzyme
activity) with COPD risk.
| Study group | N | Extremely slow vs.
normal OR (95% CI) | P-value | P-value for
heterogeneity | I2
(%) | Slow vs. normal OR
(95% CI) | P-value | P-value for
heterogeneity | I2
(%) |
|---|
| Total | 15 | 1.77
(1.23–2.55) | 0.002 | <0.0001 | 76 | 1.44
(1.13–1.85) | 0.004 | <0.00001 | 74 |
| Ethnicity | | | | | | | | | |
| Asian | 6 | 1.14
(0.84–1.54) | 0.40 | 0.95 | 0 | 1.41
(0.90–2.19) | 0.13 | 0.03 | 59 |
| Caucasian | 8 | 2.64
(1.30–5.38) | 0.007 | <0.0001 | 85 | 1.31
(1.01–1.71) | 0.04 | 0.01 | 62 |
| Sample size | | | | | | | | | |
| >200 | 13 | 1.90
(1.25–2.88) | 0.002 | <0.0001 | 79 | 1.44
(1.11–1.88) | 0.007 | <0.00001 | 77 |
| <200 | 2 | 1.21
(0.65–2.24) | 0.55 | 0.90 | 0 | 1.39
(0.66–2.92) | 0.38 | 0.47 | 0 |
| Smoking status | | | | | | | | | |
| Smokers | 6 | 1.04
(0.77–1.41) | 0.80 | 0.73 | 0 | 1.49
(1.01–2.21) | 0.04 | 0.04 | 58 |
| Non-smokers | 9 | 2.95
(1.56–5.56) | 0.00008 | <0.00001 | 85 | 1.42
(1.04–1.93) | 0.03 | 0.003 | 73 |
| Controls in
HWE | 8 | 1.35
(0.93–1.96) | 0.11 | 0.01 | 61 | 1.04
(0.96–1.12) | 0.36 | 0.0005 | 73 |
| Asian | 3 | 1.12
(0.68–1.84) | 0.66 | 0.91 | 0 | 0.87
(0.51–1.47) | 0.60 | 0.13 | 51 |
| Caucasian | 4 | 1.18
(0.75–1.86) | 0.47 | 0.09 | 53 | 1.01
(0.94–1.09) | 0.72 | 0.14 | 45 |
EPHX1 139 mutant homozygote and COPD
risk
A total of 24 studies detected an association
between the 139 mutant homozygote and COPD risk. However,
meta-analysis of these studies did not suggest an association
between the 139 mutant homozygote and COPD risk (OR, 0.90; 95% CI,
0.79–1.02) and there was no heterogeneity among the studies (P=
0.67; I2, 0%). Stratifying by ethnicity, sample size,
smoking status of the controls and HWE violation of the controls
also showed no association.
EPHX1 139 mutant heterozygote and COPD
risk
A total of 24 studies detected the association
between the 139 mutant heterozygote and COPD risk. The overall OR
showed that the 139 mutant heterozygote was not significantly
associated with COPD risk (OR, 0.96; 95% CI, 0.91–1.01) and there
was a low heterogeneity among the studies (P= 0.15; I2,
3%). Stratification by ethnicity demonstrated that there was no
significant association in Caucasian populations (OR, 0.99; 95% CI,
0.88–1.11) but revealed a statistically marginally significant
association in Asian populations (OR, 0.82; 95% CI, 0.68–0.99).
Putative EPHX1 enzyme activity and
COPD risk
In order to evaluate the association of these two
functional polymorphisms and their enzyme activity with COPD risk,
the association of EPHX1 enzyme activity predicted by the genotype
combination of T113C and A139G polymorphisms with COPD risk was
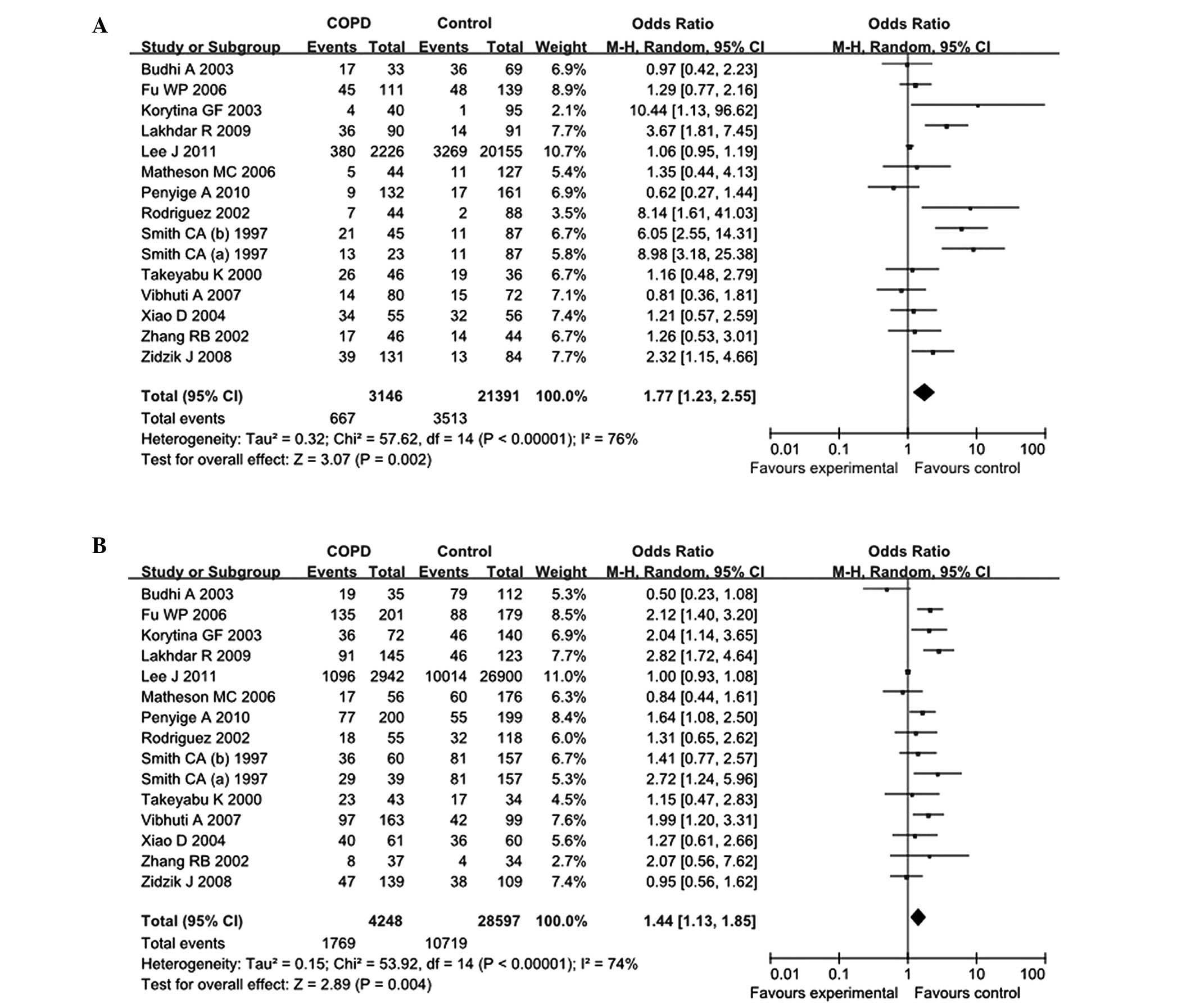
analyzed. In the overall comparisons with the putative normal EPHX1
enzyme activity, the extremely slow EPHX1 enzyme activity (OR,
1.77; 95% CI, 1.23–2.55) and slow EPHX1 enzyme activity (OR, 1.44;
95% CI, 1.13–1.85) increased the COPD risk significantly, while the
fast EPHX1 enzyme activity did not affect the COPD risk (OR, 1.03;
95% CI, 0.87–1.21; Table III and
Fig. 3).
In the analyses stratified by ethnicity, the
increased COPD risk of extremely slow and slow EPHX1 enzyme
activity was observed in the Caucasian populations (extremely slow
enzyme activity: OR, 2.64; 95% CI, 1.30–5.38; slow enzyme activity:
OR, 1.31; 95% CI, 1.01–1.71), but not in the Asian populations
(extremely slow enzyme activity: OR, 0.1.14; 95% CI, 0.84–1.54;
slow enzyme activity: OR, 1.41; 95% CI, 0.90–2.19). In the further
assessments of the analyses stratified by sample size, smoking
status of the controls and HWE violation of the controls, the
extremely slow enzyme activity showed an increased COPD risk in the
>200 subjects subgroup (OR, 1.90; 95% CI, 1.25–2.88) and the
subgroup with the non–smokers as controls (OR, 2.95; 95% CI,
1.56–5.56). For the stratified analyses using the genetic model of
slow enzyme activity vs. normal enzyme activity, the increased COPD
risk was also observed in the >200 subjects subgroup (OR, 1.44;
95% CI, 1.11–1.88). Notably, the increased COPD risk of the slow
enzyme activity was observed not only in the subgroup with the
non-smokers as controls (OR, 1.42; 955CI=1.04–1.93) but also in the
subgroup with the smokers and ex-smokers as controls (OR, 1.49; 95%
CI, 1.01–2,21; Table III).
Publication bias
The Begg’s funnel plots and Egger’s tests were
performed to assess the potential publication bias. The shape of
the Begg’s funnel plots appeared to be symmetrical in the 113
mutant homozygote vs. wildtype homozygote (113 CC vs. TT) genetic
model (Fig. 4). Neither the Begg’s
test (P= 0.907) nor Egger’s test (P= 0.158) indicated any
statistically significantly evidence of publication bias. Moreover,
evaluations of publication bias for the other genetic models did
not reveal any significant results (data not shown).
Discussion
The EPHX1 gene encodes a xenobiotic metabolizing
enzyme that detoxifies the reactive epoxides produced by smoking to
form water and soluble dihydrodiol compounds and is widely
expressed in the bronchial epithelium. Thus it has been a
significant focus for studies concerning genetic susceptibility to
COPD over the past decade. The genetic polymorphisms T113C and
A139G in the coding regions have been observed to alter the enzyme
activity and thus, are considered to affect the individual’s
susceptibility to COPD or risk of developing COPD. However, studies
of genetic polymorphisms of EPHX1 in COPD have provided different
and inconclusive results.
The explanations for this may be the different
ethnic populations used in the various studies, relatively small
sample sizes lacking the statistical power to produce a reliable
conclusion for any individual study or mismatching for age, gender
and smoking history in the controls. A perfect COPD genetic
association study would be large and longitudinal, using the
cumulative reduction in lung function in relation to cumulative
smoking as a main outcome measure. Such a study would take decades
to complete and require tens of thousands of subjects. Thus, to
avoid these error sources, meta-analysis studies appear to be an
essential tool for summarizing case-control studies.
The present meta-analysis provides the most
comprehensive and up-to-date evidence on the EPHX1 genetic
polymorphisms T113C and A139G, enzyme activity resulting from these
two genetic polymorphisms and the risk of developing COPD. A total
of 25 case-control association studies with 8,259 COPD patients and
42,883 controls were included in the present meta-analysis. Through
systematic analyses, the 113 mutant homozygote was demonstrated to
be significantly associated with an increased risk of COPD. This
result contrasted with the study by Brøgger et al, in which
a reduced risk of COPD was detected in the 113 mutant homozygotes
(17). The analyses stratified by
ethnicity showed the significant increase in risk existed only in
the Caucasian populations and not in the Asian populations. These
results were the opposite of those of the study by Hu et al,
in which an increased risk of COPD was observed in Asian
populations, not in Caucasian populations (33). The results are also inconsistent
with the study by Lee et al, in which the 113 mutant
homozygote was not associated with CODP either in Asian or
Caucasian populations (26). The
reasons for this discrepancy may be the different studied
populations and inclusion criteria of COPD phenotypes in the
previous meta-analyses. As shown in Table II, the association was only
demonstrated in analyses stratified by combining studies with a
sample size >200 subjects, which emphasized the importance of
having sufficient power with a large enough sample size. In the
analyses stratified by the cigarette smoking status of the
controls, a modest difference in the risk of COPD was detected
between cigarette smokers and non-smokers. Notably, the increased
risk of COPD was only detected in the studies with non-smokers as
controls. As is known, smoking is the main risk factor for COPD and
associated types of cancer, such as lung cancer. However, these
diseases are the outcome of genes and environmental interactions
and the pathogeneses are complex and remain unclear. In a previous
study, these T113C and A139G polymorphisms of the EPHX1 gene varied
from being risk factors to protective factors according to the
cigarette smoking status of the lung cancer patients, whereby the
low activity genotype of the EPHX1 gene was a risk factor for
non-smokers, but a protective factors for heavy smokers (41). In the study by Xiao et al,
the authors also noted that when the COPD patients were
non-smokers, the low activity genotype was a risk factor, but
became a protective factor when the COPD patients were smokers
(15). Thus in the present study,
whether the observed association was real or spurious could not be
decided due to the lack of detailed data, such as the number of
packs smoked per year and smoking history of the COPD patients and
controls in the included studies. To confirm this susceptibility
association, more studies containing the above information for
patients and controls are required in the future. The 113 mutant
heterozygote was not associated with an increased COPD risk. The
139 mutant heterozygote was significantly associated with a
decreased risk of COPD in Asian populations, but not in the
Caucasian population.
When evaluating the EPHX1 enzyme activity and the
risk of COPD, the extremely slow activity phenotype and slow
activity phenotype of EPHX1 were revealed to be significantly
associated with an increased risk of COPD. The stratified analyses
demonstrated the association in Caucasian populations, studies with
>200 subjects and studies with non-smokers as the controls.
These findings are consistent with the known role of the EPHX1
enzyme in the detoxification of harmful epoxides from smoking and
its involvement in the first-pass metabolism of smoking-induced
highly reactive epoxide intermediates (42). Regardless of the 113 mutant
homozygote, extremely slow activity or slow activity, they all
exhibited decreased EPHX1 enzyme activity and thus increased the
risk of developing COPD in populations. In summary, these results
suggest that the two polymorphisms, T113C and A139G, of EPHX1 were
associated with the risk of COPD not only in statistical
calculations, but also at physiological function levels.
Although meta-analysis is a powerful statistical
method, the inherent limitations of the present study should be
addressed. Thus, in interpreting the results, caution should be
taken. First, relatively large heterogeneity existed in the 113
mutant homozygote vs. wildtype homozygote meta-analysis. Analyses
stratified by ethnicity, sample size, cigarette smoking status of
the controls and HWE violation of the controls did not
significantly reduce the heterogeneity. The heterogeneity may be
caused by the different criteria in the selection of COPD patients
and controls, including age and gender distributions and lifestyle
factors. However, the lack of original data for the included
studies limited any further evaluation. Second, only studies
written in English and Chinese were included and related studies in
other languages were not included, which may bias the conclusion of
the present study. Third, publication bias may not be excluded,
although the tests showed negative results. Studies reporting
significant associations would be more readily published, while
studies with non-significant associations would be more difficult
to publish. Fourth, although half of the included studies were
performed in Asian populations, the total size of the Asian
populations was relatively small compared with the Caucasian
populations. In the analyses stratified by ethnicity, the detected
associations were only demonstrated in the Caucasian populations,
not in the Asian populations. As the scale of the effect of EPHX1
variants on the risk of COPD may depend on ethnicity (43), more studies performed in Asian
populations are required to test the present conclusions. Fifth,
gene-gene and gene-environment interactions may affect an
individual’s susceptibility to COPD. In a complex polygenic disease
such as COPD, it is likely that multiple genes are operating and
the genetic susceptibility may be dependent on the coincidence of
several genetic polymorphisms acting together. The polymorphism of
each gene may therefore confer only a small relative risk of COPD
and it is likely that the coincidence of numerous polymorphisms are
important in its pathogenesis, although there was not enough data
to eliminate these interfering factors. In addition, although
smoking is a significant risk factor for developing COPD, only a
limited amount of detailed information such as packs per year and
smoking history, was available in the studies so the corresponding
stratified analyses were not performed. These may bias the present
conclusions. Well-designed studies containing this information are
required. Sixth, as the majority of studies did not mention
potential population stratification of patients and controls, it is
not possible to rule out a role for population structure in the
observed association.
In conclusion, the present meta-analysis is the most
comprehensive and up-to-date appraisal of the two EPHX1 genetic
polymorphisms, T113C and A139G, enzyme activity and the risk of
developing COPD. The results indicated that they were genetic
factors for the susceptibility of an individual to COPD. Further
well-designed studies of various ethnic populations should be
performed to evaluate these associations.
Acknowledgements
The authors would like to acknowledge
Dr Liang Xie’s help in revising the manuscript draft. This study
was supported by grants from the National Natural Science
Foundation of China (31000955, 30900812 and 90919007) and from the
Science and Technology Fund of Southeast University
(KJ2010439).
References
|
1
|
Pauwels RA, Buist AS, Calverley PM,
Jenkins CR and Hurd SS; GOLD Scientific Committee: Global strategy
for the diagnosis, management, and prevention of chronic
obstructive pulmonary disease. NHLBI/WHO Global Initiative for
Chronic Obstructive Lung Disease (GOLD) Workshop summary. Am J
Respir Crit Care Med. 163:1256–1276. 2001. View Article : Google Scholar
|
|
2
|
Fabbri L, Caramori G, Beghe B, Papi A and
Ciaccia A: Chronic obstructive pulmonary disease international
guidelines. Curr Opin Pulm Med. 4:76–84. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Løkke A, Lange P, Scharling H, Fabricius P
and Vestbo J: Developing COPD: a 25 year follow up study of the
general population. Thorax. 61:935–939. 2006.PubMed/NCBI
|
|
4
|
Silverman EK, Chapman HA, Drazen JM, et
al: Genetic epidemiology of severe, early-onset chronic obstructive
pulmonary disease. Risk to relatives for airflow obstruction and
chronic bronchitis. Am J Respir Crit Care Med. 157:1770–1778. 1998.
View Article : Google Scholar
|
|
5
|
Foreman MG, DeMeo DL, Hersh CP, Reilly JJ
and Silverman EK: Clinical determinants of exacerbations in severe,
early-onset COPD. Eur Respir J. 30:1124–1130. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Oesch F, Glatt H and Schmassmann H: The
apparent ubiquity of epoxide hydratase in rat organs. Biochem
Pharmacol. 26:603–607. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Omiecinski CJ, Aicher L, Holubkov R and
Checkoway H: Human peripheral lymphocytes as indicators of
microsomal epoxide hydrolase activity in liver and lung.
Pharmacogenetics. 3:150–158. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hassett C, Aicher L, Sidhu JS and
Omiecinski CJ: Human microsomal epoxide hydrolase: genetic
polymorphism and functional expression in vitro of amino acid
variants. Hum Mol Genet. 3:421–428. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Smith CA and Harrison DJ: Association
between polymorphism in gene for microsomal epoxide hydrolase and
susceptibility to emphysema. Lancet. 350:630–633. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takeyabu K, Yamaguchi E, Suzuki I,
Nishimura M, Hizawa N and Kamakami Y: Gene polymorphism for
microsomal epoxide hydrolase and susceptibility to emphysema in a
Japanese population. Eur Respir J. 15:891–894. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yim JJ, Park GY, Lee CT, et al: Genetic
susceptibility to chronic obstructive pulmonary disease in Koreans:
combined analysis of polymorphic genotypes for microsomal epoxide
hydrolase and glutathione S-transferase M1 and T1. Thorax.
55:121–125. 2000. View Article : Google Scholar
|
|
12
|
Rodriguez F, Jardi R, Costa X, et al:
Detection of polymorphisms at exons 3 (Tyr113-->His) and 4
(His139-->Arg) of the microsomal epoxide hydrolase gene using
fluorescence PCR method combined with melting curves analysis. Anal
Biochem. 308:120–126. 2002.PubMed/NCBI
|
|
13
|
Korytina GF, Ianbaeva DG and Viktorova TV:
Role of polymorphic variants of cytochrome P450 genes (CYP1A1,
CYP2E1) and microsomal epoxide hydrolase (mEPHX) in pathogenesis of
cystic fibrosis and chronic respiratory tract diseases. Mol Biol
(Mosk). 37:784–792. 2003.(In Russian).
|
|
14
|
Cheng SL, Yu CJ, Chen CJ and Yang PC:
Genetic polymorphism of epoxide hydrolase and glutathione
S-transferase in COPD. Eur Respir J. 23:818–824. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xiao D, Wang C, Du MJ, et al: Relationship
between polymorphisms of genes encoding microsomal epoxide
hydrolase and glutathione S-transferase P1 and chronic obstructive
pulmonary disease. Chin Med J (Engl). 117:661–667. 2004.
|
|
16
|
Hersh CP, Demeo DL, Lange C, et al:
Attempted replication of reported chronic obstructive pulmonary
disease candidate gene associations. Am J Respir Cell Mol Biol.
33:71–78. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Brøgger J, Steen VM, Eiken HG, Gulsvik A
and Bakke P: Genetic association between COPD and polymorphisms in
TNF, ADRB2 and EPHX1. Eur Respir J. 27:682–688. 2006.PubMed/NCBI
|
|
18
|
Fu WP, Sun C, Dai LM, Yang LF and Zhang
YP: Relationship between COPD and polymorphisms of HOX-1 and mEPH
in a Chinese population. Oncol Rep. 17:483–488. 2007.PubMed/NCBI
|
|
19
|
Vibhuti A, Arif E, Deepak D, Singh B and
Qadar Pasha MA: Genetic polymorphisms of GSTP1 and mEPHX correlate
with oxidative stress markers and lung function in COPD. Biochem
Biophys Res Commun. 359:136–142. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang RB, Zhang AZ, He QY and Lu BB:
Microsomal epoxide hydrolase gene polymorphism and susceptibility
to chronic obstructive pulmonary disease in Han nationality of
North China. Zhonghua Nei Ke Za Zhi. 41:11–14. 2002.(In
Chinese).
|
|
21
|
Zheng Q and Zheng W: The association
between the microsomal epoxide hydrolase gene polymorphism and
host’s susceptibility to COPD. Chin J Prim Med Pham (Chin).
16:1779–1780. 2009.(In Chinese).
|
|
22
|
Zidzik J, Slabá E, Joppa P, et al:
Glutathione S-transferase and microsomal epoxide hydrolase gene
polymorphisms and risk of chronic obstructive pulmonary disease in
Slovak population. Croat Med J. 49:182–191. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chappell S, Daly L, Morgan K, et al:
Genetic variants of microsomal epoxide hydrolase and
glutamate-cysteine ligase in COPD. Eur Respir J. 32:931–937. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lakhdar R, Denden S, Knani J, et al:
Microsomal epoxide hydrolase gene polymorphisms and susceptibility
to chronic obstructive pulmonary disease in the Tunisian
population. Genet Test Mol Biomarkers. 14:857–863. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Penyige A, Poliska S, Csanky E, et al:
Analyses of association between PPAR gamma and EPHX1 polymorphisms
and susceptibility to COPD in a Hungarian cohort, a case-control
study. BMC Med Genet. 11:1522010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee J, Nordestgaard BG and Dahl M: EPHX1
polymorphisms, COPD and asthma in 47,000 individuals and in
meta-analysis. Eur Respir J. 37:18–25. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen CZ, Wang RH, Lee CH, Lin CC, Chang HY
and Hsiue TR: Polymorphism of microsomal epoxide hydrolase is
associated with chronic obstructive pulmonary disease and
bronchodilator response. J Formos Med Assoc. 110:685–689. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Budhi A, Hiyama K, Isobe T, et al: Genetic
susceptibility for emphysematous changes of the lung in Japanese.
Int J Mol Med. 11:321–329. 2003.PubMed/NCBI
|
|
29
|
Park JY, Chen L, Wadhwa N and Tockman MS:
Polymorphisms for microsomal epoxide hydrolase and genetic
susceptibility to COPD. Int J Mol Med. 15:443–448. 2005.PubMed/NCBI
|
|
30
|
Yoshikawa M, Hiyama K, Ishioka S, Maeda H,
Maeda A and Yamakido M: Microsomal epoxide hydrolase genotypes and
chronic obstructive pulmonary disease in Japanese. Int J Mol Med.
5:49–53. 2000.PubMed/NCBI
|
|
31
|
Matheson MC, Raven J, Walters EH, Abramson
MJ and Ellis JA: Microsomal epoxide hydrolase is not associated
with COPD in a community-based sample. Hum Biol. 78:705–717. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Park SS, Kim EJ, Son CY, et al: Genetic
polymorphism of epoxide hydrolase and GSTM1 in chronic obstructive
pulmonary disease. Tuberc Respir Dis (Seoul). 55:88–97. 2003.
|
|
33
|
Hu G, Shi Z, Hu J, Zou G, Peng G and Ran
P: Association between polymorphisms of microsomal epoxide
hydrolase and COPD: results from meta-analyses. Respirology.
13:837–850. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cochran WG: The combination of estimates
from different experiments. Biometrics. 10:101–129. 1954.
View Article : Google Scholar
|
|
35
|
Kavvoura FK and Ioannidis JP: Methods for
meta-analysis in genetic association studies: a review of their
potential and pitfalls. Hum Genet. 123:1–14. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Colhoun HM, McKeigue PM and Davey Smith G:
Problems of reporting genetic associations with complex outcomes.
Lancet. 361:865–872. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mantel N and Haenszel W: Statistical
aspects of the analysis of data from retrospective studies of
disease. J Natl Cancer Inst. 22:719–748. 1959.PubMed/NCBI
|
|
38
|
DerSimonian R and Laird N: Meta-analysis
in clinical trials. Control Clin Trials. 7:177–188. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Begg CB and Mazumdar M: Operating
characteristics of a rank correlation test for publication bias.
Biometrics. 50:1088–1101. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhou W, Thurston SW, Liu G, et al: The
interaction between microsomal epoxide hydrolase polymorphisms and
cumulative cigarette smoking in different histological subtypes of
lung cancer. Cancer Epidemiol Biomarkers Prev. 10:461–466.
2001.
|
|
42
|
Maekawa K, Itoda M, Hanioka N, et al:
Non-synonymous single nucleotide alterations in the microsomal
epoxide hydrolase gene and their functional effects. Xenobiotica.
33:277–287. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Smolonska J, Wijmenga C, Postma DS and
Boezen HM: Meta-analyses on suspected chronic obstructive pulmonary
disease genes: a summary of 20 years’ research. Am J Respir Crit
Care Med. 180:618–631. 2009.
|