Introduction
Advances have been made with regards to the
diagnosis and treatment (surgery and concurrent chemoradiotherapy)
of lung cancer. However, treatment results have not been effective.
In the last 5 years, the survival rate for patients with lung
cancer has been lower than 15% (1).
Therefore, finding a new and effective lung cancer treatment
strategy is crucial. Following 20 years of research, tumor gene
therapy has become a feasible alternative for the treatment of lung
cancer, besides surgery and concurrent chemoradiotherapy.
Tumor proliferation mainly depends on the tumor stem
cells (TSCs) within the tumor. The self-renewal, infinite
proliferation and potential for differentiation of the TSCs is of
vital importance for the occurrence, development and metastasis of
tumors. Recent studies have demonstrated that the HiWi gene is
involved in the self-renewal of TSCs. The HiWi gene is an important
factor that affects cell differentiation and proliferation. Its
overexpression can lead to the excessive proliferation of stem
cells, and cause a tumor. However, the expression and effect of the
HiWi gene in lung cancer TSCs remains uncertain. Therefore, the aim
of this study was to investigate the effect of HiWi gene silencing
on lung cancer tumor stem cell proliferation and apoptosis as well
as whether the HiWi gene serves as a molecular target for the
inhibition of lung TSCs.
Materials and methods
Lung cancer TSCs
The lung adenocarcinoma cell strain, SPC-A1 cell
line was purchased from the Cell Bank of Shanghai Institute of Life
Sciences, China. The cell line was cultivated in serum-free medium
until spheres formed. Flow cytometry was applied to these spheroid
cells using an Aldefluor kit (Bath, UK) to separate the SSCloALDEbr
cells and obtain the TSCs, as described in a previous study
(2).
Short hairpin RNA (shRNA) eukaryotic
expression vector pGenesil-2 plasmid
The shRNA eukaryotic expression vector pGenesil-2
plasmid was purchased from Jingsai Shenggong Biological Co., Ltd.,
China.
Construction of shRNA eukaryotic
expression vector targeting the HiWi gene
As described in a previous study (3), shRNA sequences were selected using
software from Ambion Co. (Grand Island, NY, USA). The two shRNA
sequences and a negative control shRNA sequence were designed
according to the HiWi gene mRNA sequence in GenBank. The sequences
were then transferred into the pGenesil-2 vector. shRNA eukaryotic
expression vectors, pGenesil-2-HiWi, pGenesil-2-HiWi2263 and
pGenesil-2-control, targeting the HiWi gene were constructed. These
vectors were identified using enzyme restriction cuts and
sequencing analysis. In brief, the culture liquid was obtained,
plasmid DNA as a small volume of culture was removed using a
pipette small volume kit (McMurray, PA, USA) according to the
manufacturer’s instructions, and the extracted plasmid was cut
using an SalI enzyme. The plasmid DNA was eluted in
deionized (1 μl, 10X buffer H 1 μl, SalI 1 μl) and incubated
in 37°C water for 3 h. Reactant (5 μl) was pipetted to perform 1%
agarose gel electrophoresis. The fully-constructed recombinant
plasmid was sent to Shanghai Yingjun Biotechnology Co., Ltd. for
the sequencing test.
Plasmid transfection
The plasmid was immediately transfected using PEI
(Sigma Co., St. Louis, MO, USA). SSCloALDEbr cells
(1×104 and 1×105 per well) were inoculated
onto 96- and 24-well plates, respectively, one day prior to
transfection. When the cell density reached 70–80%, the cells were
transfected. A total of 0.2 μg plasmid, 10 μl 1X PBS and 0.8 μg PEI
were added to the 96-pore plate, while a total of 2 μg plasmid, 100
μl 1X PBS and 8 μg PEI were added to the 24-pore plate. The cells
were mixed and left to rest for 15 min. The supernatant was
discarded and 10 and 100 μl PEI/DNA and 90 and 900 μl complete
medium were added to each pore of the 96- and 24-pore plates,
respectively. The plates were cultivated until analysis.
Experimental group
The experimental groups comprised pGenesil-2-HiWi1,
pGenesil-2-HiWi2263, pGenesil-2-control and PBS (no cells, only
nutrient solution), which was used as the blank for zeroing. There
were four wells in each group at each time, and 1×104
cells was added to each well.
Proliferation of lung cancer TSCs using
the MTT assay
The nutrient solution was removed 24, 48 and 72 h
following transfection. A total of 10 μl MTT (5 mg/ml) was added to
the pores, and placed into a CO2 culture box for 4 h.
The nutrient solution was removed, and 100 μl DMSO was added into
each pore. The solution was agitated to inhibit crystallization. In
order to test the light absorbance, the absorbance of each pore was
reduced in enzyme immunoassay instrument at a wavelength of 490 nm.
The cell proliferation inhibition rate was calculated using the
formula: Cell proliferation inhibition ratio (%) = (absorbance in
the control wells − absorbance in the test wells) / (absorbance in
the control wells − absorbance in the blank wells) × 100.
Statistical analyses were conducted simultaneously.
Detection of lung cancer TSCs and
apoptosis with Annexin V staining
Between 48 and 72 h after transfection in each
group, the cells were digested into mono-cell suspension by
pancreatin (0.125%), 4°C, 1000 rpm × 5 min, PBS (0.01 M, pH 7.2).
Cell suspension occurred at 4°C, 1000 rpm × 5 min. The supernatant
was discarded and 500 μl suspension liquid was added to the
centrifugal tube bottom for resuspension. The solution was removed
and 5 μl Annexin V-FITC (ShangHai Biotechnology Co., Ltd., China)
labeled solution was added. The solution was fully blended, and
flow cytometry was initiated 5 min later.
Statistical analysis
The statistical analysis was performed using
statistical software SPSS 13.0. P<0.05 was considered to
indicate a statistically significant difference. The Chi square
test and Fisher’s exact test were used to compare the counted data
between the groups.
Results
Identification of enzyme-cut recombinant
plasmid
Within the structure of the recombinant plasmid, the
short hairpin RNA (shRNA) sequence is the designed RNA interference
target sequence, and the multiple cloning site (MCS) is the clone
starting point (Fig. 1). The
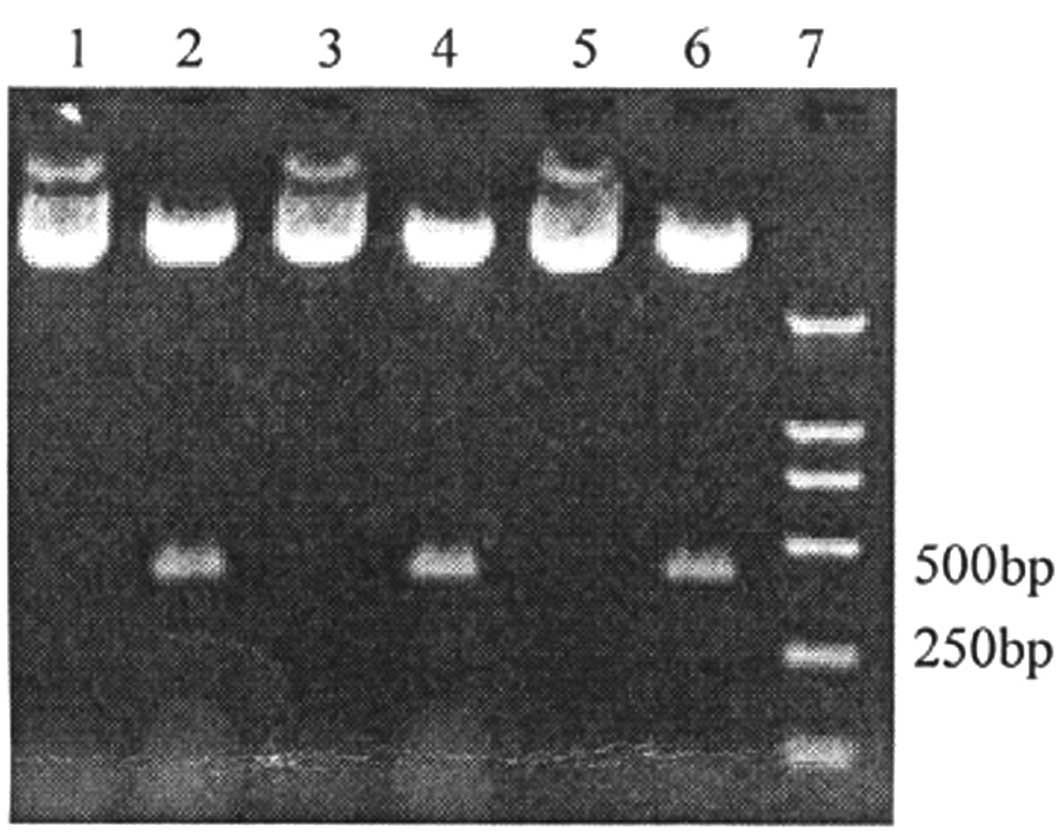
results of electrophoresis showed that the enzyme cut the
recombinant plasmid at the position point of SalI in the
gene segment. The restriction enzyme SalI was used to cut
400 bp-long segments within the recombinant plasmid. Segments (400
bp) were cut within the pGenesil-2HiWi1, pGenesil-2HiWi2263 and
blank plasmid (Fig. 2). Therefore,
the designed shRNA interference segments were inserted successfully
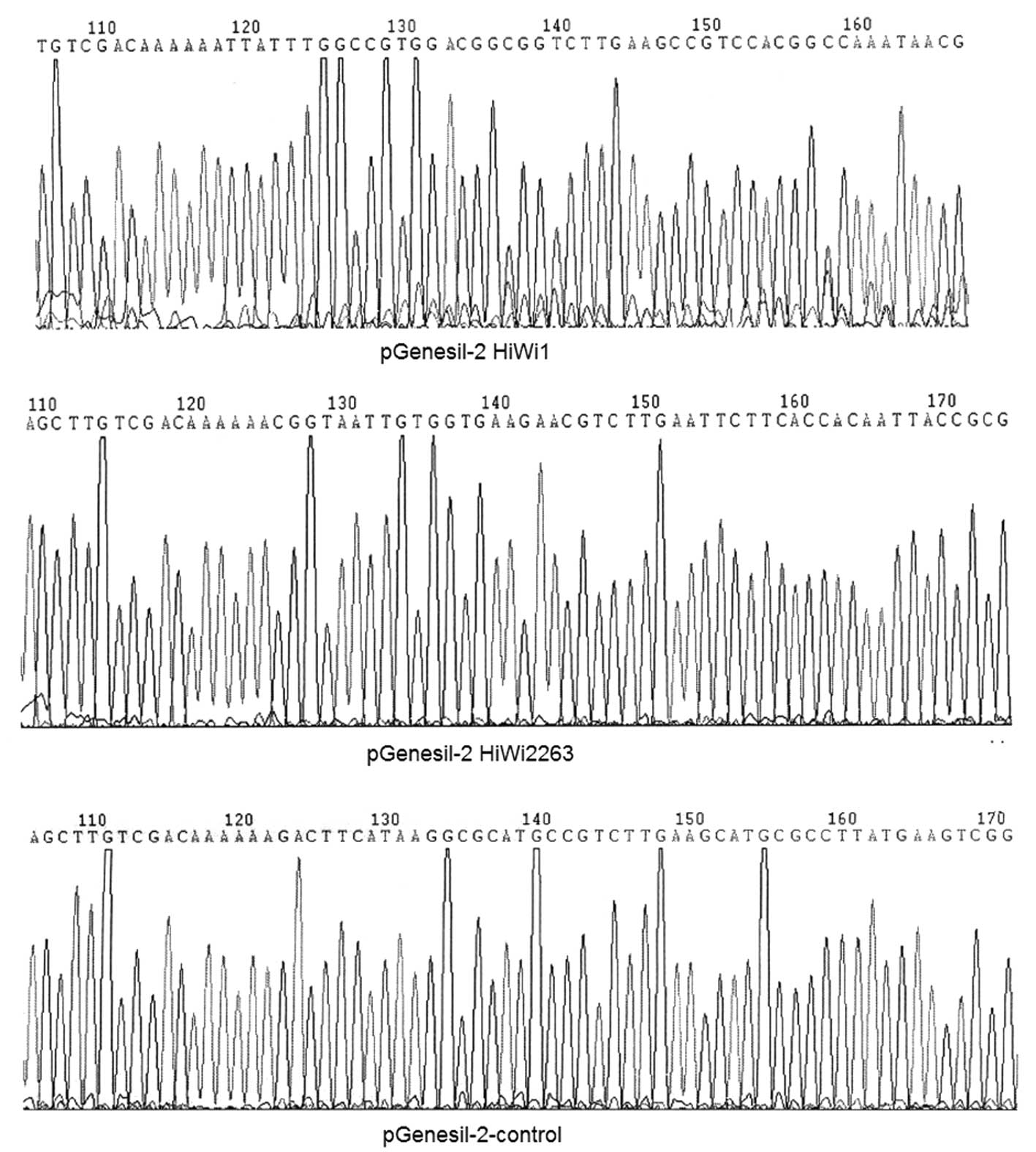
into the pGenesil-2 plasmid vector. The results from the sequencing
analysis are shown in Fig. 3. There
was a C to G base mutation in the HiWi inserted sequences.
According to the principle of RNA interference, the bases in the
siRNA sequence play vital roles and the mutated base is not located
in the siRNA sequence; therefore this recombinant plasmid could
still be used. This enabled other sequences to be inserted
correctly.
Proliferation of lung cancer TSCs using
the MTT assay
Our results showed that 24 h after transfection, the
lung cancer TSC inhibition rate for the pGenesil-2-HiWi2263,
pGenesil-2-HiWi1 and pGenesil-2-control groups were 81.62, 73.16
and 8.54%, respectively. After 48 h, the rates of the first two
groups, 62.42 and 56.96%, respectively, decreased as compared to
the increase in percentage of the pGenesil-2-control groups,
12.38%. Similarly, after 72 h, the rates of the first two groups
decreased to 47.19 and 44.26%, respectively, as compared to the
control groups, 10.37%. This finding demonstrates that the lung
cancer TSC proliferation inhibition rate was increased (P<0.01),
with the inhibition rate following 24 h being the highest. This
observation revealed that RNA-related interference of the HiWi gene
is effective in inhibiting the growth of lung cancer TSCs (Table I).
 | Table IRecombinant plasmid cell proliferation
inhibitory rates following 24, 48 and 72 h of transfection. |
Table I
Recombinant plasmid cell proliferation
inhibitory rates following 24, 48 and 72 h of transfection.
| Cell proliferation
inhibitory rate (%) |
|---|
|
|
|---|
| Group | 24 h | 48 h | 72 h |
|---|
| A
(pGenesil-2-HiWi1) | 73.16a | 56.96a | 44.26a |
| B
(pGenesil-2-HiWi2263) | 81.62a | 62.42a | 47.19a |
| C
(pGenesil-2-control) | 8.54 | 12.38 | 10.37 |
| A vs. C |
χ2=34563.451, P<0.01 |
χ2=17548.646, P<0.01 |
χ2=11569.834, P<0.01 |
| B vs. C |
χ2=43143.228, P<0.01 |
χ2=21390.388, P<0.01 |
χ2=13228.323, P<0.01 |
Annexin V staining on lung cancer TSCs
induced apoptosis
Results from the Annexin V staining on lung cancer
TSC apoptosis demonstrate that: the apoptotic rates of
pGenesil-2-HiWi1 and pGenesil-2-HiWi2263 were 26.16±1.21 and
28.06±1.78%, respectively. No stastically significant difference
was observed (P>0.05%). The apoptotic rates of the
pGenesil-2-control were found to be (2.25±0.058)% and (2.86±0.09)%.
Again, there was no statistical significant difference (P>0.05).
However, the rates in the pGenesil-2-HiWi1 and pGenesil-2-HiWi2263
groups were higher compared to those in the PBS group and the empty
plasmid group (P<0.01) (Fig.
4).
Discussion
RNA interference technology is a common method used
to control gene expression. Post-transcriptional gene silencing
occurs following the induction of double-stranded RNA, which causes
the breaking and degrading of messenger RNA, which is meditated by
double-strand RNA. During tumor gene therapy, RNA is capable of
specifically inhibiting the overexpression of cancer,
cancer-related and mutator genes, confining them to a state of
dormancy or silencing them. Therefore, RNA may be used as a new
tumor treatment strategy.
In lung cancer gene therapy with RNAi, the choice of
targeted gene silencing is crucial. Lung cancer is caused by a
series of complex signaling pathways, involving a number of cell
factors. Thus, selection of a cell target is critical when cancer
occurs as the treatment target is crucial. In 1997, Lin and
Spradling identified the PiWi gene (4) in the reproductive stem cells of fruit
flies. This gene is involved in the process of stem cell division
and is correlated with stem cell self-renewal, gamete formation,
RNA interference and translation regulation. Overexpression of PiWi
has been shown to lead to cell differentiation disorders and
ultimately cancer generation (5).
Lee et al (6) identified
PiWi12 expression in tumors including, renal cell carcinoma,
prostate, endometrial and breast cancer, and gastroenteric and
ovarian tumor. Gao (7) demonstrated
that the PiWi12 gene was important for TSC development in the
generation of tumor cells. The PiWi gene is homologous to the human
HiWi gene, which is located in the long arm of chromosome 12. It
was first cloned from the cDNA library of testes. Sharma et
al (8) identified that HiWi was
highly expressed in human prostate, brain, liver and other tissues,
but was highly expressed in fetal kidney and adult testes. The HiWi
gene also had a regulatory role in the division of hematopoietic
stem cells. Grochola et al (9) found that an extremely high or
extremely low expression of HiWi gene was correlated with male
survival in patients with pancreatic cancer. In their study, Liu
et al (10) found that
expression of the HiWi gene in different stomach cancer cell lines
correlated with the degree of malignancy. The study revealed that
upon inhibition of the expression of the HiWi gene, the growth of
stomach cancer cells was inhibited by RNA interference. Therefore,
these authors considered that the HiWi gene could be used as a
marker of tumor proliferation and as an evaluation index of cancer
prognosis (11). Huang et al
(12) revealed that tumor growth
and proliferation in the body depended on cancer stem cells.
Moreover, the ability of self-renewal, differentiation and
proliferation of TSCs was correlated with the HiWi gene.
Overexpression of the HiWi gene is the significant cause of
malignancy and tumor. Therefore, it is essential to study the
involvement of the HiWi gene in TSCs.
The purpose of RNA interference technology is to
create siRNA segments. These are short strands of double-stranded
RNA, which are able to degrade homologous complementary sequences
of mRNA, known as the RNA interference pathway. Some of the most
common methods of siRNA preparation include chemical synthesis,
in vitro transcription, degradation of long sequences of RNA
and PCR expression and production of siRNA in the cells. siRNA may
be generated by encoding its carrier, which is capable of
self-renewal as well as the inhibition of the expression of target
genes in the cells over a long period of time. Thus, this method is
convenient and cost-effective. In this study, the shRNA eukaryotic
expression vector of targeting Hiwi gene was constructed using this
method. Complying with the design requirements of shRNA carriers,
this carrier employed U6 to initiate shRNA transcription and pol
III to terminate transcription. The salI site is located
within the PGenesil-2 carrier and so the same enzyme site is
designed in the shRNA fragments. If the fragment is inserted
correctly, then it can be cut into 400-bp sections by the
salI enzymes. From the results from the salI
restriction enzyme digestion analysis, it can be observed that the
plasmid enzyme-cut sections are consistent with the expected
sections, and there are no sequence base mutations.
Results from the MTT assay and flow cytometry
revealed that the growth of the lung cancer TSCs was inhibited, and
the proliferation rate was increased following HiWi gene silencing.
Immediately following transfection, the cell growth density was
expected to reach 70–90%, however, 48 and 72 h following
transfection the 96-pore plate was saturated with cells. Therefore,
the difference between the long-time inhibition effect of the RNA
interference group and the control group could not be identified.
Nevertheless, it was observed that following gene silencing the
apoptotic rates in the lung cancer TSCs were higher than those in
the control group. Thus, results of this study have demonstrated
that the HiWi gene is involved in the differentiation and
proliferation of lung cancer TSCs. These results are also
significant for the potential to inhibit lung cancer TSC growth and
target apoptosis.
This study suggests that following silencing of the
HiWi gene, the pathway regulating lung cancer TSC growth and
inhibition may have been activated. Therefore, we hypothesize that
abnormal function of the HiWi gene may be the key to the growth,
inhibition and apoptosis of lung cancer TSCs. The HiWi gene may be
crucial in the proliferation and differentiation of lung cancer
TSCs. Overexpression of the HiWi gene may be the cause of viscious
proliferation of lung cancer TSCs and ultimately lung cancer. We
consider that the HiWi gene has the potential to become a molecular
target for the inhibition of lung cancer TSCs. Silencing the HiWi
gene may have potential for the treatment of lung cancer, thereby
setting the basis for treatment surgeries.
Acknowledgements
This study was supported by the Heilongjiang Youth
Science Foundation (Fund Code: QC2009C95).
References
|
1
|
Jemal A, Siegel R and Ward E: Cancer
statistics. CA Cancer J Clin. 56:106–130. 2006.
|
|
2
|
Dong L and Shi Y: Aldehyde dehydrogenase-1
is a specific marker for stem cells in human lung adenocarcinoma.
Med Oncol. View Article : Google Scholar
|
|
3
|
Chen Q-H, Lu J, Wang X-Q, et al:
Construction and identification of shRNA eudaryotic expression
vectors targeting to HIWI gene. Exp Diagn Chin. 12:42–46. 2008.
|
|
4
|
Lin H and Spradling AC: A novel group of
pumilio mutations affects the asymmetric division of germline stem
cells in the Drosophila ovary. Development. 124:2463–2476.
2007.PubMed/NCBI
|
|
5
|
Qiao D, Zeeman AM, Deng W, et al:
Molecular characterization of hiwi, a human member of the piwi gene
family whose overexpression is correlated to seminomas. Oncogene.
25:3988–3999. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee JH, Schutte D, Wulf G, et al:
Stem-cell protein Piwil2 is widely expressed in tumors and inhibits
apoptosis through activation of Stat3/Bcl-XL pathway. Hum Mol
Genet. 15:201–211. 2006.PubMed/NCBI
|
|
7
|
Gao JX: Cancer stem cells: the lessons
from pre-cancerous stemcells. J Cell Mol Med. 12:67–96.
2008.PubMed/NCBI
|
|
8
|
Sharma AK, Nelson MC, Brandt JE, et al:
Human CD34(+) stem cells express the hiwi gene, a human homologue
of the Drosophila gene piwi. Blood. 97:426–434. 2001.
|
|
9
|
Grochola LF, Greither T, Taubert H, et al:
The stem cell-associated Hiwi gene in human adenocarcinoma of the
pancreas: expression and risk of tumour-related death. Br J Cancer.
99:1083–1088. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dong X, Liu Z, Ning M, et al: Significance
of Hiwi gene in tumor formation. Chin Community Doc. 287:10–11.
2011.
|
|
11
|
Liu X, Sun Y, Guo J, et al: Expression of
hiwi gene in human gastric cancer was associated with proliferation
of cancer cells. Int J Cancer. 118:1922–1929. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang EH, Heidt DG, Li CW, et al: Cancer
stem cells: a new paradigm for understanding tumor progression and
therapeutic resistance. Surgery. 141:415–419. 2007. View Article : Google Scholar : PubMed/NCBI
|