Introduction
The inhibitor of growth (ING) family of type II
tumor suppressors comprises five conserved genes, ING1-5, which
share 32–76% DNA sequence homology (1–7). ING
proteins contain a conserved plant homeodomain (PHD) in the
C-terminal region, a nuclear localization signal (NLS) in the
middle region and a novel conserved region (NCR) of unknown
function at the N-terminus (8).
Results of previous studies showed that ING genes are involved in
DNA repair, chromatin remodeling, cell cycle control, senescence
and apoptosis (9,10).
The deregulation of ING genes was frequently
detected in various types of cancer (9,11).
ING1 gene expression was observed to be downregulated or lost in
various types of cancer including breast, gastric, esophageal, lung
and brain (2,12–16).
Previous studies investigating ING2 in cancer have suggested that a
reduction in expression is involved in the initiation of melanoma
and the progression of hepatocellular carcinoma (HCC) (17,18).
Certain studies have reported that ING4 expression was
significantly suppressed in brain tumors, HCC, breast cancer and
head and neck squamous cell carcinoma (HNSCC) (16,19–21).
Our previous study also reported the decreased expression of ING5
gene in HCC (22). Therefore, the
deregulation of ING genes may contribute to tumorigenesis (8).
The ING3 gene, which encodes a 46.8 kDa protein, has
been associated with the modulation of p53-mediated transcription,
cell cycle control and apoptosis (5). Findings of previous studies
demonstrated that the ectopic expression of ING3 in RKO cells
decreased colony formation and the number of cells in the S phase.
Although physical association with p53 is required for the function
of the other ING members, ING3 does not appear to interact with p53
(8). In melanoma cells, ING3
overexpression promotes UV-induced apoptosis through a
Fas/caspase-8-dependent pathway in a p53-independent manner
(23). ING3 has been reported to be
a tumor suppressor in melanoma and HNSCC (24,25).
Low levels of ING3 mRNA may indicate an aggressive head and neck
carcinoma. In melanoma, ING3 nuclear expression is reduced and may
be an independent prognostic factor (21).
In this study, the expression of ING3 was evaluated
in tissues at different stages of HCC using the reverse
transcription-polymerase chain reaction (RT-PCR) and an
immunohistochemical assay of tissue microarray (TMA). ING3 was
significantly downregulated in malignant HCC tissue. Moreover, it
was demonstrated that ING3 suppressed HCC cell proliferation,
colony formation and inhibited cell migration. This suggests that
the deregulation of ING3 is involved in the tumorigenesis and
metastasis of HCC.
Materials and methods
Tissue specimens and cell lines
The tumor and normal liver specimens were obtained
from patients who had provided informed consent. HepG2, Hep3B,
Huh7, Bel-7402, Bel-7404, Bel-7405, PLC, PCL/PRF/5, LM3, LM6,
QCY-7701, SNU398, MHCC-H, MHCC-L, YY-8103, SK-HEP, SMMC-7721 and
Focus were the 18 liver tumor-derived cell lines used in this
study. The study and the protocol for the use of human tissues for
this study were approved by the ethics committee of the Chinese
National Human Genome Center (Shanghai, China).
Plasmids and antibodies
The entire open reading frame of human ING3 was
subcloned into pcDNA3.0 (Invitrogen, Carlsbad, CA, USA) mammalian
cell expression vectors. pGEX5x-1-ING3 was constructed to produce
the GST-ING3 fusion protein for generating antibodies against human
ING3. Rabbit polyclonal anti-ING3 antibodies were raised against
the GST-ING3 fusion protein and purified from anti-serum with
protein G sepharose beads (Roche Diagnostics, Mannheim, Germany).
The specificity of the ING3 antibody was verified by western blot
analysis with the protein samples from the cells transfected with
plasmids expressing ING1-5 (data not shown). Mouse anti-actin
antibody was purchased from Sigma (St. Louis, MO, USA).
RNA extraction and real-time RT-PCR
Total RNA was extracted using TRIzol solution
(Invitrogen) in accordance with the manufacturer’s instructions.
Reverse transcription was performed in a 20 μl reaction system with
2 μg total RNA treated with M-MLV reverse transcriptase to
synthesis first-strand cDNA (Promega, Madison, WI, USA). Real-time
quantitative RT-PCR was performed with specific primers for ING3
and GAPDH served as an internal control. The sequences of the sense
and antisense primers were as follows: ING3:
5′-ACCTGAGTGGAGGGAAGAGC-3′ (F) and 5′-CTGGTTTGCCAACTGAACCT-3′ (R);
GAPDH: 5′-GAAGGTGAAGGTCGGAGTC-3′ (F) and 5′-GAAGATGGTGATGGGATTTC-3′
(R).
Immunohistochemical analysis
Slides containing 121 HCC specimens with adjacent
noncancerous tissues (Shanghai Biochip Company Ltd., Shanghai,
China) were used to evaluate ING3 expression via
immunohistochemistry. The slides were incubated overnight at 4°C
with rabbit anti-ING3 polyclonal antibody (1:200 dilution),
followed by incubation with a horseradish peroxidase-conjugated
anti-rabbit secondary antibody (Dako Japan Ltd., Kyoto, Japan) at
37°C for 30 min. Normal rabbit IgG was used as a negative control.
The signals were visualized using 3,3′-diaminobenzidine
tetrahydrochloride (DAB). The slides were counterstained with
hematoxylin, dehydrated through gradient alcohols and mounted for
observation. The total ING3 immunostaining was calculated as the
sum of the relative positivity of stained tumor cells and the
staining intensity. The relative positivity was scored as weakly,
moderately and strongly stained. The final ING3 expression levels
were defined as: ‘upregulated’, ‘equal’ and ‘downregulated’ in HCC
samples compared with noncancerous livers.
Cell culture and transfection
Cell lines were cultured in DMEM-high supplemented
with 10% fetal bovine serum (Gibco, Carlsbad, CA, USA) in a 5%
CO2 atmosphere at 37°C. The expression vector pcDNA3.0
and the pcDNA3.0-ING3 plasmids were transfected by lipofectamine
2000 (Invitrogen) according to the manufacturer’s instructions.
Cell proliferation and colony
formation
To observe cell proliferation, the Hep3B cells
transfected with ING3 were seeded in 96-well plates at 3,000
cells/well and cultured for 6 days. Cell viability was measured
using the Cell Counting Kit-8 (Dojindo Laboratories, Kunamoto,
Japan). HCC cells transfected with pcDNA3.0-ING3 were cultured on
100-mm plates and selected with G418 (Life Technologies, Inc.,
Carlsbad, CA, USA) at a final concentration of 0.6 to 1 mg/ml for
colony formation.
Wound-healing assay
The transfected cells were plated in 60-mm dishes at
the same density, scratch wounded with a micropipette tip, washed
to remove detached cells, provided with fresh medium including 2%
FBS and were incubated at 37°C during image capture. The wounds
were marked under the dish with a felt tip pen and images were
captured at these sites using a ×10 objective at 0 h and again at
24 and 48 h. Cell motility was evaluated using the formula: Wound
closure = (distance24 or 48 h − distance0
h)/distance0 h.
Western blot analysis
Following transfection, the cells were obtained via
lysis buffer [25 mmol/l Tris (pH 6.8), 1% SDS, 5 mmol/l EDTA,
protease inhibitor cocktail (Sigma)]. The protein samples were
resolved via SDS-PAGE and transferred onto a nitrocellulose
membrane which was blocked in 5% skimmed milk in phosphate-buffered
saline (PBS) containing Tween (PBS-Tween) and probed with the
indicated antibodies. The membrane was scanned on an Odyssey
infrared imaging system (LI-COR) at a wavelength of 700 or 800 nm.
Rabbit anti-ING3 and mouse anti-β-actin (Sigma) antibodies were
used in this study.
Statistical analysis
Quantitative values are shown as the mean ± standard
deviation (SD) or median (range). A paired samples t-test was used
to determine the difference between HCC and noncancerous livers. A
χ2 test was performed to analyze the correlation between
ING3 expression and various clinicopathological characteristics.
The SPSS (Chicago, IL, USA) 13.0 software was used for all
statistical analyses. P<0.05 was considered to indicate a
statistically significant result.
Results
Decreased expression of ING3 in HCC
Previous studies have suggested that most members of
the ING family, including ING1, ING2, ING4 and ING5, are
downregulated in HCC (16,18,26,27).
In the present study, the relative expression levels of ING1-5 were
investigated in 18 pairs of human HCC specimens by real-time RT-PCR
and it was observed that the mRNA levels of the five ING family
members were significantly downregulated in HCC tumor tissues
(22). To understand the
correlation of ING3 expression with HCC, the mRNA expression level
of ING3 in an additional 31 human HCC specimens was observed. ING3
was markedly downregulated in all 49 tumor tissues compared with
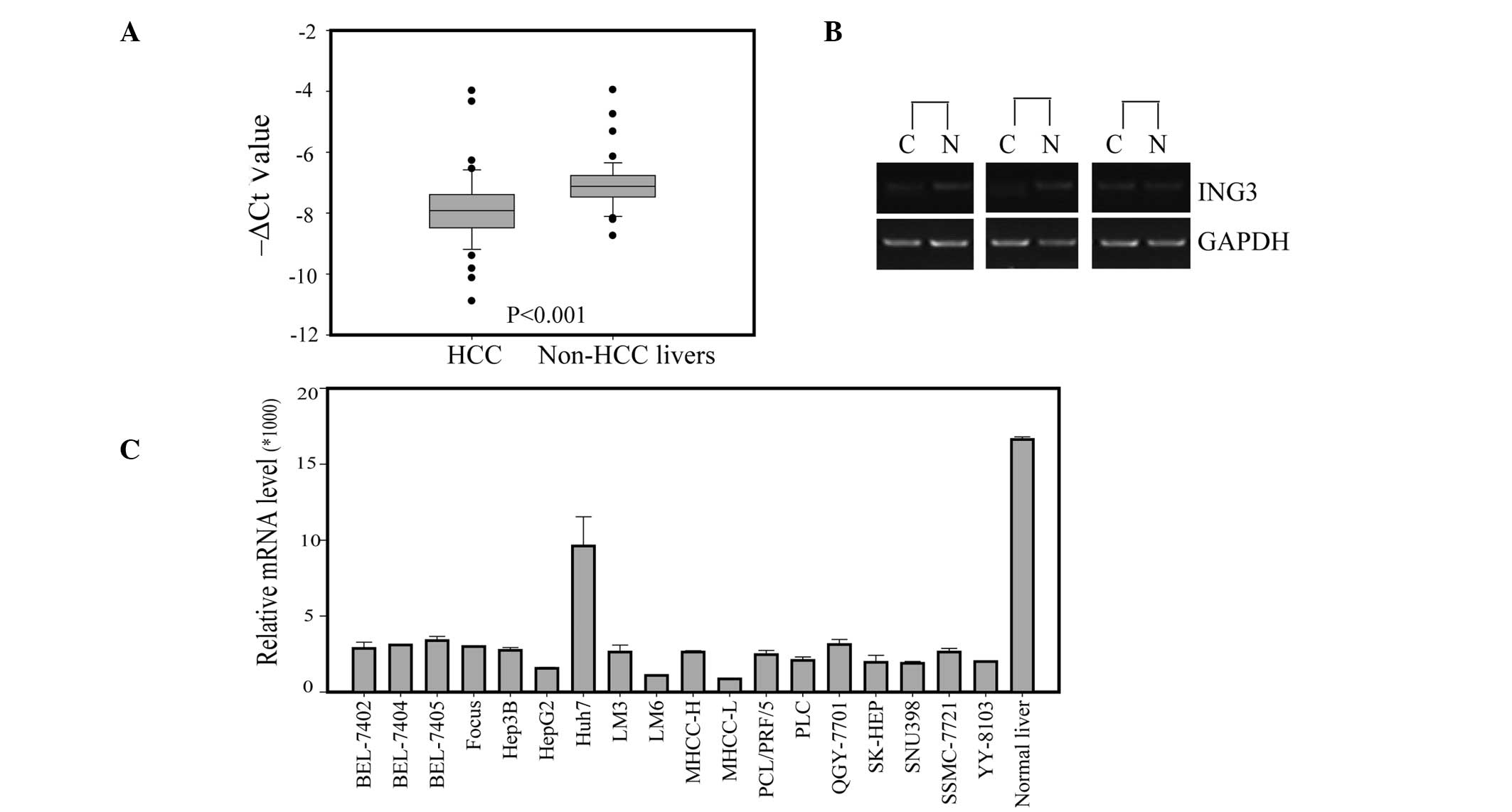
adjacent noncancerous livers (Fig.
1A). The semiquantitative RT-PCR of 3 pairs of randomly
selected human HCC specimens confirmed the observation of a
decreased ING3 expression in HCC tissues (Fig. 1B). The corresponding mRNA expression
in 18 typical HCC cell lines was also examined by RT-PCR (Fig. 1C). The cultured HCC cells expressed
various levels of ING3, which were lower than those of normal liver
tissue.
Immunohistochemistry and
clinicopathological analysis
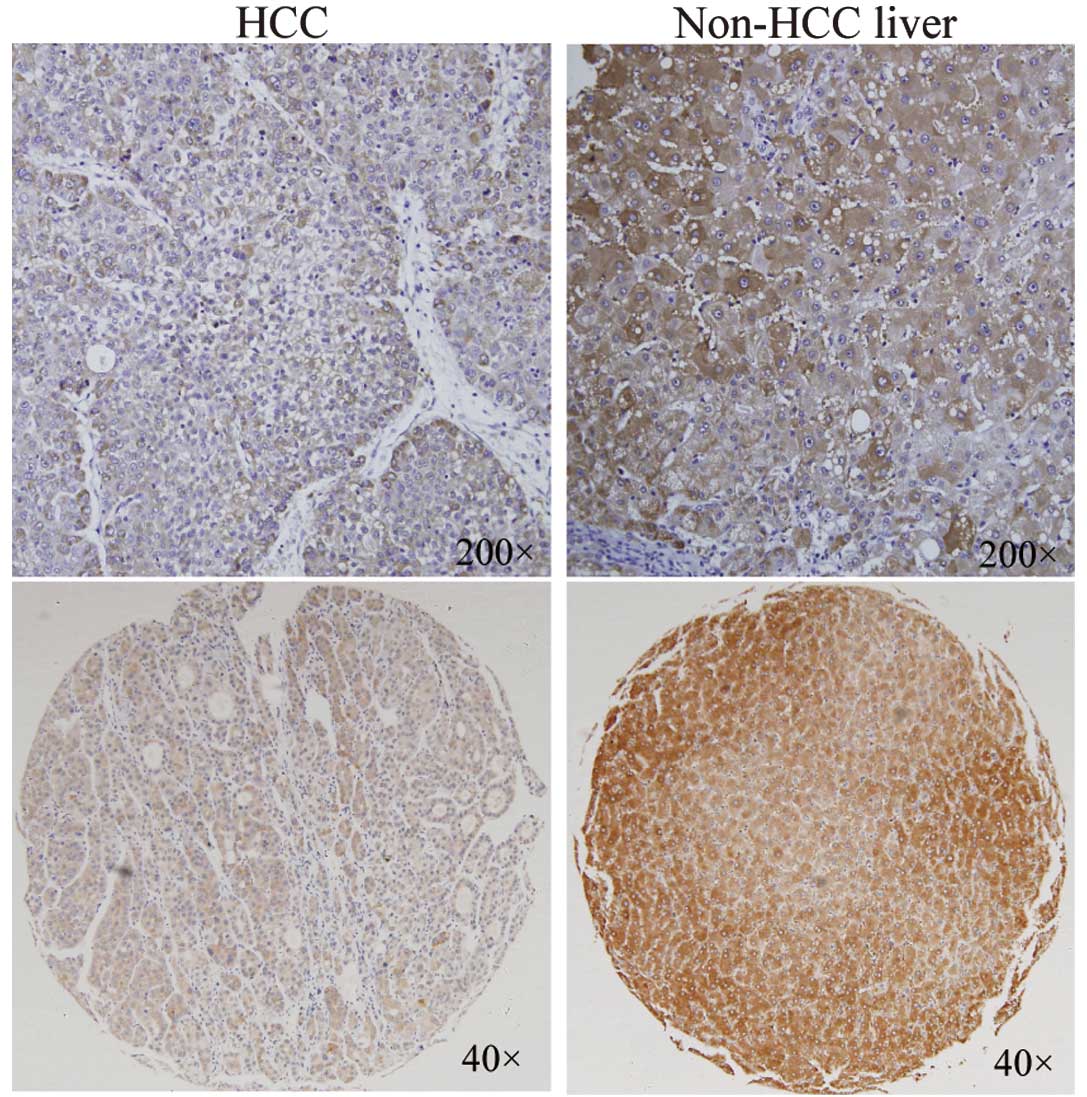
To investigate the ING3 protein expression in human
HCC tissues, a tissue array containing 112 pairs of HCC specimens
was examined by immunohistochemical staining with an ING3 antibody
(Fig. 2). The expression of the
ING3 protein was downregulated in 57.14% (64/112) HCC tissues
compared with the corresponding noncancerous livers, while the
upregulation of ING3 was detected in 21.42% (24/112) samples of HCC
and the remaining 24 HCC tissues exhibited almost the same level of
ING3 expression (Table I). To
understand the correlation of ING3 levels with clinicopathological
features, the χ2 test was used to analyze the
correlations between the staining intensity of the ING3 protein and
the clinicopathological variables of HCC (Table I). The ING3 protein staining
intensity showed no significant correlation with gender or age
(p>0.05; Table I). However, the
downregulation ratio of the ING3 protein was significantly greater
in the HCC samples with Edmondson-Steiner grades II/III than in
those with Edmondson-Steiner grades I/II (Table I; p=0.004), suggesting that a low
expression level of ING3 in HCC is associated with tumor
differentiation and classification.
 | Table ICorrelations between ING3 expression
and clinicopathological variables of 112 cases of HCC. |
Table I
Correlations between ING3 expression
and clinicopathological variables of 112 cases of HCC.
| Clinicopathological
variables | Number of
patients | ING3 expression level
(Ca compared with Nb) | P-value | χ2 |
|---|
|
|---|
| Upregulated n
(%) | Equal n (%) | Downregulated n
(%) |
|---|
| All cases | 112 | 24 (21.42) | 24 (21.42) | 64 (57.14) | | |
| Age (years) | | | | | 0.182 | 3.412 |
| ≥50 | 79 | 20 (25.32) | 18 (22.78) | 41 (51.90) | | |
| <50 | 33 | 4 (12.12) | 6 (18.18) | 23 (69.70) | | |
| Gender | | | | | 0.982 | 0.037 |
| Male | 102 | 22 (21.57) | 22 (22.57) | 58 (56.86) | | |
| Female | 10 | 2 (20.00) | 2 (20.00) | 6 (60.00) | | |
| Edmondson-Steiner
grade | | | | | 0.004c | 11.000 |
| Low I/II | 28 | 12 (42.85) | 6 (21.43) | 10 (35.71) | | |
| High II/III | 84 | 12 (14.29) | 18 (21.43) | 54 (64.29) | | |
ING3 inhibits cell proliferation and
colony formation
To investigate the effect of ING3 on HCC cells in
vitro, recombinant pcDNA3.0-ING3 plasmids were used to induce
the transient overexpression of ING3 in hepatoma cells. The
expression level of ING3 in Hep3B and Huh7 cells transfected with
the indicated plasmids was detected with western blot analysis
(Fig. 3A). The tumor cell growth
was examined daily for 6 days using the CCK-8 assay and the ability
to form colonies was measured following the treatment of the cells
with G418. As shown in Fig. 3B, the
overexpression of ING3 significantly suppressed cell proliferation
of Hep3B cells (p<0.05). In addition, the colony formation was
also markedly inhibited by ING3 in Hep3B cells (Fig. 3C; p<0.01). In Huh7 cells,
although no effect on the cell proliferation was detected (data not
shown), ING3 greatly decreased the colony formation (Fig. 3D; p<0.05). These data suggest
that as a tumor suppressor, ING3 inhibited the hepatoma cell growth
in vitro.
ING4 inhibits hepatoma cell
migration
Cell motility plays a significant role during tumor
progression and tumor metastasis. The present study also showed
that the downregulation of ING3 was correlated with the grade of
HCC. The effect of ING3 on hepatoma cell migration was then
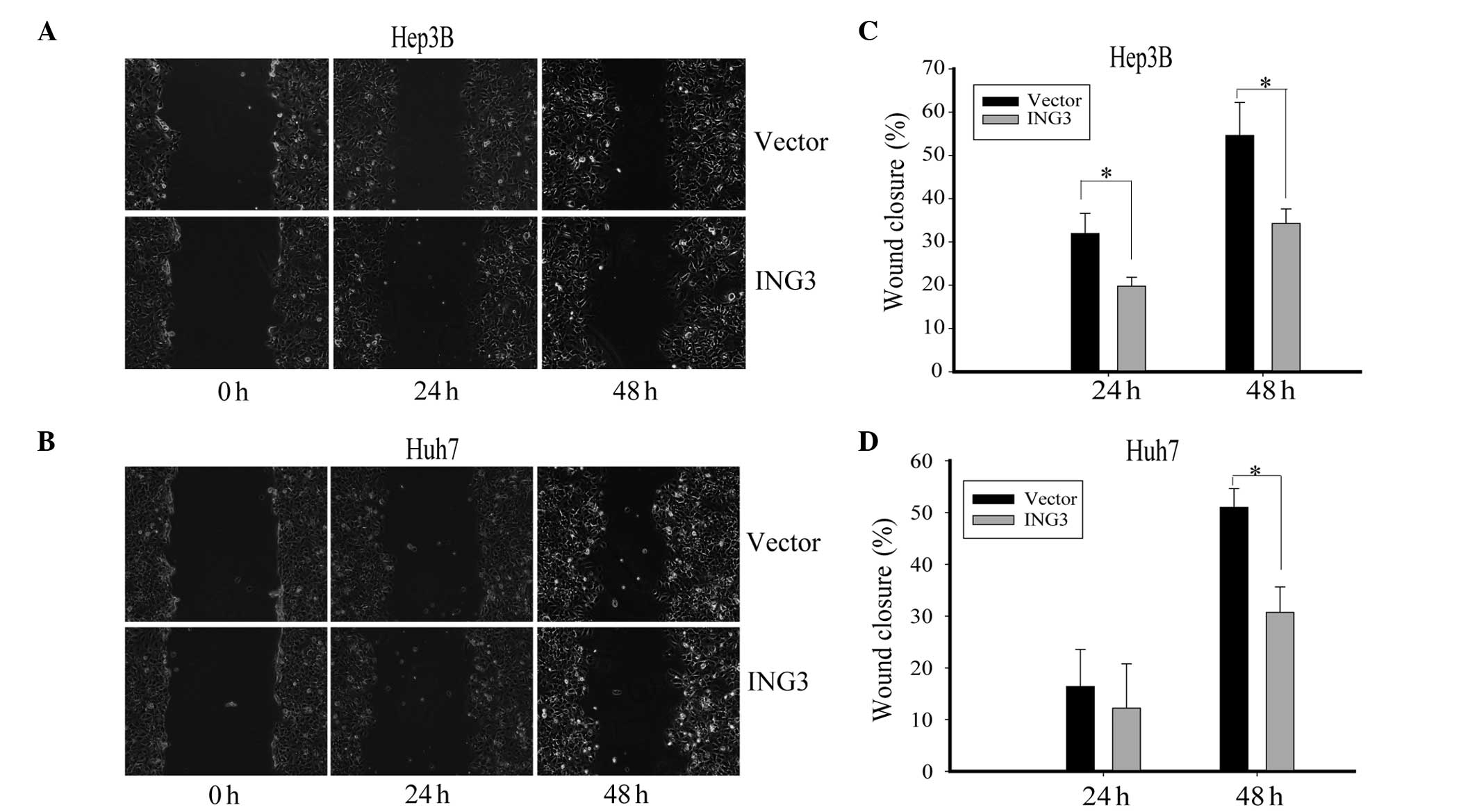
investigated using the wound-healing assay. The Hep3B and Huh7
cells with ING3 overexpression were scratched and monitored for
wound healing with time. Notably, the hepatoma cells in which ING3
was overexpressed migrated markedly slower than the control cells
(Fig. 4A and B). The wound healing
rate of the Hep3B and Huh7 cells was significantly decreased by
ING3 overexpression (Fig. 4C and D,
p<0.05). These data indicate that ING3 impaired the migration
ability of hepatoma cells, implying that ING3 plays an inhibitory
role in HCC metastasis.
Discussion
The ING family members are type II tumor suppressor
genes and have been reported to be involved in multiple cell
processes, including cell growth and proliferation, senescence, DNA
repair, oncogenesis, apoptosis, angiogenesis and tumorigenesis
(8,9). Previous studies have indicated that
the ING family is involved in HCC pathogenesis. The expression of
ING1 variants was markedly reduced in HCC samples. ING1 inhibited
the hepatoma cell growth via stabilization and activation of p53 by
interacting with Mdm2 and p14ARF (26,27).
As a candidate tumor suppressor gene, ING2 was involved in the
progression of HCC (18). The
downregulation of ING2 has been observed in 52.8% of HCC and is
associated with tumor size, histopathological classification and
serum α-fetoprotein. Similarly, ING4 expression was found to be
significantly reduced in HCC and the expression level also
correlated with patient prognosis and the metastatic potential of
HCC (16). In a recent study, we
reported that the suppression of ING5 was correlated with the
progression of HCC (22). However,
the role of ING3 in human HCC remains unknown. In this study, the
mRNA expression of ING3 in 49 paired HCC samples and 18 HCC cell
lines was investigated by RT-PCR. The results of this study showed
that approximately 83.67% (33/49) HCC samples expressed lower
levels of ING3 mRNA than the adjacent noncancerous tissues and the
expression of ING3 was decreased in the HCC cell lines.
Immunohistochemical staining of the tissue array consistently
indicated that the ING3 protein was also downregulated in HCC
compared with the corresponding noncancerous liver specimens.
Notably, the downregulation of ING3 was more frequent in moderate
and high Edmondson-Steiner grades of tumor. Previous studies have
reported that Edmondson-Steiner grades were closely associated with
tumor progression and differentiation. Therefore, the decreased
ING3 expression may be associated with tumor progression.
A previous study has shown that ING3 regulated the
cell cycle and apoptosis. Nagashima et al indicated that
ING3 overexpression may reduce the number of RKO cells in S phase
and induce p53-dependent apoptosis (5). Wang et al noted that ING3
significantly promoted UV-induced apoptosis through the activation
of the Fas/caspase-8 pathway in melanoma cells (23). In the present study, cell
proliferation, colony formation and wound-healing assays were
performed to investigate the functional role of ING3 in HCC cells.
The ectopic expression of ING3 in Hep3B and Huh7 cells inhibited
colony formation and cell migration significantly, suggesting that
ING3 is involved in tumorigenesis and metastasis. These findings
were consistent with the immunohistochemical staining and tissue
array results. However, the cell proliferation results of these HCC
cells demonstrated a different effect of ING3. Overexpression of
ING3 suppressed the cellular growth in Hep3B cells but not in Huh7
cells, suggesting that the cellular inhibitory effect of ING3 in
HCC depends on various mechanisms.
Previous studies have suggested that ING3 was
downregulated in HSNCC and may play a suppressing role in melanoma
tumorigenesis. The degradation of ING3 protein by SCF-mediated
ubiquitin-proteasome system in melanoma results in progression of
the tumor (28). Another study
showed that the nuclear-to-cytoplasmic translocation of ING3
resulted in a decreased nuclear expression and was involved in
melanoma initiation and tumor progression (25). However, the potential role of ING3
as a tumor suppressor in other types of cancer remains unclear. In
this study, downregulation of ING3 was found to be correlated with
tumorigenesis and the progression of HCC. Moreover, ING3 may
inhibit the tumorigenesis through suppressing cell proliferation
and colony formation. Furthermore, ING3 may impede tumor metastasis
by inhibiting cell migration. The role played by the molecular
mechanism of ING3 in the suppression of HCC requires further
clarification; however, the present study indicated that ING3 is a
potential therapeutic target of HCC. The function of ING3 in HCC
tumorigenesis and progression therefore merits further
investigation.
Acknowledgements
The study was supported by Chinese High-Tech
Research and Development Program Grants (2006AA02A305), the
National Natural Science Foundation of China (81071842) and the
Shanghai Commission for Science and Technology (10JC1411900).
References
|
1
|
He GH, Helbing CC, Wagner MJ, Sensen CW
and Riabowol K: Phylogenetic analysis of the ING family of PHD
finger proteins. Mol Biol Evol. 22:104–116. 2005.PubMed/NCBI
|
|
2
|
Toyama T, Iwase H, Watson P, et al:
Suppression of ING1 expression in sporadic breast cancer. Oncogene.
18:5187–5193. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nagashima M, Shiseki M, Miura K, et al:
DNA damage-inducible gene p33ING2 negatively regulates cell
proliferation through acetylation of p53. Proc Natl Acad Sci USA.
98:9671–9676. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shimada Y, Saito A, Suzuki M, Takahashi E
and Horie M: Cloning of a novel gene (ING1L) homologous to ING1, a
candidate tumor suppressor. Cytogenet Cell Genet. 83:232–235. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nagashima M, Shiseki M, Pedeux RM, et al:
A novel PHD-finger motif protein, p47ING3, modulates p53-mediated
transcription, cell cycle control, and apoptosis. Oncogene.
22:343–350. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shiseki M, Nagashima M, Pedeux RM, et al:
p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity.
Cancer Res. 63:2373–2378. 2003.PubMed/NCBI
|
|
7
|
Zhang X, Xu LS, Wang ZQ, et al: ING4
induces G2/M cell cycle arrest and enhances the chemosensitivity to
DNA-damage agents in HepG2 cells. FEBS Lett. 570:7–12. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Coles AH and Jones SN: The ING gene family
in the regulation of cell growth and tumorigenesis. J Cell Physiol.
218:45–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gong W, Suzuki K, Russell M and Riabowol
K: Function of the ING family of PHD proteins in cancer. Int J
Biochem Cell Biol. 37:1054–1065. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Russell M, Berardi P, Gong W and Riabowol
K: Grow-ING, Age-ING and Die-ING: ING proteins link cancer,
senescence and apoptosis. Exp Cell Res. 312:951–961. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Campos EI, Chin MY, Kuo WH and Li G:
Biological functions of the ING family tumor suppressors. Cell Mol
Life Sci. 61:2597–2613. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tokunaga E, Maehara Y, Oki E, et al:
Diminished expression of ING1 mRNA and the correlation with p53
expression in breast cancers. Cancer Lett. 152:15–22. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Oki E, Maehara Y, Tokunaga E, Kakeji Y and
Sugimachi K: Reduced expression of p33(ING1) and the relationship
with p53 expression in human gastric cancer. Cancer Lett.
147:157–162. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hara Y, Zheng Z, Evans SC, et al: ING1 and
p53 tumor suppressor gene alterations in adenocarcinomas of the
esophagogastric junction. Cancer Lett. 192:109–116. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Okano T, Gemma A, Hosoya Y, et al:
Alterations in novel candidate tumor suppressor genes, ING1
and ING2 in human lung cancer. Oncol Rep. 15:545–549.
2006.PubMed/NCBI
|
|
16
|
Fang F, Luo LB, Tao YM, Wu F and Yang LY:
Decreased expression of inhibitor of growth 4 correlated with poor
prognosis of hepatocellular carcinoma. Cancer Epidemiol Biomarkers
Prev. 18:409–416. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu F, Dai DL, Martinka M, Ho V and Li G:
Nuclear ING2 expression is reduced in human cutaneous melanomas. Br
J Cancer. 95:80–86. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang HK, Pan K, Wang H, et al: Decreased
expression of ING2 gene and its clinicopathological significance in
hepatocellular carcinoma. Cancer Lett. 261:183–192. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Garkavtsev I, Kozin SV, Chernova O, et al:
The candidate tumour suppressor protein ING4 regulates brain tumour
growth and angiogenesis. Nature. 428:328–332. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim S, Chin K, Gray JW and Bishop JM: A
screen for genes that suppress loss of contact inhibition:
identification of ING4 as a candidate tumor suppressor gene in
human cancer. Proc Natl Acad Sci USA. 101:16251–16256. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gunduz M, Nagatsuka H, Demircan K, et al:
Frequent deletion and down-regulation of ING4, a candidate tumor
suppressor gene at 12p13, in head and neck squamous cell
carcinomas. Gene. 356:109–117. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lu ML, Chen F, Wang QW, Han ZG and Zhang
X: Study on expression of ING5 in hepatocellular carcinoma. Wei
Chang Bing Xue Za Zhi Tou Gao Xu Zhi. 15:86–89. 2010.
|
|
23
|
Wang Y and Li G: ING3 promotes UV-induced
apoptosis via Fas/caspase-8 pathway in melanoma cells. J Biol Chem.
281:11887–11893. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gunduz M, Beder LB, Gunduz E, et al:
Downregulation of ING3 mRNA expression predicts poor prognosis in
head and neck cancer. Cancer Sci. 99:531–538. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Y, Dai DL, Martinka M and Li G:
Prognostic significance of nuclear ING3 expression in human
cutaneous melanoma. Clin Cancer Res. 13:4111–4116. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ohgi T, Masaki T, Nakai S, et al:
Expression of p33(ING1) in hepatocellular carcinoma: relationships
to tumour differentiation and cyclin E kinase activity. Scand J
Gastroenterol. 37:1440–1448. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu Z, Luo Z, Li Y, Ni C, Li H and Zhu M:
Human inhibitor of growth 1 inhibits hepatoma cell growth and
influences p53 stability in a variant-dependent manner. Hepatology.
49:504–512. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen G, Wang Y, Garate M, Zhou J and Li G:
The tumor suppressor ING3 is degraded by SCF(Skp2)-mediated
ubiquitin-proteasome system. Oncogene. 29:1498–1508. 2010.
View Article : Google Scholar : PubMed/NCBI
|