Introduction
The Wnt family is a group of glycoproteins
characterized by a number of conserved cysteine residues. These Wnt
proteins are extensively involved in embryonic development and the
regulation of adult cell proliferation, differentiation and
apoptosis (1). The secreted
Frizzled-related protein family (SFRPs), including five members
ranging from secreted frizzled-related protein 1 (SFRP1) to SFRP5,
are Wnt antagonists. These members possess a characteristic
cysteine-rich domain (CRD), which is extremely similar to the CRD
of the Wnt receptor Frizzled protein family. Consequently, SFRPs
can also combine with Wnt through the CRD to dampen cellular signal
transduction (2). Downregulated
expression of the Wnt antagonists can lead to the combination of
more Wnt proteins with the receptor, Frizzled. This event results
in reduced degradation of GSK3 on its substrate β-catenin, which
translocates into the nucleus and has an impact on the
transcription factor TCF/LEF. In turn, this regulates the
transcription of the target genes c-myc and cyclinD1,
causing excessive cell proliferation and thereby inducing
tumorigenesis. Downregulated expression of SFRP1 occurs in
several types of cancer. For example, Zou et al reported
that in esophageal cancers, SFRP1 and other Wnt antagonists
are widely methylated (3), while
Lodygin et al found that the SFRP1 gene was also
methylated in prostate cancer (4).
These studies demonstrate that SFRP1 gene methylation is the
primary mechanism underlying the repression of SFRP1 in these
tumors.
The mechanisms underlying the pathogenesis of
bladder cancer, the most common urinary tract cancer, remain
unclear. Previous studies have shown that abnormal Wnt signaling is
also involved in the pathogenesis of bladder cancer. For example,
it has been demonstrated that the expression of the Wnt family
member, Wnt7b, is significantly higher in superficial bladder
cancer than in normal bladder tissue (5). Similarly, the overexpression of Wnt5A
has been demonstrated in a bladder cancer cell line (6). Furthermore, the expression of Wnt
antagonists, including Wnt inhibitory factor 1 (WIF1), is
downregulated in bladder cancer via a gene promoter
methylation-related mechanism (7).
In the present study, we investigated the mechanisms underlying
SFRP1 expression in bladder cancer and explored the role of
altered SFRP1 expression in the pathogenesis of bladder
cancer.
Materials and methods
Materials
Samples were obtained from the Department of
Urology, Shengjing Hospital affiliated to China Medical University.
Among the 45 bladder cancer tissue samples, 28 were from males and
17 from females, with an age range of 45–66 years old and mean age
of 52 years old. Selected patients with a solitary bladder tumor
underwent partial cystectomy and postoperative pathology led to a
diagnosis of urothelial carcinoma. Exclusion criteria involved
multiple bladder tumors and bladder cancer recurrence following
surgery. A total of 45 normal tumor-adjacent tissues, corresponding
to each bladder cancer specimen, were also used in this study, with
the inclusion criterion that the normal bladder incisal edge was ≥2
cm from the tumor margin. The samples were frozen in liquid
nitrogen and stored at −80°C until use. The study was approved by
the Ethics Committee of China Medical University and consent was
obtained for all of the specimens, from patients or their family
members. The human bladder cancer cell lines T24, 5637 and SCaBER
were purchased from Shanghai Kunken Biochemical Engineering Co.,
Ltd., China.
Cell culture
The human bladder cancer cell lines T24, 5637 and
SCaBER were cultured in RPMI-1640 culture medium and placed in an
incubator with 5% CO2 at 37°C. The medium contained 10%
fetal bovine serum, penicillin (100 IU/ml) and streptomycin (100
μg/ml).
Methylation-specific PCR
The Genmed gene methylation detection kit (Shanghai
Genmed Gene Pharmaceutical Technology Co., Ltd., Shanghai, China)
was used to determine the methylation status of the SFRP1
gene. Specific steps included a transformation experiment and
methylation-specific PCR (MSR) detection. The PCR conditions were
as follows: 95°C for 5 min for 1 cycle and 95°C, 30 sec; 58°C, 30
sec; 72°C, 30 sec; for 35 cycles. Methylation detection included
detection by a methylated (M) reaction and unmethylated reaction
(U). The primer sequences used were: M, forward:
5′-TGTAGTTTTCGGAGTTAGTGTCGCGC-3′; and reverse:
5′-CCTACGATCGAAAACGACGCGAACG-3′; forward:
5′-GTTTTGTAGTTTTTGGAGTTAGTGTTGTGT-3′; and reverse,
5′-CTCAACCTACAATCAAAAACAACACAA ACA-3′.
RNA extraction and reverse transcription
PCR (RT-PCR)
TRIzol (Takara Bio Inc., Shiga, Japan) was used for
RNA extraction and the specific steps involved are described in the
instruction manual. Nine random primers and AMV reverse
transcriptase (Takara Bio, Inc.) were used for cDNA synthesis. The
total reaction volume was 10 μl, with reverse transcription
reaction conditions consisting of: 30°C, 10 min; 42°C, 25 min;
99°C, 5 min; and 5°C, 5 min for 1 cycle. The total volume for the
PCR was 40 μl, with the following reaction conditions: 94°C, 2 min
for 1 cycle and 94°C, 30 sec; 58°C, 30 sec; and 72°C, 2 min for 30
cycles. The PCR conditions used were: 94°C, 2 min for 1 cycle,
94°C, 30 sec; 60°C, 30 sec; 72°C, 2 min, for a total of 30 cycles.
Primer sequences used were: SFRP1, forward:
5′-TCTACACCAAGCCACCTCAG-3′; and reverse:
5′-CAGTCACCCCATTCTTCAGG-3′; and for the GADPH gene, forward:
5′-GGGAAACTGTGGCGTGAT-3′; and reverse:
5′-AAAGGTGGAGGAGTGGGT-3′.
Western blotting
Lysis buffer (50 mmol/l Tris-HCl, 100 mmol/l DTT, 2%
SDS, 0.1% bromophenol blue and 10% glycerol) was used for cell
lysis, protein sample collection and SDS-PAGE gel electrophoresis.
SFRP1 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA)
polyclonal antibodies and horseradish peroxidase-conjugated rabbit
anti-goat antibodies (Santa Cruz Biotechnology, Inc.) were diluted
at 1:1000. β-actin (Santa Cruz Biotechnology, Inc.) was used as the
loading control.
Statistical analysis
The extent of methylation in bladder cancer tissue
and tumor-adjacent tissue was compared using χ2 tests.
mRNA expression (gray value) was compared using Student’s t-tests.
P<0.05 was considered to indicate a statistically significant
result. SPSS 11.5 software (Chicago, IL, USA) was used for
statistical analysis.
Results
Methylation and abnormal expression of
the SFRP1 gene in bladder cancer and normal tumor-adjacent
tissue
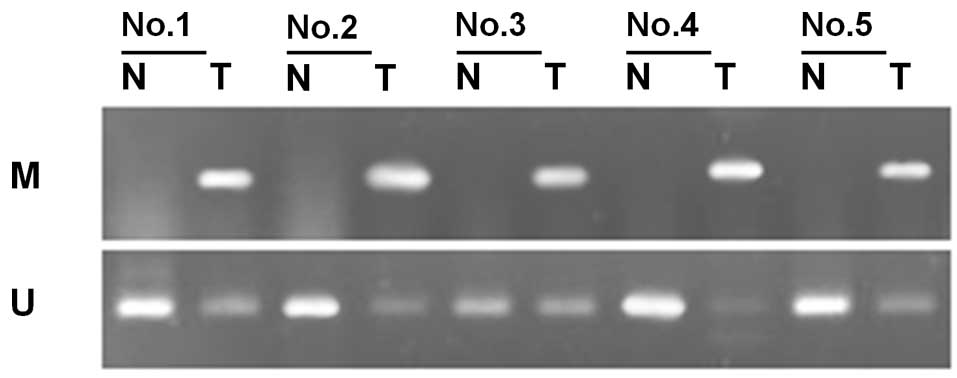
Through methylation-specific PCR, we determined the
methylation status of SFRP1 in 45 cases of bladder cancer
and their corresponding normal tumor-adjacent tissues. Of the 45
patients with bladder cancer, methylation of SFRP1 was
detected in 28 tumor samples (62.2%). In the corresponding normal
tumor-adjacent tissues, SFRP1 was methylated in only 6 cases
(13.3%). The frequency of SFRP1 methylation in bladder
cancer tissues was significantly higher than that in the adjacent
tissues (P<0.01, χ2=22.88, Fig. 1, Table
I).
 | Table IMethylation status of the SFRP1
gene in bladder cancer and normal tumor-adjacent tissues. |
Table I
Methylation status of the SFRP1
gene in bladder cancer and normal tumor-adjacent tissues.
| Tissue | n | Methylation (+) | Methylation (−) |
|---|
| Bladder cancer | 45 | 28 | 17 |
| Normal
tumor-adjacent | 45 | 6 | 39 |
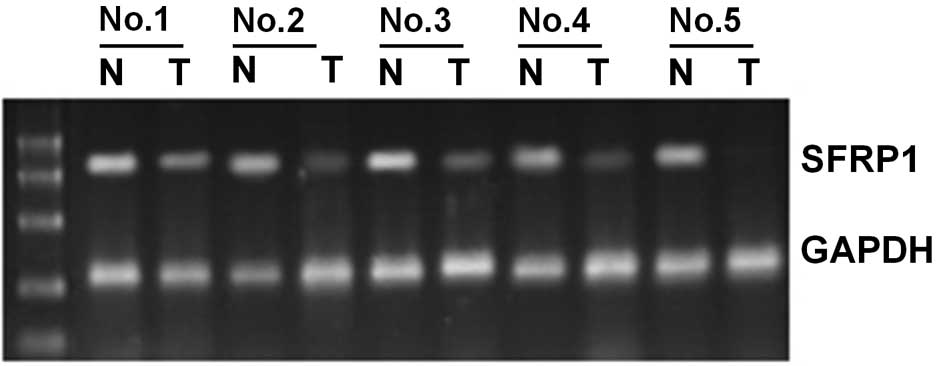
The expression of SFRP1 mRNA was analyzed by
RT-PCR in 10 bladder cancer specimens and the corresponding normal
tumor-adjacent tissues. SFRP1 was methylated in 5 of the 10
bladder cancer tissues, but was unmethylated in the remaining five.
In the five bladder cancer tissues demonstrating methylation of the
SFRP1 gene, it was found that SFRP1 mRNA expression
was significantly lower than that in the corresponding normal
tissues (P<0.01, Fig. 2). In the
five bladder cancer tissues that did not demonstrate methylation of
the SFRP1 gene, there was no significant difference in
SFRP1 mRNA expression compared to normal tumor-adjacent
tissues.
Methylation and abnormal expression of
the SFRP1 gene in bladder cancer cell lines
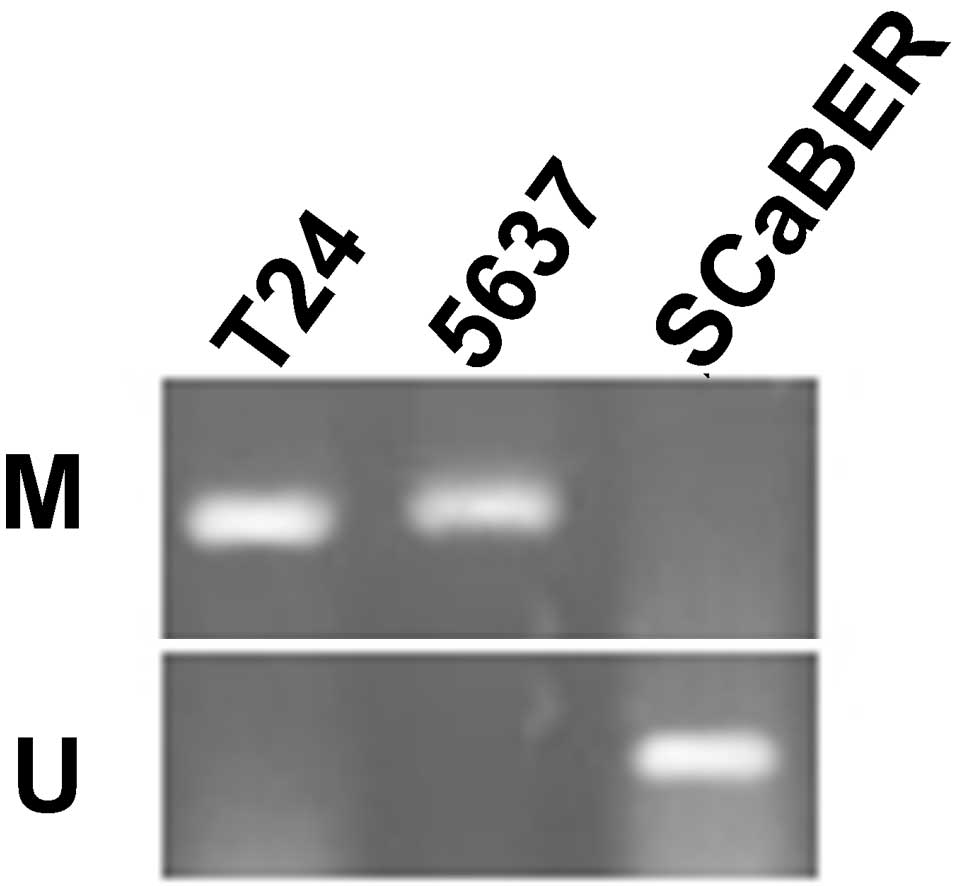
Using MSR, the methylation status of the
SFRP1 gene was analyzed in the bladder cancer cell lines
T24, 5637 and SCaBER. The data revealed that SFRP1 is
methylated in T24 and 5637 cells, but unmethylated in SCaBER cells
(Fig. 3).
RT-PCR was used to determine the expression of
SFRP1 mRNA in bladder cancer cell lines. We found that
SFRP1 mRNA was not expressed in T24 and 5637 cells, in which
SFRP1 was methylated, but was expressed in SCaBER cells,
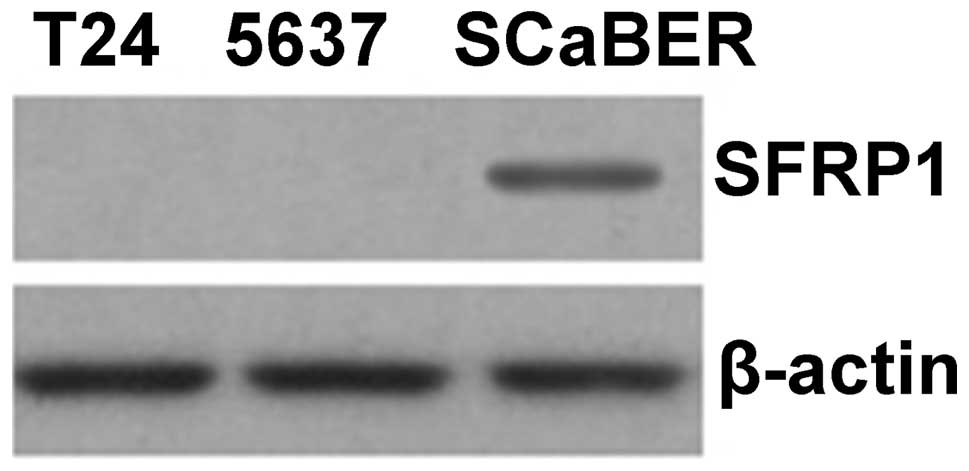
which do not demonstrate methylation of SFRP1 (Fig. 4). The expression of SFRP1 protein
was then analyzed in bladder cancer cell lines by western blotting.
Similar to our findings at the mRNA level, it was found that the
SFRP1 protein was not expressed in T24 and 5637 cells, where
SFRP1 is methylated, but was expressed in SCaBER cells,
which do not demonstrate methylation of SFRP1 (Fig. 5).
Effect of a demethylating reagent on
expression of the SFRP1 gene in bladder cancer cell lines
The bladder cancer cell lines, T24 and 5637, in
which the SFRP1 gene is normally methylated, were treated
for 6 h with the demethylating agent 5′-aza-deoxycytidine acid
(DAC, 1 μg/ml). Following treatment, methylation of the
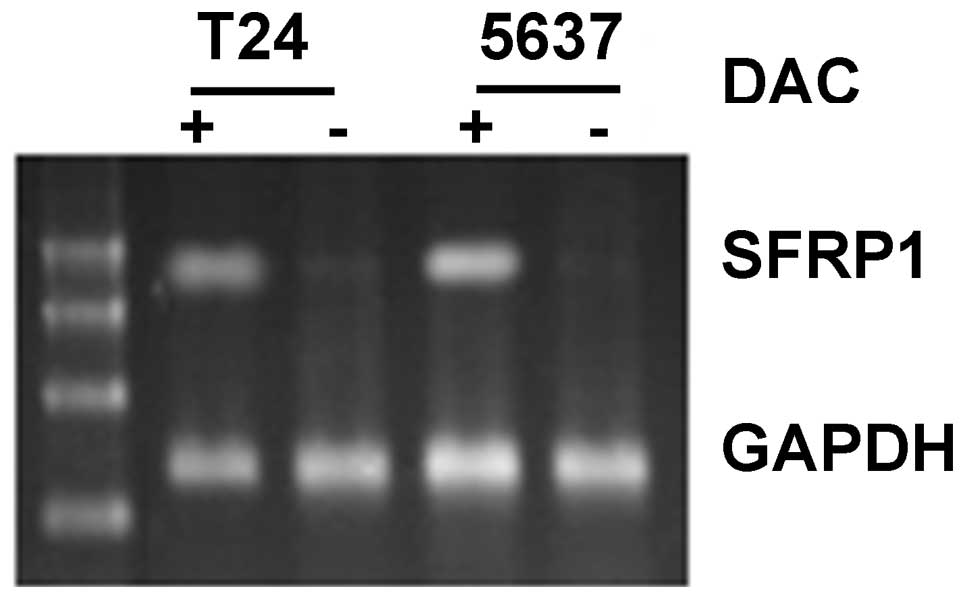
SFRP1 gene in the two cell lines had disappeared (Fig. 6). Furthermore, using RT-PCR, it was
found that SFRP1 mRNA expression increased in DAC-treated
T24 and 5637 cells (Fig. 7) and
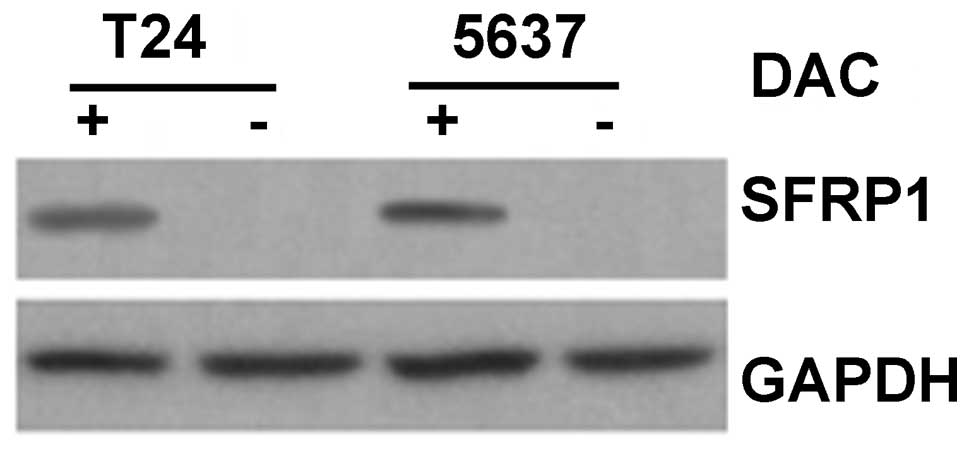
that this was accompanied by an increase in SFRP1 protein, as
determined by western blotting (Fig.
8).
Discussion
The pathogenesis of cancer is a complicated process,
involving a series of hereditary and certain epigenetic changes
that ultimately result in uncontrolled cell growth. Epigenetic
changes, including abnormal DNA methylation, are significant
tumor-inducing mechanisms through their effects on oncogene
activation or anti-oncogene inactivation (8). DNA methylation occurs though the
interaction of three types of DNA methyltransferases, DNMT1, DNMT3A
and DNMT3B, leading to the addition of a methyl group to the fifth
carbon atom of cytosine. Methylation typically occurs in CG
repetitive sequences within the CpG islands of gene promoters,
although the majority of these sequences are not methylated under
normal circumstances. However, under the influence of pathogenic
factors, CG sequences within gene promoters can become methylated,
thereby suppressing transcription. The mechanism involved may be
related to the fact that the methylation of cytosine prevents
transcription factors from combining with recognition sites within
CG sequences. Since it was first discovered approximately 20 years
ago that cytosine is methylated in tumors, an increasing number of
gene promoters have been identified that may become methylated,
leading to transcriptional suppression. Although the mechanism of
methylation is not fully understood, it is likely to be associated
with a deficiency of protective factors that compete with the
methyl transferase for binding sites on the cytosine.
The Wnt gene was discovered in 1982 and since
then a large body of research has revealed that the Wnt gene
family is widely involved in essential developmental processes in
embryos, including Drosophila segmentation, murine nerve
development, axis formation and toad muscle production among
others. The finding of the Wnt signaling pathway antagonist
heralded a new era in the study of the Wnt gene and its
effects. Currently known Wnt antagonists include SFRPs and Dickkopf
(Dkk). The SFRP family consists of five members, SFRP1-5. Similar
to the other members of the SFRP family, SFRP1 is one of the
secretory proteins of 36 kDa and is made up of over 300 amino
acids. SFRP1 contains a signal peptide, a CRD near the amino
terminal and a hydrophilic carboxyl terminus. Consisting of
approximately 110 amino acid residues and 10 highly conserved
cysteines, the CRD area has 30–40% homology with the Frizzled
extracellular CRD. Studies have revealed that SFRP1 is capable of
stimulating apoptosis and suppressing cell proliferation (9–11).
Since SFRP1 is able to negatively regulate the Wnt
signal transduction pathway and stimulate apoptosis, downregulation
of SFRP1 expression generally causes overactivation of the
Wnt signal transduction pathway, increased cell proliferation and
tumorigenesis. In recent years, the Dkk and SFRP family members,
which are SFRP1 antagonists, were recognized to be downregulated in
tumors. Since the SFRP1 gene is located on chromosome
8p11.2, which is incomplete in many types of tumors, the
SFRP1 gene is suspected to be a tumor suppressor gene.
In this study, 45 specimens of bladder cancer with
their corresponding normal tumor-adjacent tissues were analyzed and
it was found that SFRP1 was methylated in 62.2% of bladder
cancer specimens and in 13.3% of normal tumor-adjacent tissues.
Statistical analysis revealed that methylation of the SFRP1
gene in cancer tissues occurs more frequently than in its
corresponding normal tumor-adjacent tissues. Analysis of
SFRP1 mRNA expression in 10 specimens of bladder cancer and
their normal adjacent tissues by RT-PCR revealed that methylation
of the SFRP1 gene is significantly lower than that in normal
tumor-adjacent tissues. Conversely, SFRP1 mRNA expression in
cancer specimens that do not demonstrate methylation of the
SFRP1 gene is not markedly different to that in adjacent
normal tissues. Thus, our data prove that SFRP1 methylation
is common in bladder cancer and suggest that the downregulated
SFRP1 expression is likely to be due to the methylation of
the SFRP1 gene.
To analyze the methylation and expression status of
the SFRP1 gene in bladder cancer, three types of bladder
cancer cell lines were used. It was found that the SFRP1
gene was methylated in the T24 and 5637 cell lines and that there
was also a reduced expression of SFRP1 mRNA and protein in
these cells. Furthermore, upon treatment with a demethylating
reagent, SFRP1 mRNA and protein expression re-occurred in
these cells. These results provide further evidence that
SFRP1 expression is frequently downregulated in bladder
cancer and that this downregulation is mainly due to the abnormal
methylation of SFRP1.
According to our findings, we hypothesize that
methylation of the SFRP1 gene and the consequent
downregulation of protein expression is partially involved in the
pathogenesis of bladder cancer. The mechanism involved is likely to
be related to the over-activation of the Wnt signaling pathway.
Since the Wnt signaling pathway is divided into the classical
β-catenin and other non-classical pathways, and β-catenin was not
detected in the cytoplasm, the specific molecular mechanisms by
which downregulation of SFRP1 expression is involved in the
pathogenesis of bladder cancer remains unclear. To fully define the
role of abnormal SFRP1 expression in the pathogenesis of
bladder cancer, the involvement of genes, including c-myc and
cyclinD1, that may be modulated by the downregulation of
SFRP1 expression should be analyzed.
Acknowledgements
This study was supported by grants from Liaoning
Natural Science Foundation (20092138).
References
|
1
|
Katanaev VL: The Wnt/Frizzled GPCR
signaling pathway. Biochemistry (Mosc). 75:1428–1434. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Esteve P and Bovolenta P: The advantages
and disadvantages of sfrp1 and sfrp2 expression in pathological
events. Tohoku J Exp Med. 221:11–17. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zou H, Molina JR, Harrington JJ, et al:
Aberrant methylation of secreted frizzled-related protein genes in
esophageal adenocarcinoma and Barrett’s esophagus. Int J Cancer.
116:584–591. 2005.PubMed/NCBI
|
|
4
|
Lodygin D, Epanchintsev A, Menssen A,
Diebold J and Hermeking H: Functional epigenomics identifies genes
frequently silenced in prostate cancer. Cancer Res. 65:4218–4227.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bui TD, O’Brien T, Crew J, Cranston D and
Harris AL: High expression of Wnt7b in human superficial bladder
cancer vs invasive bladder cancer. Br J Cancer. 77:319–324. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhuang L, Zhang Z and Guo Y: Aberrant
expression of growth factor Wnt-5A in six urinary malignant cell
lines. Chin Med J (Engl). 112:251–255. 1999.PubMed/NCBI
|
|
7
|
Urakami S, Shiina H, Enokida H, et al:
Epigenetic inactivation of Wnt inhibitory factor-1 plays an
important role in bladder cancer through aberrant canonical
Wnt/beta-catenin signaling pathway. Clin Cancer Res. 12:383–391.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tycko B: Cancer epigenetics and targeted
therapies. Oncology (Williston Park) 25. 228:2312011.PubMed/NCBI
|
|
9
|
Ezan J, Leroux L, Barandon L, et al:
FrzA/sFRP-1, a secreted antagonist of the Wnt-Frizzled pathway,
controls vascular cell proliferation in vitro and in vivo.
Cardiovasc Res. 63:731–738. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Häusler KD, Horwood NJ, Chuman Y, et al:
Secreted frizzled-related protein-1 inhibits RANKL-dependent
osteoclast formation. J Bone Miner Res. 19:1873–1881.
2004.PubMed/NCBI
|
|
11
|
Bodine PV, Zhao W, Kharode YP, et al: The
Wnt antagonist secreted frizzled-related protein-1 is a negative
regulator of trabecular bone formation in adult mice. Mol
Endocrinol. 18:1222–1237. 2004. View Article : Google Scholar : PubMed/NCBI
|