Introduction
Circulating DNA in serum or plasma is increasingly
being recognized as a biomarker for cancer as it has been shown to
bear the same genetic and epigenetic changes as the tumor tissues,
indicating the possibility of creating minimally invasive
diagnostic tests based on tumor-specific DNA markers (1,2).
Circulating DNA exists in plasma/serum as free unbound DNA, DNA
complexed with histones as nucleosomes or DNA included in apoptotic
bodies (3). Associating with
proteins protects DNA against digestion by plasma and serum
nucleases (4). The results of
previous studies showed that patients with various types of tumors
had DNA fragments mainly of 150–200, 400, 600 and 800 bp as mono-
and oligonucleosomes (5–7).
Previous studies have investigated circulating
nucleosomes (cNUCs) for their potential as diagnostic and
prognostic biomarkers or usefulness in therapy monitoring (for
review see ref. 3). The results of
these studies have revealed that, although cancer patients have a
generally higher level of cNUCs compared to healthy individuals,
its diagnostic value is limited as various benign diseases were
also often associated with an elevated serum level of nucleosomes.
The prognostic value of pretherapeutic nucleosome concentrations
has been demonstrated in univariate analyses (8,9). cNUCs
have been shown to be valuable for monitoring for the early
estimation of efficacy of cytotoxic cancer therapy (3).
Another potential use of cNUCs involves its utility
as a diagnostic modality in disease-associated quantitative changes
of modified histone tails in blood circulation. In a previous
study, we showed that methylated histone marks may be detected on
cNUCs (10). In a subsequent study,
we focused on two methyl marks, the trimethylation of H3 lysine 9
(H3K9me3) and H4 lysine 20 (H4K20me3), which are hallmarks of
pericentric heterochromatin. Of these methyl marks, H4K20me3 was
previously reported to be reduced in certain primary tumors and
tumor cell lines (11). Our
previous results have provided evidence that H3K9me3 may be reduced
in the circulating plasma of patients with colorectal cancer (CRC)
when compared with healthy subjects or patients with multiple
myeloma (MM) (12). In the present
study, our aim was to analyze the correlation between cNUCs and two
histone methyl marks.
Materials and methods
Study population
The study group comprised patients with
histologically confirmed CRC (N=25), MM (N=17) and healthy
volunteers (N=15). Blood samples were obtained from CRC patients at
surgery and from patients with MM prior to chemotherapy. The blood
plasma was immediately separated from the cells using
Ficoll-gradient centrifugation and stored in aliquots at −80°C.
Patient characteristics with regard to age and gender are shown in
Table I and the clinical
characteristics of the CRC patients are shown in Table II. The study was approved by the
Institutional Review Board of the Institute of Oncology of Istanbul
University.
 | Table ICharacteristics of the study
population. |
Table I
Characteristics of the study
population.
| | Gender | Age (years) |
|---|
| |
|
|
|---|
| N | Male | Female | Mean | Range |
|---|
| Controls | 15 | 8 | 7 | 41 | 24–65 |
| Colorectal
cancer | 25 | 15 | 10 | 64 | 48–83 |
| Multiple myeloma | 17 | 6 | 11 | 62 | 46–82 |
 | Table IIClinical characteristics of colorectal
cancer and multiple myeloma patients. |
Table II
Clinical characteristics of colorectal
cancer and multiple myeloma patients.
| N |
|---|
| Colorectal
cancer |
| Tumor
localization |
| Colon | 8 |
| Rectum | 17 |
| Stage (UICC) |
| I–II | 7 |
| III–IV | 18 |
| Multiple myeloma |
| Stage
(Durie-Salmon) |
| I–II | 7 |
| III | 10 |
Quantitation of cNUCs
The concentrations of cNUCs in blood plasma were
determined using the Cell-Death Detection ELISA kit (Roche
Diagnostics, Mannheim, Germany), as previously reported (10). Briefly, we applied 20 μl of plasma
twice and the mean signal values, measured in optical density (OD),
were considered to be the relative plasma concentrations.
Chromatin immunoprecipitation (ChIP) from
blood plasma
The ChIP assay was performed as previously reported
by our laboratory (12). Briefly,
agarose beads were blocked with BSA and, following washing, the
beads were pre-incubated with antibodies against the H3K9me3 and
H4K20me3 (Millipore, Temecula, CA, USA) for 4 h at 4°C.
Subsequently, 200 μl of plasma was diluted into 800 μl of the ChIP
dilution buffer and was then added to the pelleted agarose beads
that were pre-incubated with antibodies. Following overnight
incubation at 4°C, the beads were washed with low salt, high salt,
LiCl and Tris/EDTA buffers. Finally, the chromatin was eluted by
incubating the beads at 65°C and proteins were removed by treatment
with proteinase K. ChIP DNA was then purified using an appropriate
purification kit and stored at −20°C.
Quantitative real-time PCR (qPCR)
H3K9me3- or H4K20me3-related ChIP plasma DNA was
amplified using qPCR with satellite 2 as the target sequence, as
previously reported (12). The PCR
amplifications were performed twice and the mean values were
calculated. The comparative ΔCt method was used for quantitation. A
linear standard curve was generated through serial dilutions of
human genomic DNA with a linear amplification (correlation
coefficient = 0.99). These dilution series were co-amplified in
each PCR session and the relative concentration of H3K9me3- or
H4K20me3-related nucleosomal DNA from a given sample was derived
from the cross-over threshold (Ct) values using this dilution
standard.
Statistical analysis
The Pearson’s correlation test was used to evaluate
the correlation between cNUCs and histone methyl marks. The
differences between the respective nucleosomes or histone methyl
marks of the groups were compared in a univariate analysis using
the Mann-Whitney U test. P<0.05 was considered to indicate a
statistically significant result.
Results and Discussion
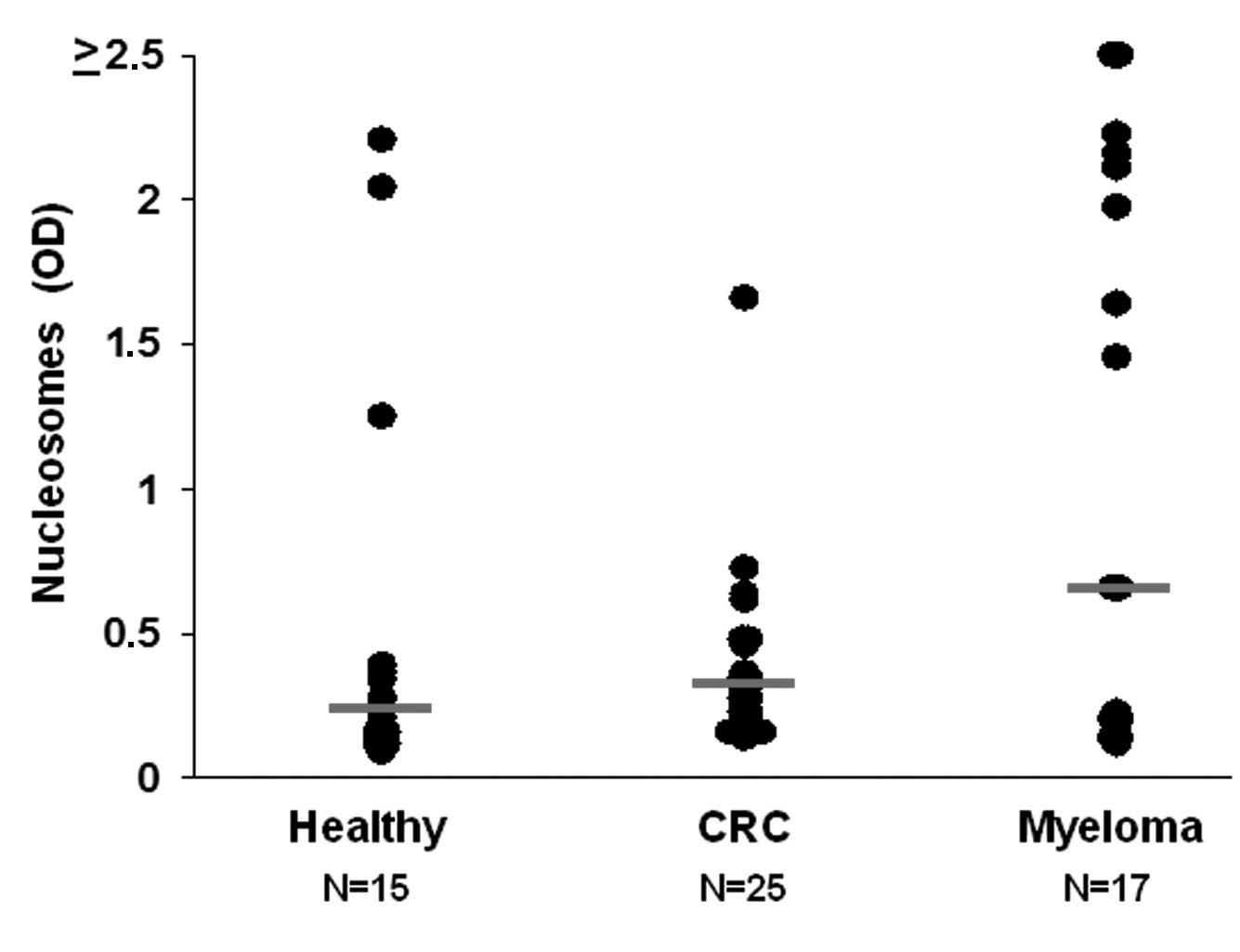
We measured the concentration of cNUCs in the plasma
of individual study participants (Fig.
1A) and detected a high variation, with a range of 0.098–2.5.
The healthy controls had the lowest relative levels of cNUCs
(median, 0.194), the CRC patients intermediate (median, 0.25) and
the MM patients the highest levels (median, 0.648). However, the
difference between the healthy individuals and the CRC patients did
not reach statistical significance (p=0.5), while for the
difference between the healthy group and MM patients there was a
trend towards significance (p=0.075). The finding that CRC and MM
patients have higher levels of cNUCs than healthy subjects is
consistent with previous reports (3,13).
ChIP assays were performed to precipitate H3K9me3
and H4K20me3-related cNUCs and qPCR to quantitate satellite 2. The
number of methyl marks should be normalized since the levels of
cNUCs vary highly between individual samples. The method of data
normalization has a major impact on the quality of ChIP analyses
(14). In ChIP assays within the
context of cell or tissues, ‘input’ controls (e.g., total
chromatin) or pan antibodies for histones (e.g., histone H3 or H4)
are used to normalize the target protein or modification (15). This process, however, is not
appropriate for blood fluids (serum, plasma) as not all the
circulating DNA is associated with histones in the form of
nucleosomes (3). Thus, using the
total plasma DNA for normalization may lead to inaccurate results.
Similarly, the pool of cNUCs is likely to include modified and
unmodified forms of the histone residue of interest, hampering the
normalization of the target modification. Therefore, we considered
total nucleosomes for normalizing the circulating levels of methyl
marks.
To ascertain whether cNUCs are suitable for that
purpose we first determined whether there was a correlation between
cNUCs and histone marks. Figs. 1B and
C show the correlation curves for the whole study group (N=57).
Analysis with the Pearson’s test revealed a significant positive
correlation between cNUCs and H3K9me3 or H4K20me3 (p<0.001 for
both histone marks). This correlation indicates that histone marks
may be normalized using the values of cNUCs. However, when we
studied the correlation between cNUCs and histone marks in
individual study groups, the correlation between cNUCs and H3K9me3
in the CRC patients was found to deviate from that of the whole
study group and was at the limit of significance (p=0.046). This
reveals that the amount of H3K9me3 in circulation may be modified
in CRC patients.
Figs. 2A and B show
H3K9me3 and H4K20me3 levels normalized by cNUCs, respectively. In
agreement with the weak correlation between cNUCs and H3K9me3 in
CRC patients, H3K9me3 levels were lowest in this group (0.047),
whereas the values were 0.06 in healthy subjects and 0.2 in MM
patients. The difference between the healthy group and MM patients
was, however, not significant (p=0.38). The distribution of H3K9me3
in the CRC patients was different from that of the MM patients
(p=0.044). For H4K20me3, the median values were 0.022 in the
healthy subjects, 0.052 in the CRC patients (p=0.07) and 0.056 in
the MM patients (p=0.36).
In conclusion, this pilot study is the first to
assess the correlation between cNUCs and histone methyl marks. Our
findings indicate that there is a marked positive correlation
between these parameters. In addition, the normalizing circulating
levels of H3K9me3 and H4K20me3 confirm our previous finding that
H3K9me3 may be reduced in CRC patients (12). Studies with larger sample sizes are
required to confirm and validate the potential of these findings in
CRC.
Acknowledgements
This study was supported by the Scientific Research
Projects Coordination Unit of Istanbul University (project number
17758).
References
|
1
|
Beck J, Urnovitz HB, Mitchell WM and
Schütz E: Next generation sequencing of serum circulating nucleic
acids from patients with invasive ductal breast cancer reveals
differences to healthy and nonmalignant controls. Mol Cancer Res.
8:335–342. 2010. View Article : Google Scholar
|
|
2
|
Vlassov VV, Laktionov PP and Rykova EY:
Circulating nucleic acids as a potential source for cancer
biomarkers. Curr Mol Med. 10:142–165. 2010.PubMed/NCBI
|
|
3
|
Holdenrieder S and Stieber P: Clinical use
of circulating nucleosomes. Crit Rev Clin Lab Sci. 46:1–24. 2009.
View Article : Google Scholar
|
|
4
|
Ng EK, Tsui NB, Lam NY, Chiu RW, Yu SC,
Wong SC, Lo ES, Rainer TH, Johnson PJ and Lo YM: Presence of
filterable and nonfilterable mRNA in the plasma of cancer patients
and healthy individuals. Clin Chem. 48:1212–1217. 2002.PubMed/NCBI
|
|
5
|
Giacona MB, Ruben GC, Iczkowski KA, Roos
TB, Porter DM and Sorenson GD: Cell-free DNA in human blood plasma:
length measurements in patients with pancreatic cancer and healthy
controls. Pancreas. 17:89–97. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jahr S, Hentze H, Englisch S, Hardt D,
Fackelmayer FO, Hesch RD and Knippers R: DNA fragments in the blood
plasma of cancer patients: quantitations and evidence for their
origin from apoptotic and necrotic cells. Cancer Res. 61:1659–1665.
2001.PubMed/NCBI
|
|
7
|
Deligezer U, Yaman F, Erten N and Dalay N:
Frequent copresence of methylated DNA and fragmented nucleosomal
DNA in plasma of lymphoma patients. Clin Chim Acta. 335:89–94.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Holdenrieder S, Stieber P, von Pawel J,
Raith H, Nagel D, Feldmann K and Seidel D: Circulating nucleosomes
predict the response to chemotherapy in patients with advanced
non-small cell lung cancer. Clin Cancer Res. 10:5981–5987. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kremer A, Holdenrieder S, Stieber P,
Wilkowski R, Nagel D and Seidel D: Nucleosomes in colorectal cancer
patients during radiochemotherapy. Tumor Biol. 27:235–242. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Deligezer U, Akisik EE, Erten N and Dalay
N: Sequence-specific histone methylation is detectable on
circulating nucleosomes in plasma. Clin Chem. 54:1125–1131. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fraga MF, Ballestar E, Villar-Garea A,
Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S,
Petrie K, et al: Loss of acetylation at Lys16 and trimethylation at
Lys20 of histone H4 is a common hallmark of human cancer. Nat
Genet. 37:391–400. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Deligezer U, Akisik EZ, Akisik EE,
Kovancilar M, Bugra D, Erten N, Holdenrieder S and Dalay N:
H3K9me3/H4K20me3 ratio in circulating nucleosomes as potential
biomarker for colorectal cancer. Circulating Nucleic Acids in
Plasma and Serum. Gahan PB: Springer Books; Amsterdam: pp. 97–103.
2011
|
|
13
|
Holdenrieder S, von Pawel J, Dankelmann E,
Duell T, Faderl B, Markus A, Siakavara M, Wagner H, Feldmann K,
Hoffmann H, et al: Nucleosomes and CYFRA 21–1 indicate tumor
response after one cycle of chemotherapy in recurrent non-small
cell lung cancer. Lung Cancer. 63:128–135. 2009.
|
|
14
|
Haring M, Offermann S, Danker T, Horst I,
Peterhansel C and Stam M: Chromatin immunoprecipitation:
optimization, quantitative analysis and data normalization. Plant
Methods. 3:112007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jayani RS, Ramanujam PL and Galande S:
Studying histone modifications and their genomic functions by
employing chromatin immunoprecipitation and immunoblotting. Methods
Cell Biol. 98:35–56. 2010. View Article : Google Scholar : PubMed/NCBI
|