Introduction
Cholangiocarcinoma (CCA) is a highly malignant
cancer with a poor prognosis which arises from the epithelial
lining of intra- or extrahepatic bile ducts (1). CCA is the second most common primary
liver malignancy and epidemiological studies have reported rising
incidence rates in Western countries (2,3). Since
clinical presentation and diagnosis at locally advanced and/or
metastatic tumor stages beyond potential surgical cure are common,
the reported 5-year survival rates rarely exceed 5–10%.
Notwithstanding recent progress in treatment options for advanced
CCA, alternative routes to improve the prognosis may include the
identification and characterization of individuals at high risk of
the disease, thus allowing for tumor surveillance and early
diagnosis (4–6). Clinical risk factors include, among
others, primary sclerosing cholangitis, viral hepatitis and liver
fluke infections in Asian countries (7,8).
However, knowledge of genetic susceptibility factors, with its
potential to elucidate novel carcinogenic pathways and define
high-risk populations, is limited, reflecting difficulties in
establishing study cohorts of informative sample sizes for this
rare cancer. We previously reported the increased genetic risk of
CCA in α1 antitrypsin Z allele carriers, which was in
accordance with previously published pathology-based data (9–11).
Although the distinct molecular pathogenesis underlying malignant
cholangiocyte transformation is poorly understood, chronic
cholestasis and inflammation are considered to be key elements in
CCA initiation (12). Recent
studies have highlighted the carcinogenic role of the activation of
nuclear factor κB (NF-κB) signaling in conferring resistance to
apoptosis to malignant cholangiocytes; these findings substantiate
the concept of CCA as an inflammation-driven cancer (13).
The p53 tumor suppressor network is a central
component of cellular stress responses and is critical for the
maintenance of genomic stability in response to various stresses,
including DNA damage, metabolic perturbation and oncogene
activation (14). Notably, p53 has
been suggested to be a negative regulator of the proinflammatory
transcription factor NF-κB (15,16).
Deregulation of the p53 pathway by either overexpression or somatic
mutation is a common feature in a number of types of human cancer,
including CCA (17–19). Furthermore, germline p53
alteration has been linked to CCA formation in animal models
(20). Notably, common genetic
variations, relevant to the p53 network and involved in cell
cycle control and apoptosis regulation, have been identified
(21). The single nucleotide
polymorphism (SNP) c.309 T>G in the promoter/enhancer site of
the murine double minute 2 (mdm2) gene, which is a
critical negative regulator of p53 and is itself transcriptionally
regulated by p53, affects the binding efficiency of the
transcription factor SP1. Consequently, the SNP309 G allele has
been suggested to translate into increased intracellular mdm2
levels, thereby limiting the cellular availability of functional
p53 under stressful conditions (22). The p53 SNP c.72 G>C is located in
a proline-rich domain and may affect the molecular structure of the
p53 protein. This has been reported to have a functional impact, as
the C allele is thought to induce apoptosis and suppress
oncogene-induced transformation less efficiently (23).
Although these two genetic variants are among the
most widely studied SNPs in genetic cancer research (21), no data are currently available for
CCA. In this respect, the aim of the current study was to assess
the potential contribution of the common functional gene variants
p53 (SNP72 G/C) and mdm2 (SNP309 T/G) to the genetic
susceptibility to bile duct cancer.
Patients and methods
Patient recruitment and
characteristics
In total, 182 European CCA patients (103 males and
79 females) and 350 controls (151 males and 199 females) were
recruited at equal case-control proportions at three academic
medical centers in Germany and Romania. Specific aspects of the
study population were reported in our previous study (9). Briefly, the clinical CCA diagnosis and
classification criteria were in accordance with the recommendations
of the British Society of Gastroenterology (24). There was a tissue diagnosis of CCA
in 84% of the subjects, while in 16% of the patients the CCA
diagnosis was based on clinical assessment, taking into account
imaging studies, follow-up and exclusion of alternative diagnoses,
including postoperative biliary strictures. The control subjects
were recruited following either negative colorectal cancer
screening (n=290) or normal liver ultrasound (n=60). Written
informed consent was obtained from all participants. The study was
approved by the respective ethics committees and was in accordance
with the revised Declaration of Helsinki.
Genomic DNA isolation and genotyping
Following venipuncture and sampling of 5 ml whole
blood, genomic DNA was extracted by standard techniques using a
commercial kit (QIAamp, Qiagen, Hilden, Germany). Following
appropriate spectrophotometric calibration of the DNA
concentrations (NanoDrop ND-1000, NanoDrop Technologies,
Wilmington, DE, USA), rs1042522 (p53 SNP72) and
rs2279744 (mdm2 SNP309) were genotyped using
solution-phase hybridization reactions with 5′-nuclease and
fluorescence detection (TaqMan assays, Applied Biosystems, Foster
City, CA, USA) on an ABI 7500 Fast Real-Time polymerase chain
reaction (PCR) system (Applera, Norwalk, CT, USA). Specifically,
the PCR assays contained 5 μl 2X Genotyping Mastermix, 0.25 μl 40X
Genotyping mix, 3.75 μl H2O and 5–50 ng/μl DNA. The
amplification conditions were as follows: 95°C for 10 min, 40
cycles of 95°C for 10 sec and 60°C for 1 min.
Statistical analysis
The data were analysed using the SPSS software
package (version 17.0; SPSS, Munich, Germany). The age and gender
structure of the study groups were analysed using t-tests. Data are
shown as the mean ± SD. Genotypic data for the two SNPs were
assessed for consistency with the Hardy-Weinberg equilibrium (HWE)
in controls (P>0.05) using an exact test (http://ihg.gsf.de/snps.html). The allele and genotype
frequencies of the study groups were compared using
χ2-tests for 2×2 and 2×3 contingency tables. Fisher’s
exact tests were used as appropriate, in particular for small
sample sizes. Differences in allele and genotype distributions,
including exploratory data analyses stratifying for gender, tumor
localization, early onset and genotypic interactions, were assessed
using odds ratios (ORs) and 95% confidence intervals (CIs).
P<0.05 was considered statistically significant.
To assess for adequate sample size in our study, we
performed power calculations using PS: Power and Sample Size
Calculation v.3.0 (http://biostat.mc.vanderbilt.edu/wiki/Main/PowerSampleSize).
Results
Clinical parameters of the patients
Allelic discrimination for rs1042522 and
rs2279744 was successful in all study subjects, yielding
call rates of 100% for the two SNPs. The genotype distributions in
the controls did not deviate from the Hardy-Weinberg equilibrium
(P>0.05 for both SNPs). The detected allele frequencies were
compatible with the Entrez SNP database entries (http://www.ncbi.nlm.nih.gov/snp). Of the cancer
group, 57% of the patients were male, compared with 43% in the
control group (P<0.01). The median age at CCA diagnosis/study
entry was 65.8±11.4 years for cancer patients and 61.0±11.1 years
for the control subjects (P<0.01). There were more patients with
tumors arising from the extrahepatic biliary system than from
intrahepatic localizations (141 extrahepatic vs. 41 intrahepatic
localizations). The demographic and clinical data of the study
population reflecting the epidemiology of CCA are shown in Table I.
 | Table IDemographic and clinical parameters of
the study population. |
Table I
Demographic and clinical parameters of
the study population.
| Parameters | CCA | Controls | P-value |
|---|
| Subjects (n) | 182 | 350 | |
| Age, years (mean ±
SD) | 65.8±11.4 | 61.0±11.1 | <0.01 |
| Gender, male/female
(%) | 103/79 (57/43) | 151/199 (43/57) | <0.01 |
| Localization,
intra-/extrahepatic (%) | 41/141 (23/77) | | |
| Diagnosis,
tissue/clinical (%) | 152/30 (84/16) | | |
Effect of allelic and genotypic
distributions in the study groups
A comparison of the allelic distributions yielded no
significant differences in either SNP between the study groups
(SNP72, 27.2 vs. 26.9%; SNP309, 37.4 vs. 36.1% in CCA patients vs.
controls, respectively). Similarly, no significant difference or
marked trend was noted when analyzing genotypic distributions in
the total study groups (Table II).
We then evaluated the potential of specific SNP effects in terms of
parameters such as gender, tumor localization (extra- vs.
intrahepatic) and early onset CCA, arbitrarily defined as CCA
diagnosis <60 years. Similarly, in this exhaustive exploratory
data analysis no association signal was obtained for either SNP
(data not shown).
 | Table IIAllele and genotype frequencies of
p53 SNP72 and mdm2 SNP309 in CCA vs. controls. |
Table II
Allele and genotype frequencies of
p53 SNP72 and mdm2 SNP309 in CCA vs. controls.
| Frequency type | | CCA n (%) | Controls n (%) | P-value |
|---|
| Alleles |
| p53
SNP72 | G | 265 (72.8) | 512 (73.1) | 0.92 |
| C | 99 (27.2) | 188 (26.9) | (0.98)a |
| | 364 (100) | 700 (100) | [0.74–1.31]b |
| mdm2
SNP309 | T | 228 (62.6) | 447 (63.9) | 0.70 |
| G | 36 (37.4) | 253 (36.1) | (0.95)a |
| | 364 (100) | 700 (100) | [0.73–1.23]b |
| Genotypes |
| p53
SNP72 | GG | 94 (51.7) | 190 (54.3) | 0.49 |
| GC | 77 (42.3) | 132 (37.7) | (1.42)c |
| CC | 1 (6.0) | 28 (8.0) | |
| | 182 (100) | 350 (100) | |
| mdm2
SNP309 | TT | 74 (40.7) | 141 (40.3) | 0.62 |
| TG | 80 (44.0) | 165 (47.1) | (0.97)c |
| GG | 28 (15.3) | 44 (12.6) | |
| | 182 (100) | 350 (100) | |
Effects of epistatic SNP
As the two variants under investigation are known to
be functionally coupled, we explored the epistatic SNP effects. The
results revealed a trend for the overrepresentation of SNP309 TT in
carriers of the variant SNP72 genotype CC (81.8 vs. 50.0%; Fisher’s
exact test P=0.24; Table III). In
agreement with this trend, the allelic distributions reached
borderline significance for T allele positivity in SNP72 CC
homozygotes (90.9 vs. 69.6%; OR, 4.36; 95% CI, 0.92–20.77; P=0.049;
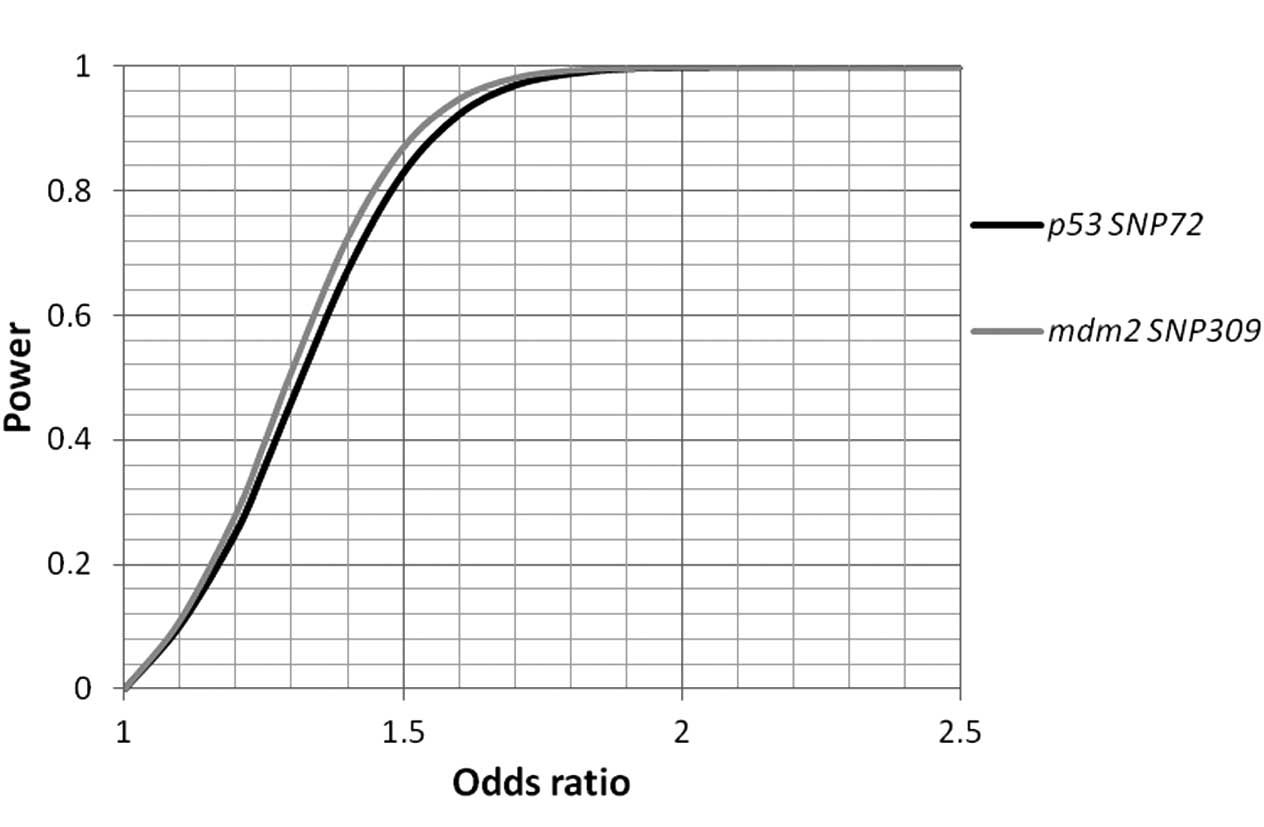
Table IV). The statistical power
was calculated, and sufficient power (>97% each) to pinpoint
allelic association signals beyond ORs >1.7 for the two SNPs was
identified (Fig. 1).
 | Table IIIGenotypic interaction between
p53 and mdm2 variants. |
Table III
Genotypic interaction between
p53 and mdm2 variants.
| Genotypes | | CCA n (%) | Controls n (%) | P-value |
|---|
| p53
SNP72 |
| GG | SNP309 | 94 (100) | 190 (100) | 0.74 |
| TT | 34 (36.2) | 69 (36.3) | (0.6)a |
| TG | 44 (46.8) | 95 (50.0) | |
| GG | 16 (17.0) | 26 (13.7) | |
| GC | SNP309 | 77 (100) | 132 (100) | 0.66 |
| TT | 31 (40.3) | 58 (43.9) | (0.83)a |
| TG | 34 (44.1) | 59 (44.7) | |
| GG | 12 (15.6) | 15 (11.4) | |
| CC | SNP309 | 11 (100) | 28 (100) | 0.24b |
| TT | 9 (81.8) | 14 (50.0) | |
| TG | 2 (18.2) | 11 (39.3) | |
| GG | 0 (0) | 3 (10.7) | |
| mdm2
SNP309 |
| TT | SNP72 | 74 (100) | 141 (100) | 0.85 |
| GG | 34 (36.2) | 69 (49.0) | (0.32)a |
| GC | 31 (46.8) | 58 (41.1) | |
| CC | 9 (17.0) | 14 (9.9) | |
| TG | SNP72 | 80 (100) | 165 (100) | 0.29 |
| GG | 44 (40.3) | 95 (57.5) | (2.47)a |
| GC | 34 (44.1) | 59 (35.8) | |
| CC | 2 (15.6) | 11 (6.7) | |
| GG | SNP72 | 28 (100) | 44 (100) | 0.38b |
| GG | 16 (81.8) | 26 (59.1) | |
| GC | 12 (18.2) | 15 (34.1) | |
| CC | 0 (0) | 3 (6.8) | |
 | Table IVAllelic interaction between
p53 and mdm2 variants. |
Table IV
Allelic interaction between
p53 and mdm2 variants.
| Genotypes | | CCA n (%) | Controls n (%) | P-value |
|---|
| p53
SNP72 |
| GG | SNP309 | | | |
| T | 112 (59.6) | 223 (58.7) | 0.86 |
| G | 76 (40.4) | 147 (38.7) | (1.03)a |
| | 188 (100) | 380 (100) | [0.72–1.47]b |
| GC | SNP309 | | | |
| T | 96 (62.3) | 175 (66.3) | 0.41 |
| G | 58 (37.7) | 89 (33.7) | (0.84)a |
| | 154 (100) | 264 (100) | [0.56–1.27]b |
| CC | SNP309 | | | |
| T | 20 (90.9) | 39 (69.6) | 0.049 |
| G | 2 (9.1) | 17 (30.4) | (4.36)a |
| | 22 (100) | 56 (100) |
[0.92–20.77]b |
| mdm2
SNP309 |
| TT | SNP72 | | | |
| G | 99 (66.9) | 196 (69.5) | 0.58 |
| C | 49 (33.1) | 86 (30.5) | (0.89)a |
| | 148 (100) | 282 (100) | [0.58–1.36]b |
| TG | SNP72 | | | |
| G | 122 (76.3) | 249 (75.5) | 0.84 |
| C | 38 (23.7) | 81 (24.5) | (0.96)a |
| | 160 (100) | 330 (100) | [0.62–1.49]b |
| GG | SNP72 | | | |
| G | 44 (78.6) | 67 (76.1) | 0.74 |
| C | 12 (21.4) | 21 (23.9) | (0.87)a |
| | 56 (100) | 88 (100) | [0.39–1.95]b |
Discussion
Given the relative rarity of bile duct cancer and
the ensuing lack of informative study populations, there is a
paucity of information concerning genetic risk factors for CCA
development. As yet, CCA is one of the few cancer types for which
no genome-wide association study (GWAS) is available (25). Using a multi-institutional approach
we have established a cohort comprising 182 cancer patients, which
is currently the largest European-based cohort. However, this
sample size may still be regarded as limited with respect to
genetic association studies (26).
In consideration of our overall negative association signals for
either SNP, power estimations were important to ascertain adequate
sample size. In our study, the study power exceeded 97% for the two
variants with an OR set at 1.7, thus reasonably excluding major SNP
effects in CCA susceptibility. The study power to firmly refute
associations within distinct exploratory subgroups was limited,
meaning that specific risk modulations for subsets of patients may
not have been detectable in our data set. For example, there are
data indicating gender-specific effects of SNP309 in the
mdm2 promoter, which has been suggested to be regulated by
estrogens (27–29). Moreover, the two SNPs have been
reported to modulate the age of onset in certain types of cancer,
e.g., in head and neck and hereditary non-polyposis colorectal
cancer (HNPCC) (30,31). Bile duct cancer is characterized by
late onset, with a median age at diagnosis of >70 years, which
is compatible with the age structure of our study population
(32).
However, we observed a potential genotype-specific
interaction between p53 SNP72 and mdm2 SNP309.
Specifically, in the presence of the apoptosis-deficient p53
variant genotype SNP72 CC, the mdm2 SNP309 major allele T increased
the genetic risk of CCA (P=0.049; OR, 4.36; 95% CI, 0.92–20.77).
Thus, it appears that in this specific genotypic context the
relative attenuation of cellular mdm2 availability (related to the
T allele of SNP309) with increased degradation of variant p53 in
SNP72 CC homozygous individuals may result in the SNP309 T variant
becoming a risk allele. In this respect, it should be noted that,
beyond its low-apoptotic capacity, the C allele of p53 SNP72
has been reported to be a stronger inducer of transcription and
cell-cycle arrest. These are potential pro-oncogenic effects that
may become relevant in a genetic setting of less effective p53
suppression mediated by the mdm2 SNP309 T variant (33,34).
However, the association signal was weak and based on a limited
number of individuals. With regard to such complex gene-gene
interactions, further confirmatory studies in larger cohorts are
necessary to ascertain such epistatic SNP effects.
In conclusion, our data do not support the
hypothesis of a prominent role of p53 SNP72 and mdm2
SNP309 in the genetic architecture of bile duct cancer. A subtle
risk modulation by specific genotype-specific interactions of these
functionally coupled genes may modify CCA susceptibility and this
requires confirmatory studies in different cohorts.
Acknowledgements
This study was supported by HOMFOR (to V.Z.) and
preliminary results were presented, in part, at the Annual Meeting
of the American Association for the Study of Liver Diseases (AASLD)
in Boston, MA, USA, in October 2009.
References
|
1
|
Khan SA, Thomas HC, Davidson BR and
Taylor-Robinson SD: Cholangiocarcinoma. Lancet. 366:1303–1314.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shaib Y and El-Serag HB: The epidemiology
of cholangiocarcinoma. Semin Liver Dis. 24:115–125. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cardinale V, Semeraro R, Torrice A, et al:
Intra-hepatic and extra-hepatic cholangiocarcinoma: new insight
into epidemiology and risk factors. World J Gastrointest Oncol.
2:407–416. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Valle J, Wasan H, Palmer DH, et al:
Cisplatin plus gemcitabine versus gemcitabine for biliary tract
cancer. N Engl J Med. 362:1273–1281. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hezel AF, Deshpande V and Zhu AX: Genetics
of biliary tract cancers and emerging targeted therapies. J Clin
Oncol. 28:3531–3540. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ortner MA: Photodynamic therapy for
cholangiocarcinoma: overview and new developments. Curr Opin
Gastroenterol. 25:472–476. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gatto M, Bragazzi MC, Semeraro R, et al:
Cholangiocarcinoma: update and future perspectives. Dig Liver Dis.
42:253–260. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Blechacz BR and Gores GJ:
Cholangiocarcinoma. Clin Liver Dis. 12:131–150. 2008. View Article : Google Scholar
|
|
9
|
Mihalache F, Hoblinger A, Grunhage F, et
al: Heterozygosity for the alpha1-antitrypsin Z allele may confer
genetic risk of cholangiocarcinoma. Aliment Pharmacol Ther.
33:389–394. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou H, Ortiz-Pallardo ME, Ko Y and
Fischer HP: Is heterozygous alpha-1-antitrypsin deficiency type PIZ
a risk factor for primary liver carcinoma? Cancer. 88:2668–2676.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hoblinger A and Lammert F: Genetics of
biliary tract diseases: new insights into gallstone disease and
biliary tract cancers. Curr Opin Gastroenterol. 24:363–371. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Blechacz B and Gores GJ:
Cholangiocarcinoma: advances in pathogenesis, diagnosis, and
treatment. Hepatology. 48:308–321. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Grivennikov SI, Greten FR and Karin M:
Immunity, inflammation, and cancer. Cell. 140:883–899. 2010.
View Article : Google Scholar
|
|
14
|
Vogelstein B, Lane D and Levine AJ:
Surfing the p53 network. Nature. 408:307–310. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Komarova EA, Krivokrysenko V, Wang K, et
al: p53 is a suppressor of inflammatory response in mice. FASEB J.
19:1030–1032. 2005.PubMed/NCBI
|
|
16
|
Gudkov AV and Komarova EA: Dangerous
habits of a security guard: the two faces of p53 as a drug target.
Hum Mol Genet. 16(Spec No 1): R67–R72. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Khan SA, Taylor-Robinson SD, Carmichael
PL, Habib N, Lemoine NR and Thomas HC: Analysis of p53 mutations
for a mutational signature in human intrahepatic
cholangiocarcinoma. Int J Oncol. 28:1269–1277. 2006.PubMed/NCBI
|
|
18
|
Khan SA, Thomas HC, Toledano MB, Cox IJ
and Taylor-Robinson SD: p53 Mutations in human cholangiocarcinoma:
a review. Liver Int. 25:704–716. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tannapfel A, Weinans L, Geissler F, et al:
Mutations of p53 tumor suppressor gene, apoptosis, and
proliferation in intrahepatic cholangiocellular carcinoma of the
liver. Dig Dis Sci. 45:317–324. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Farazi PA, Zeisberg M, Glickman J, Zhang
Y, Kalluri R and DePinho RA: Chronic bile duct injury associated
with fibrotic matrix microenvironment provokes cholangiocarcinoma
in p53-deficient mice. Cancer Res. 66:6622–6627. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Whibley C, Pharoah PD and Hollstein M: p53
polymorphisms: cancer implications. Nat Rev Cancer. 9:95–107. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bond GL, Hu W, Bond EE, et al: A single
nucleotide polymorphism in the MDM2 promoter attenuates the p53
tumor suppressor pathway and accelerates tumor formation in humans.
Cell. 119:591–602. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dumont P, Leu JI, Della Pietra AC III,
George DL and Murphy M: The codon 72 polymorphic variants of p53
have markedly different apoptotic potential. Nat Genet. 33:357–365.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Khan SA, Davidson BR, Goldin R, et al:
Guidelines for the diagnosis and treatment of cholangiocarcinoma:
consensus document. Gut. 51(Suppl 6): 1–9. 2002.PubMed/NCBI
|
|
25
|
Hartman M, Loy EY, Ku CS and Chia KS:
Molecular epidemiology and its current clinical use in cancer
management. Lancet Oncol. 11:383–390. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hattersley AT and McCarthy MI: What makes
a good genetic association study? Lancet. 366:1315–1323. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bond GL, Hirshfield KM, Kirchhoff T, et
al: MDM2 SNP309 accelerates tumor formation in a gender-specific
and hormone-dependent manner. Cancer Res. 66:5104–5110. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bond GL and Levine AJ: A single nucleotide
polymorphism in the p53 pathway interacts with gender,
environmental stresses and tumor genetics to influence cancer in
humans. Oncogene. 26:1317–1323. 2007. View Article : Google Scholar
|
|
29
|
Zimmer V, Widmann T, Muller M, et al:
Genotypic interaction and gender specificity of common genetic
variants in the p53/mdm2 network in Crohn’s disease. Digestion.
81:246–251. 2010.PubMed/NCBI
|
|
30
|
Jones JS, Chi X, Gu X, Lynch PM, Amos CI
and Frazier ML: p53 polymorphism and age of onset of hereditary
nonpolyposis colorectal cancer in a Caucasian population. Clin
Cancer Res. 10:5845–5849. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shen H, Zheng Y, Sturgis EM, Spitz MR and
Wei Q: P53 codon 72 polymorphism and risk of squamous cell
carcinoma of the head and neck: a case-control study. Cancer Lett.
183:123–130. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Welzel TM, Graubard BI, El-Serag HB, et
al: Risk factors for intrahepatic and extrahepatic
cholangiocarcinoma in the United States: a population-based
case-control study. Clin Gastroenterol Hepatol. 5:1221–1228. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Thomas M, Kalita A, Labrecque S, Pim D,
Banks L and Matlashewski G: Two polymorphic variants of wild-type
p53 differ biochemically and biologically. Mol Cell Biol.
19:1092–1100. 1999.PubMed/NCBI
|
|
34
|
Pim D and Banks L: p53 polymorphic
variants at codon 72 exert different effects on cell cycle
progression. Int J Cancer. 108:196–199. 2004. View Article : Google Scholar : PubMed/NCBI
|