Introduction
Lung cancer is one of the most common types of
malignancy worldwide, which has a complex pathogenesis.
Epidemiological and etiological studies have revealed that risk of
lung cancer can be affected by a complex interaction between a
number of genetic and environmental factors (e.g., tobacco smoke,
radiation and infectious agents) (1). Further research has identified that
there are polymorphisms in susceptibility genes, including those
coding for DNA repair enzymes.
The incidence of lung cancer is associated with the
alteration of DNA by external environmental factors and endogenous
carcinogens (2). If the alteration
cannot be repaired, it may cause genetic instability, mutation and
cell death. There are at least four pathways involved in DNA
repair, including base excision repair, DNA double-strand break
repair, mismatch repair and nucleotide excision repair (NER). The
expression of genes involved in the different DNA repair pathways
varies greatly with the age of the individual. The NER system is
one of the most significant repair pathways. A number of enzymes
are involved in the NER pathway, including xeroderma pigmentosum
complementary group A (XPA), replication protein A (RPA), xeroderma
pigmentosum complementary group C (XPC), xeroderma pigmentosum
complementary group D (XPD), excision repair cross-complementing 1
(ERCC1) and xeroderma pigmentosum complementary group F (XPF)
(3). A number of studies have
examined the role of ERCC1 (4).
However, whether XPD polymorphisms correlate with different lung
cancer pathologies remains to be determined.
Cytidine deaminase (CDA) is one of the enzymes
involved in the pyrimidine rescue pathway, which catalyzes the
deamination of cytidine and deoxycytidine to form uracil
derivatives (5). Recent studies on
CDA have focused on the association between cytarabine (Ara-c) and
CDA in leukemia patients, while other studies have investigated the
effect of gemcitabine, one of the first-line treatments for
non-small cell lung cancer (NSCLC), on CDA polymorphisms in lung
cancer (14). However, there are
few studies that have examined the correlation between CDA
polymorphisms and lung cancer risk and pathogenesis.
To determine the susceptibility genes which are most
relevant for screening the Chinese population for lung cancer, we
investigated the frequency distributions of the XPD and CDA genes
in lung cancer patients compared to healthy controls.
Patients and methods
Patients
The study included 103 patients with lung cancer and
103 healthy control subjects. All patients were diagnosed with
NSCLC or small-cell lung carcinoma at the Department of Respiratory
Medicine Ruijin Hospital School of Medicine, JiaoTong University,
Shanghai, China, between January 2006 and August 2010. Each
diagnosis was confirmed by histopathology or cytology. Individuals
in the control group were selected following a medical examination
at our hospital. All subjects lived in South East China. The
recommendations of the Declaration of Helsinki for biomedical
research involving human subjects were followed.
Clinical data on age, gender, smoking and medical
history, and data from physical examinations were systematically
recorded at the start of the study. Performance status,
pathological type and clinical stage were recorded for the lung
cancer patients.
Methods
Blood samples (3 ml) were collected for genotyping
at the start of the study, and genomic DNA was isolated from blood
by DNA extraction. Genotype distributions, including XPD Asp312Asn
and Lys751Gln, and CDA Lys27Gln and Ala70Thr, were determined using
polymerase chain reaction-restriction fragment length polymorphism
(PCR-RFLP). The primers for Asp312Asn were
5′-gcggtcccaaaagggtcagttacc ggcgtctggtgga-3′ and
5′-gcggtcccaaaagggtcagtgctcaccctgca gcacttctt-3′. The PCR product
was 143 bp, and was digested with MboII. The A/A genotype
was 143 bp, the G/G genotype was 114 bp and 29 bp, and the A/G
genotype was 114, 29 and 143 bp. The primers for Lys751Gln were
5′-cttcataagacctt ctagcaccac-3′ and 5′-ctccctcagccccatctta-3′, and
the Eam1104-restricted product of the G/G genotype was 368
bp, the G/T genotype was 130, 238 and 368 bp and the T/T genotype
was 130 and 238 bp. The primers for CDA Lys27Gln were
5′-gcggtcccaaaagggtcagtttgctcccaggaggcgaag-3′ and
5′-gcggtcccaaaagggtcagtagattctcccctcctgggt-3′, which yielded a 129
bp product. The MboII-restricted products were C/C (129 bp),
A/A (48 and 81 bp) and A/C (48, 81 and 129 bp). The primers for
Ala70Thr were 5′-tgtccttctccccaccttg-3′ and
5′-ggaagatgttggctaaagagatg-3′, which yielded a 300 bp product. The
CpoI-restricted products were A/A (300 bp), G/G (119 and 181
bp) and A/G (119, 181 and 300 bp). PCR reactions (10 μl) contained
0.3 μl template DNA, 1 μl 10× Taq buffer, 0.5 μl dNTP, 0.5 pmol/μl
primer, and 0.3 units of Taq polymerase (Bio Basic Inc., Canada).
The PCR reaction conditions were as follows: initial denaturation
for 5 min at 95°C, then 40 cycles of 30 sec at 95°C, annealing for
45 sec, extension for 60 sec at 72°C, followed by 6 min at 72°C.
The annealing temperature for the XPD Asp312Asn and CDA Ala70Thr
primers was 60°C, and the temperature for the XPD Lys751Gln and CDA
Asp312Asn primers was 58°C. PCR products were evaluated by
overnight restriction enzyme digestion at 37°C, followed by 2%
agarose gel electrophoresis (Fig.
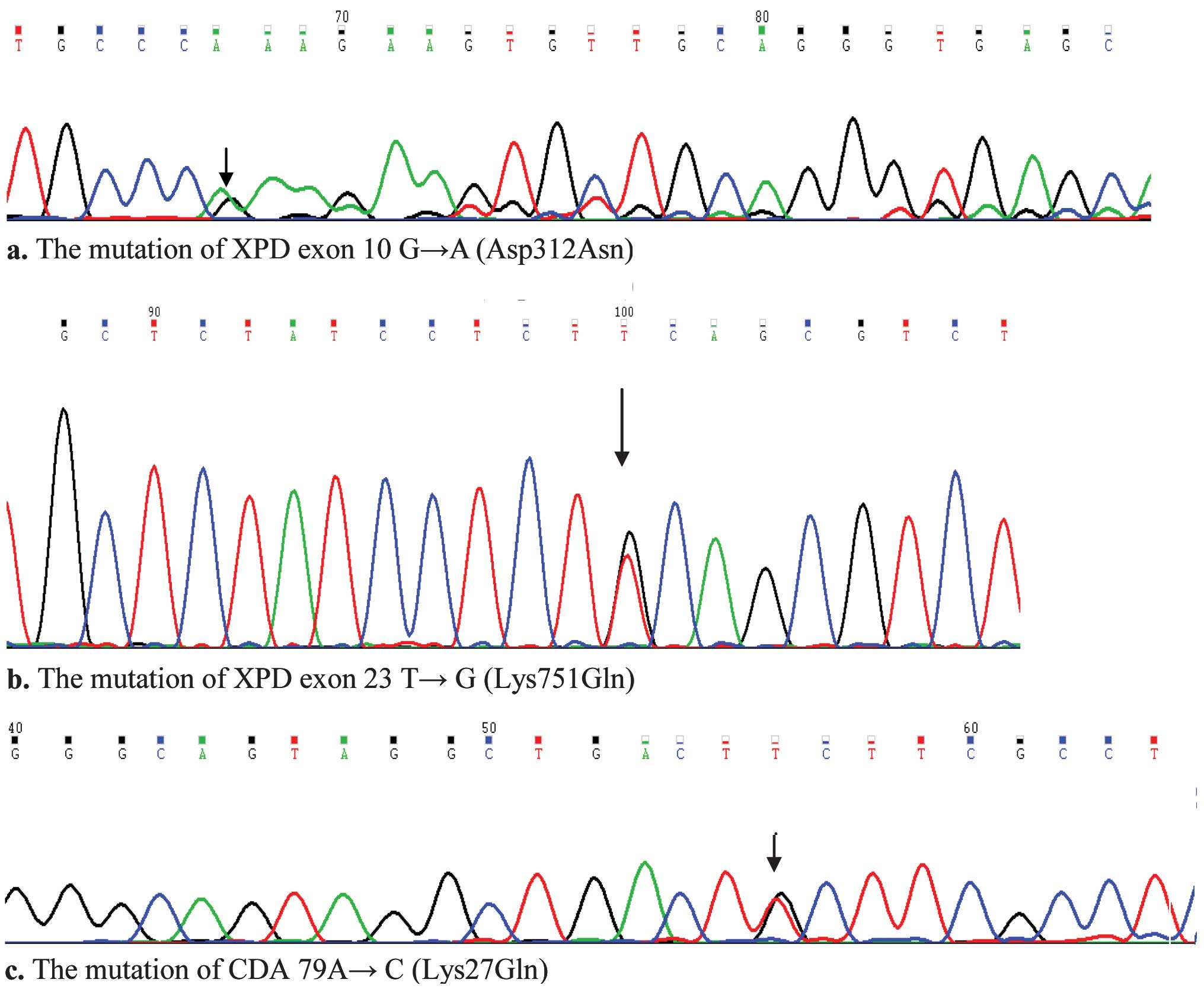
1). Mutations were identified by DNA sequencing (Fig. 2).
Statistical analysis
Clinical information and genotype frequencies were
compared between the lung cancer patients and controls using the
Chi-square test. Analysis of genotypes and their interaction with
smoking was evaluated using logistic regression. Data were analyzed
using SPSS 13.0 (IBM) software. P<0.05 was considered to
indicate a statistically significant difference.
Results
Demographic characteristics and genotype
distributions for the lung cancer patients and the controls
No significant difference was found in the average
age or gender ratio between the two groups (Table I). The XPD Asp312Asn genotypes in
the lung cancer patients were G/G 85 (82.52%), A/G 18 (17.48%) and
G/G 85 (82.52%), and in the controls the genotypes were A/G 17
(16.50%) and A/A 1 (0.98%). The XPD Lys751Gln genotypes were A/A 86
(83.49%) and A/C 17 (16.50%) in the lung cancer patients and A/A 87
(84.47%) and A/C 16 (15.53%) in the controls. No significant
difference was evident between the two groups in the two
single-nucleotide polymorphism (SNP) loci (P=0.598, 0.849). The CDA
Lys27Gln genotypes were A/A 82 (79.61%), A/C 20 (19.42%) and C/C 1
(0.98%) in the lung cancer patients, and A/A 81 (78.65%), A/C 21
(20.39%) and C/C 1 (0.98%) in the controls. No significant
differences were detected between the two groups (P=0.985). The CDA
Ala70Thr genotypes were G/G 101 (98.06%) and A/G 2 (1.94%) in the
controls. However, CDA Ala70Thr genotypes were all wild-type in the
lung cancer patients. There were no significant differences in CDA
Ala70Thr between the two groups (P=0.155).
 | Table IDemographic characteristics and
genotype distributions for lung cancer patients and controls. |
Table I
Demographic characteristics and
genotype distributions for lung cancer patients and controls.
| Characteristic | Genotype | Healthy controls | Lung cancer
patients | P-value |
|---|
| Number of cases | | 103 | 103 | |
| Age (years) | | 60.79±1.16 | 61.53±1.21 | 0.912 |
| Gender | | | | |
| Male | | 65 | 66 | 1.000 |
| Female | | 38 | 37 | |
| Smoking history | | | | |
| Smokers | | 74 (71.8%) | 81 (78.6%) | 0.462 |
| XPD Asp312Asn | G/G | 85 (82.52%) | 85 (82.52%) | 0.598 |
| A/G | 17 (16.50%) | 18 (17.48%) | |
| A/A | 1 (0.98%) | 0 | |
| XPD Lys751Gln | A/A | 87 (84.47) | 86 (83.49) | 0.849 |
| A/C | 16 (15.53) | 17 (16.50%) | |
| CDA Lys27Gln | A/A | 81 (78.65) | 82 (79.61) | 0.985 |
| A/C | 21 (20.39) | 20 (19.42) | |
| C/C | 1 (0.98%) | 1 (0.98%) | |
| CDA Ala70Thr | G/G | 101 (98.06%) | 103 (100%) | 0.155 |
| A/G | | 2 (1.94%) | 0 |
Different frequency distributions of XPD
and CDA genotypes in patients with different lung cancer
pathologies
The XPD genotypes Asp312Asn and XPD Lys751Gln, and
CDA Lys27Gln and CDA Ala70Thr were not significantly different
between the large cell lung cancer, squamous cell carcinoma,
adenocarcinoma or small cell lung cancer (Table II) (P>0.05).
 | Table IIGenotypic differences between patients
with various lung cancer pathologies. |
Table II
Genotypic differences between patients
with various lung cancer pathologies.
| Pathology | Genotype | Large cell lung
cancer | Squamous cell
carcinoma | Adenocarcinoma | Small cell lung
cancer | P-value |
|---|
| XPD Asp312Asn | G/G | 2 (1.94%) | 16 (15.53%) | 59 (57.28%) | 8 (7.77%) | 0.854 |
| A/G | | 4 (3.88%) | 13 (12.62%) | 1 (0.98%) | |
| XPD Lys751Gln | A/A | 2 (1.94%) | 16 (15.53) | 60 (58.25%) | 8 (7.77%) | 0.858 |
| A/C | | 4 (3.88%) | 12 (11.65%) | 1 (0.98%) | |
| CDA Lys27Gln | A/A | 2 (1.94%) | 15 (14.6%) | 58 (56.31%) | 7 (6.79%) | 0.070 |
| A/C | | 5 (4.85%) | 14 (13.55%) | 1 (0.98%) | |
| C/C | | 0 (0%) | 0 (0%) | 1 (0.98%) | |
| CDA Ala70Thr | G/G | 2 (1.94%) | 20 (19.42%) | 72 (69.90%) | 9 (8.74%) | 0.896 |
Using logistic regression analysis, it was revealed
that individuals with a smoking history combined with XPD
Lys751Gln, had a 4.04-fold increased risk of lung cancer (P=0.044).
Patients with XPD Lys751Gln and XPD Asp312Asn mutations had a
6.13-fold increase risk of lung cancer (P=0.047).
Discussion
Cancer is caused by a series of DNA alterations in a
single cell or a clone of a cell, which leads to loss of normal
cell function. At least four DNA repair pathways operate on
specific types of damaged DNA. The NER pathway is one of the most
significant and versatile repair systems that removes a wide range
of DNA lesions, including UV-induced ones. A number of studies have
examined the polymorphisms of genes within the NER pathway and
their correlation to lung cancer susceptibility (2).
XPD is an ATP-dependent DNA helicase, which is
involved in DNA repair in the NER pathway. XPD is a multifunctional
gene that encodes a component of the TFIIH transcription factor and
is involved in p53-mediated apoptotic responses (6). It is located on human chromosome 19
q13.3 and contains 23 exons. There are six identified SNPs in the
XPD gene, located at codons 199, 201, 312, 711 and 751. The
polymorphisms in codons 199, 201, 312 and 751 may result in amino
acid changes. Mutation of the XPD gene may affect activity of the
XPD protein. The mutation frequency of the 199 and 201 alleles is
only 4%, whereas the mutation of codons 312 and 751 is more common,
thus prompting our focus on Lys751G1n and Asp312Asn. The XPD
mutated phenotype is closely correlated to diminished DNA repair
capacity, which could increase the risk of cancer. Sequence
alignment studies have demonstrated that XPD codon 312 is highly
conserved, indicating that it may have an effect on preserving XPD
protein function, whereas codon 751 on the N-terminus is poorly
conserved. However, XPD 312 and 751 SNPs may lead to changes in DNA
repair capacity (7).
Epidemiological studies revealed that the two
polymorphisms were associated with certain malignancies, including
lung cancer, head and neck squamous cell carcinoma and basal cell
carcinoma (8–10). Hemminki et al (7) reported that 312 Asn/Asn and 715
Gln/Gln carriers were 50% less efficient in DNA repair compared to
carriers of the wild-type. Our results demonstrated no significant
difference in the mutation of XPD Asp312Asn and XPD LyS751Gln
between the lung cancer patients and the controls. However, the
risk of cancer may be increased in individuals carrying the
Lys751Gln mutation and with a history of smoking. Individuals with
XPD mutations are more likely to develop lung cancer, inferring an
interaction between these two sites.
CDA is a key enzyme in pyrimidine metabolism
(11), which is significant in the
metabolism of deoxycytidine anti-neoplatic drugs (including Ara-C),
which has a role in leukemia (12).
CDA is also the key enzyme that determines gemcitabine clearance,
since it catalyzes its degradation. Thus, CDA activity determines
the transformation efficiency of gemcitabine to
difluorodeoxyuridine and the length of exposure to gemcitabine. The
majority of studies have focused on the correlation between CDA
polymorphism or enzyme activity with gemcitabine efficacy and
toxicity (13,14). However, little is known regarding
the correlation between CDA polymorphisms and lung cancer risk. CDA
contains three nucleotide mutations; 79 A→C, 208 G→A and 435 T→C.
As the CDA 435 SNP is a nonsense mutation, this study focused on
SNPs at codons 79 and 208.
Yue et al (15) observed that CDA polymorphisms
differed between children with acute leukemia and healthy children.
Results of that study showed that the mutation rate of codon 79 was
approximately 12%, whereas that of codon 208 in the leukemia
patients and healthy children studied was 0 and 0.94%,
respectively. Our results demonstrated that the mutation rate of
CDA 79 was 20.39% in the controls, compared with 19.42% in the lung
cancer patients, indicating no significant difference between the
two groups. The mutation rate of CDA 208 in the controls was 1.94%,
while no mutations in the lung cancer group were observed; this
difference was not statistically significant. This suggests that
the mutation rate of CDA 208 is extremely low among this Chinese
population. Previous studies have suggested that the CDA 208
mutation rate varied according to ethnicity. No mutations were
observed in Europeans, while there was a mutation rate of 12.5% in
Africans. No significant difference in the mutation rate of CDA 79
and CDA 208 was observed between the lung cancer patients and the
controls. This suggests that these CDA mutations do not increase
the risk of lung cancer. CDA enzyme activity can be affected by
base mutations, thus a change in a more critical position may be
required to influence sensitivity to gemcitabine in lung
cancer.
The association between SNPs and lung cancer
susceptibility has examined, with varying results being obtained.
These differences may stem from a limited number of samples. There
may also be a range of susceptibility factors, which are difficult
to fully investigate. In addition, gene polymorphism is correlated
to ethnicity of the individual. Although there are certain
limitations in SNP detection technology, susceptibility gene
testing is important in providing early detection. Individuals who
carry certain susceptibility mutations and smoke, or those who
carry more than one mutation are increasingly susceptible to lung
cancer. Thus, screening of susceptibility genes holds promise for
the prevention and early diagnosis of lung cancer.
Acknowledgements
This study was supported in part by the Shanghai
Cancer Foundation (Shanghai, China).
References
|
1
|
Pharoah PD, Dunning AM, Ponder BA and
Easton DF: Association studies for finding cancer-susceptibility
genetic variants. Nat Rev Cancer. 4:850–860. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kiyohara C and Yoshimasu K: Genetic
polymorphisms in the nucleotide excision repair pathway and lung
cancer risk: a meta-analysis. Int J Med Sci. 4:59–71. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chiang CC, Tsai YY, Bau DT, Cheng YW,
Tseng SH, Wang RF and Tsai FJ: Pterygium and genetic polymorphisms
of the DNA repair enzymes XRCC1, XPA, and XPD. Mol Vis. 16:698–704.
2010.PubMed/NCBI
|
|
4
|
Takenaka T, Yano T, Kiyohara C, Miura N,
Kouso H, Ohba T, Kometani T, Shoji F, Yoshino I and Maehara Y:
Effects of excision repair cross-complementation group 1 (ERCC1)
single nucleotide polymorphisms on the prognosis of non-small cell
lung cancer patients. Lung Cancer. 67:101–107. 2010. View Article : Google Scholar
|
|
5
|
Fukunaga AK, Marsh S, Murry DJ, Hurley TD
and McLeod HL: Identification and analysis of single-nucleotide
polymorphisms in the gemcitabine pharmacologic pathway.
Pharmacogenomics J. 4:307–314. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lunn RM, Helzlsouer KJ, Parshad R, Umbach
DM, Harris EL, Sanford KK and Bell DA: XPD polymorphisms: effects
on DNA repair proficiency. Carcinogenesis. 21:551–555. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hemminki K, Xu G, Angelini S, Snellman E,
Jansen CT, Lambert B and Hou SM: XPD exon 10 and 23 polymorphisms
and DNA repair in human skin in situ. Carcinogenesis. 22:1185–1188.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Spitz MR, Wu X, Wang Y, Wang LE, Shete S,
Amos CI, Guo Z, Lei L, Mohrenweiser H and Wei Q: Modulation of
nucleotide excision capacity by XPD polymorphisms in lung cancer
patients. Cancer Res. 61:1354–1357. 2001.PubMed/NCBI
|
|
9
|
Jelonek K, Gdowicz-Klosok A, Pietrowska M,
Borkowska M, Korfanty J, Rzeszowska-Wolny J and Widlak P:
Association between single-nucleotide polymorphisms of selected
genes involved in the response to DNA damage and risk of colon,
head and neck, and breast cancers in a Polish population. J Appl
Genet. 51:343–352. 2010. View Article : Google Scholar
|
|
10
|
Tomescu D, Kavanagh G, Ha T, Campbell H
and Melton DW: Nucleotide excision repair gene XPD polymorphisms
and genetic predisposition to melanoma. Carcinogenesis. 22:403–408.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Laliberté J and Momparler RL: Human
cytidine deaminase: purification of enzyme, cloning, and expression
of its complementary DNA. Cancer Res. 54:5401–5407. 1994.PubMed/NCBI
|
|
12
|
Yue L, Saikawa Y, Ota K, Tanaka M,
Nishimura R, Uehara T, Maeba H, Ito T, Sasaki T and Koizumi S: A
functionnal single-nucleotide polymorphism in the human cytidine
deaminase gene contributing to ara-C sensitivity. Pharmacogenetics.
13:29–38. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rosell R, Taron M, Ariza A, Barnadas A,
Mate JL, Reguart N, Margel M, Felip E, Méndez P and García-Campelo
R: Molecular predictors of response to chemotherapy in lung cancer.
Semin Oncol. 31:20–27. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Maring JG, Wachters FM, Slijfer M, Maurer
JM, Boezen HM, Uges DR, de Vries EG and Groen HJ: Pharmacokinetics
of gemcitabine in non-small-cell lung cancer patients: impact of
the 79A>C cytidine deaminase polymorphism. Eur J Clin Pharmacol.
66:611–617. 2010. View Article : Google Scholar
|
|
15
|
Yue LJ, Chen XW, Li CR, Li CG, Shi HS and
Zhang M: Single-nucleotide polymorphisms of the cytidine deaminase
gene in childhood with acute leukemia and normal Chinese children.
Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 24:699–702. 2007.PubMed/NCBI
|