Introduction
Interleukin-23 (IL-23), a heterodimeric type 1
cytokine composed of the specific p19 subunit and the IL12/p40
subunit, is predominantly secreted by activated dendritic cells,
monocytes and macrophages and is crucial in the mucosal immune
system (1–5). The IL-23 receptor is also a
heterodimer composed of the specific IL-23R subunit and the
IL-12Rβ1 subunit. Various studies support the contribution of IL-23
to the expansion and stabilization of T helper type 17 (TH17)
cells, a subset of T cells characterized by IL-17 secretion and the
expression of the transcription factor retinoic acid-related orphan
receptor (ROR) γT and IL-23R (6).
IL-23R is also expressed on other immune cells (e.g., dendritic
cells, macrophages and eosinophils) (7–9) and is
considered to modulate inflammation via these cells.
Results of previous studies showed that IL-23 is
associated with carcinogenesis as well as inflammation. High levels
of IL-23 were detected in human squamous carcinoma (10). In a cutaneous carcinogenesis model,
IL-23a knockout rodents demonstrated decreased skin tumor
formation (11). The antibody
blockade of IL-23R in vivo decreased the growth rates of
certain transplanted tumors (12).
Although the precise mechanisms are unclear, IL-23 is considered to
affect tumor cells through T-cell responses via signal transducer
and activator of transcription 3 (STAT3) signaling, since IL-23
increases the activity of TH17 and regulatory T cells (Tregs),
which positively affects the activity of STAT3 in tumor cells and
promotes tumor growth (12,13).
Findings of another study showed that IL-23R is
expressed in colorectal carcinoma cells (14). The expression of IL-23R increased
progressively from normal tissue to colorectal carcinoma tissue,
and the IL-23R protein was also detected in the human colorectal
carcinoma cell line SW-480. The study suggested that IL-23 directly
regulated tumor growth and enhanced the malignant change from
adenoma to adenocarcinoma. However, important issues such as the
differences in IL-23R expression among patients or cell lines have
yet to be elucidated. Moreover, no study has directly investigated
the effects of IL-23 on colorectal carcinoma cells.
The aim of this study was to examine the
characteristics of IL-23R expression in human colorectal carcinoma
tissues and the direct effect of IL-23 on colorectal cancer
cells.
Materials and methods
Cell culture
The human colon carcinoma cell lines (MIP101, DLD-1
and KM12c) were provided by Tohoku University (Sendai, Japan). The
cells were grown in RPMI-1640 medium (Sigma-Aldrich, St. Louis, MO,
USA) supplemented with 10% heat-inactivated fetal bovine serum
(FBS) (Sigma-Aldrich) and antibiotic antimycotic solution
(Sigma-Aldrich) under an atmosphere of 95% air and 5%
CO2 at 37°C. In some experiments, the cultured cells
were stimulated with 10 μg/ml of recombinant human IL-23
(HumanZyme, Chicago, IL, USA).
Immunohistochemistry
Colorectal carcinoma tissue and normal adjacent
tissue samples were obtained from 15 patients (two cecum, two
ascending colon, two transverse colon, two descending colon, five
sigmoid colon and two rectum) undergoing colectomy or anterior
resection at Tohoku University Hospital. Informed consent was
obtained from all patients prior to the operation. The tissues were
fixed in 10% formalin, and embedded in paraffin for the
histological analysis. Immunohistochemical staining for IL-23R was
performed using the streptavidin-biotin-peroxidase complex (SAB-PO)
method, using a Histofine SAB-PO kit (Nichirei Co., Tokyo, Japan)
according to the manufacturer’s instructions. The rabbit anti-human
polyclonal antibody against IL-23R (LifeSpan Biosciences, Seattle,
WA, USA) was diluted at 1:100. The goat anti-human polyclonal
antibody against IL-23R (AbD Serotec, Oxford, UK) was diluted at
1:125. Counterstaining was carried out with hematoxylin. This study
was approved by the ethics review board of Tohoku University
Graduate School of Medicine.
Protein extraction and western blot
analysis
Protein was extracted from cells with the M-PER
mammalian protein extraction reagent (Thermo Scientific, Waltham,
MA, USA) according to the manufacturer’s protocol. The protein
concentration was determined using the Bradford method. Total
protein (20 μg) was extracted and resolved by SDS-PAGE. The
proteins in the gels were transferred electrophoretically to PVDF
membranes at a constant voltage of 30 V for 1 h. The PVDF membrane
was blocked with 5% skim milk for 1 h, incubated with the primary
antibody at 4°C overnight, and incubated in secondary antibody at
room temperature for 1 h. The rabbit anti-human polyclonal antibody
against IL-23R (LifeSpan Biosciences) was diluted at 1:1000. The
mouse anti-human monoclonal antibody against β-actin (Applied
Biological Materials, Richmond, Canada) was diluted at 1:2000. The
HRP-linked goat anti-rabbit and goat anti-mouse IgG secondary
antibodies were purchased from Epitomics (Burlingame, CA, USA) and
Santa Cruz Biotechnology (Santa Cruz, CA, USA), respectively.
Cell proliferation assay
Cell proliferation was assessed by a tetrazolium
salt (WST-8)-based colorimetric assay using the Cell Counting Kit-8
(Dojindo Laboratories, Kumamoto, Japan). Briefly, the cultured
cells were seeded onto 96-well plates at an initial density of
1×104 cells/ml with or without recombinant human IL-23
(10 μg/ml). The plates were incubated for 24 h, and CCK-8 solution
was added to each well. Following a 4-h incubation, cell
proliferation was determined by scanning the plates with a
microplate reader at a wavelength of 450 nm.
Invasion assay
The cell invasion assay was performed using BD
BioCoat Matrigel Invasion Chambers (BD Biosciences, Franklin Lakes,
NJ, USA). Briefly, the cultured cells (2×103 cells/ml in
0.5 ml serum-free medium) were added in suspension to each insert
and medium (0.8 ml, supplemented with 5% FBS) was added to each
bottom well. Recombinant human IL-23 (10 μg/ml) was added to the
upper wells. Following a 24-h incubation, invasive cells on the
under surface of the membrane were stained with Diff-Quik (Sysmex
Corporation, Kobe, Japan) and counted microscopically.
RNA extraction, reverse transcription and
real-time PCR
The mRNA expression of TGF-β1 was examined by
real-time PCR. Total RNA from cultured cells was isolated using the
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer’s protocol, and washed using an RNeasy MinElute
Cleanup Kit (Qiagen, Valencia, CA, USA). Total RNA (2 μl) was
reverse-transcribed into complementary DNA (cDNA) using the
First-strand cDNA synthesis system for the quantitative RT-PCR kit
(Marligen Biosciences, Ijamsville, MD, USA). Real-time PCR was
performed using an Applied Biosystems 7300 Sequence Detection
system (Applied Biosystems, Foster City, CA, USA). The 20 μl PCR
reaction included 1 μl of cDNA, 10 μl of 2× TaqMan gene expression
master mix and 1 μl of TaqMan gene expression assays reagent
(Applied Biosystems). The reactions were incubated in 96-well
optical plates at 95°C for 10 min, followed by 40 cycles of 95°C
for 15 sec and 60°C for 1 min. The Ct values for the reference gene
(β-actin) and target gene (TGF-β1) were determined using default
threshold settings, and relative quantification was performed
according to the 2−ΔΔCt method.
Enzyme-linked immunosorbent assay
(ELISA)
The cultured cells were grown to confluence in
RPMI-1640 medium supplemented with 10% FBS. The medium was then
replaced with FBS-free RPMI-1640 medium with or without recombinant
human IL-23 (10 μg/ml). Following 48-h incubation, the levels of
TGF-β1 in the supernatant were analyzed using the Quantikine ELISA
kit (R&D Systems, Minneapolis, MN, USA), according to the
manufacturer’s protocol. The color reaction was measured by
scanning the plates with a microplate reader at a wavelength of 450
nm. The concentration of TGF-β1 was determined via a standard curve
that was obtained using the kit’s standards.
Statistical analyses
The effects of IL-23 on the cell invasion,
proliferation and TGF-β1 production of the cultured cells were
evaluated by t-tests.
Results
IL-23R expression in colorectal carcinoma
tissue
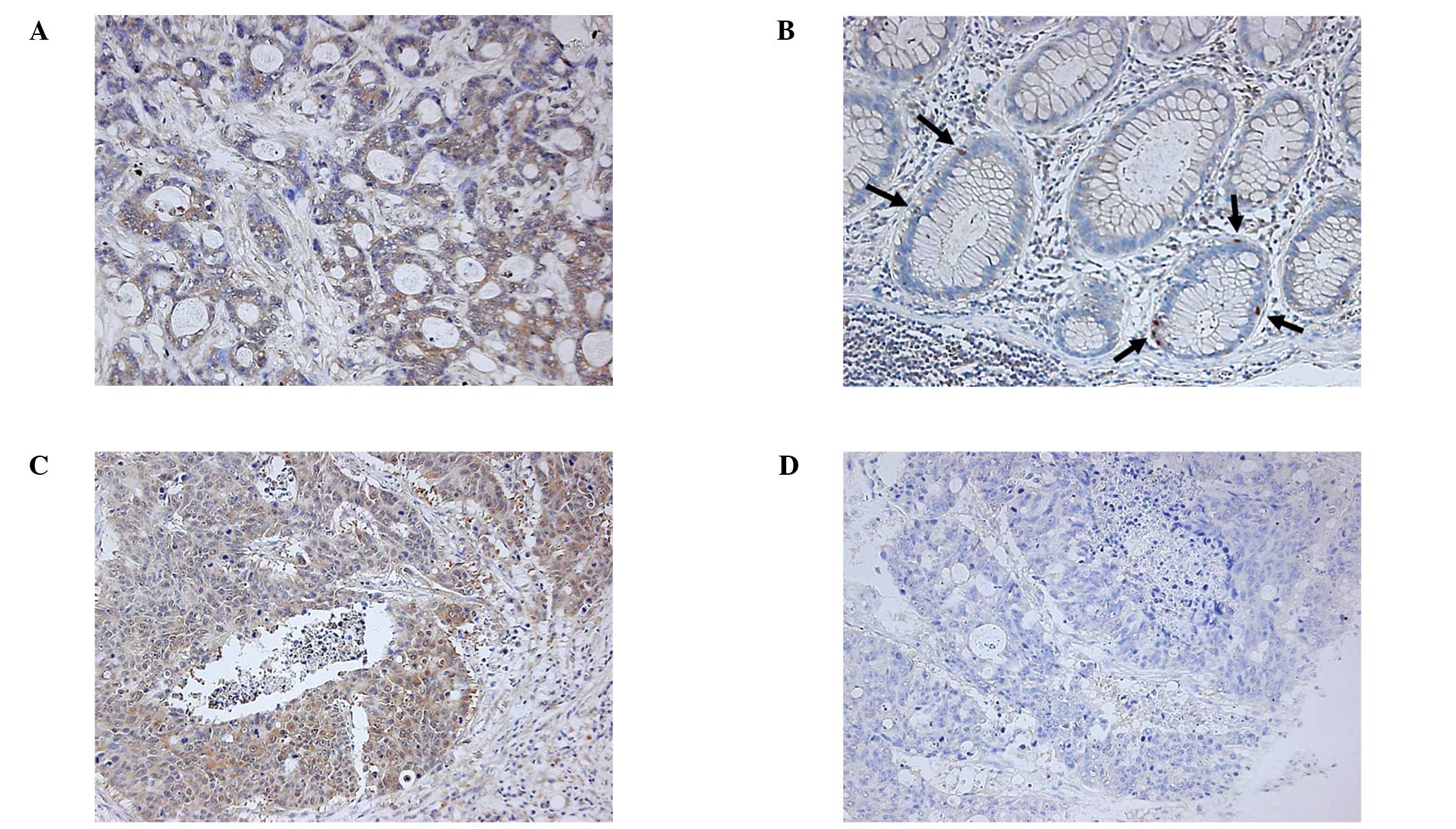
First, we performed an immunohistochemical analysis
and were able to detect the expression of IL-23R in the colorectal
carcinoma tissues in 9 out of 15 patients (Fig. 1A, Table
I). All of the TNM stage IV patients were positive for IL-23R,
but the other TNM stages and tissue types of cancer had little
correlation with the IL-23R expression. In normal sections of the
colon or rectum, a few epithelial cells expressed IL-23R,
especially at the crypt bases (Fig.
1B). We also found that the IL-23R expression of the carcinoma
tissue was relatively high at the deepest point of invasion in
certain cases (Fig. 1C and D). We
used two different anti-IL23R antibodies (rabbit and goat), and
obtained similar results (data not shown).
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| Age | Gender | Portion | TNM-Stage | Tissue type | IL-23R
expression |
|---|
| 56 | M | S | I | Tub2 | Positive |
| 62 | M | A | I | Tub2 | Positive |
| 68 | M | R | II | Tub2 | Positive |
| 73 | F | D | IIIA | Tub2+muc | Positive |
| 70 | M | R | IIIB | Tub2 | Positive |
| 64 | F | D | IIIB | Tub2+por1 | Positive |
| 72 | M | S | IV | Tub2 | Positive |
| 76 | M | S | IV | Tub2 | Positive |
| 82 | M | T | IV | Tub2 | Positive |
| 41 | F | T | I | Tub2 | Negative |
| 68 | M | S | I | Tub2 | Negative |
| 35 | M | C | II | Tub2 | Negative |
| 66 | M | C | IIIA | Por1 | Negative |
| 75 | M | A | IIIB | Tub2 | Negative |
| 26 | F | S | IIIB | Tub2 | Negative |
IL-23R expression in colon carcinoma cell
lines
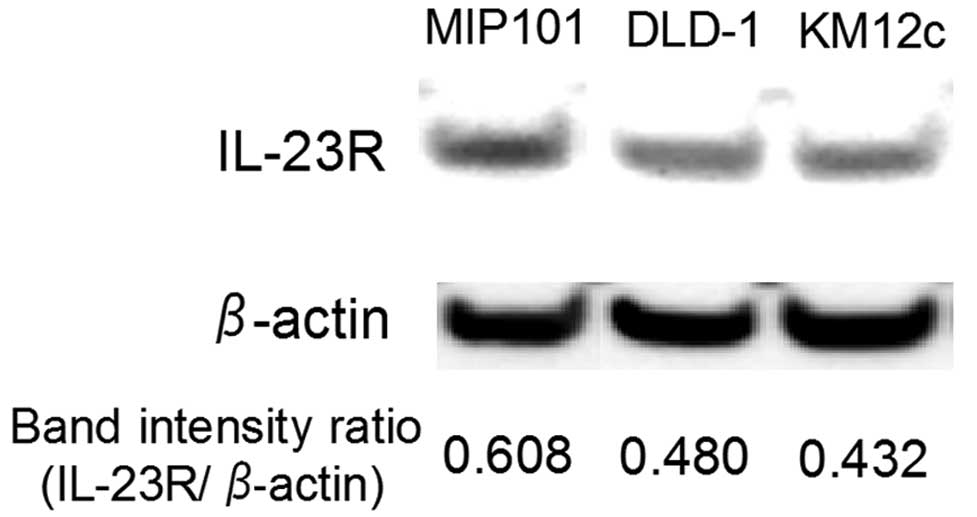
The expression of IL-23R in three types of cultured
cells derived from colon carcinoma, MIP101, DLD-1 and KM12c was
investigated. Western blot analysis revealed that all of the cell
lines express the IL-23R protein (Fig.
2). However, the signal intensity differed among the cell
lines. We also confirmed the expression of IL-23R mRNA in the cell
lines by RT-PCR (data not shown).
Cell proliferation and invasion
assays
We investigated whether IL-23 directly contributes
to the proliferation and invasive activity of the cultured colon
carcinoma cells. Following stimulation with IL-23, the
proliferative and invasive activities of DLD-1 cells were
significantly increased by approximately 20%, whereas those of
MIP101 and KM12c cells were not significantly altered (Fig. 3). The highest expression of IL-23R
was observed in MIP101 cells, followed by DLD-1 and KM12c cells
(Fig. 2). However, the changes in
the proliferative and invasive activities did not reflect the
IL-23R expression level.
TGF-β secretion and mRNA expression by
colon carcinoma cell lines
We investigated whether IL-23 contributes to TGF-β1
production by colon carcinoma cells. Following stimulation with
IL-23, the TGF-β1 concentration in the medium increased
approximately 20% in the DLD-1 cells, whereas it was not
significantly altered in the MIP101 and KM12c cells (Fig. 4A). The relative mRNA expression of
TGF-β1/β-actin in DLD-1 cells following stimulation with IL-23 for
24 or 48 h was also increased (Fig.
4B).
Discussion
Results of the present study revealed that IL-23R
was expressed in the carcinoma tissue samples in 9 out of 15 cases,
but not in the remaining 6 cases (Fig.
1A). The expression of IL-23R in colorectal carcinoma tissue
was also reported in a recent study (14), but the differences among the
patients were not described. All of the stage IV cases in the
present study were positive for IL-23R. In addition, among the
three colon carcinoma cell lines examined in the present study, the
MIP101 cells demonstrated the highest expression of IL-23R. MIP101
cells were derived from a liver metastasis of colon adenocarcinoma,
whereas DLD-1 and KM12c cells were derived from primary colon
adenocarcinoma of Dukes’ stage B and C patients without distant
metastasis, respectively. Therefore, the IL-23R expression in
carcinoma may be associated with distant metastasis, although
further studies are required to determine whether IL-23R expression
is associated with the invasion and aggressiveness of the
cancer.
Notably, the IL-23R expression was heterogeneous
even within the same tumor (Fig. 1C and
D). Our data revealed that IL-23R-positive and -negative (or
very weakly positive) cells coexisted within the same tumor.
Moreover, the IL-23R expression was relatively higher at the
deepest point of invasion in certain cases, indicating that IL-23R
expression may be correlated with the invasion of carcinoma.
Our results also indicated that there were a few
IL-23R positive cells present in the normal colon tissue (Fig. 1B). Considering their localization,
we expected that these IL-23R-positive cells were enterochromaffin
cells, and we confirmed that these IL-23R-positive epithelial cells
were positive for chromogranin A, a specific marker of
enterochromaffin cells (data not shown). The effects of IL-23 on
these IL-23R-positive enterochromaffin cells is currently under
investigation.
Notably, our results demonstrated that the
proliferative and invasive activities of DLD-1 cells increased when
the cells were stimulated with IL-23, in spite of the absence of
any immune cells, such as TH17 cells (Fig. 3). These findings indicate that IL-23
directly enhances the malignancy of the colon carcinoma cells.
However, in the cases of MIP101 and KM12c cells, IL-23 did not
change these activities in spite of their expression of IL-23R
(Fig. 2). Therefore, the effects of
IL-23 on colorectal carcinoma cells varies, and the effect is not
in accordance with the intensity of the IL-23R expression. In TH17
cells, the increased STAT3 activity induced by IL-23 promotes the
expression of cytokines such as IL-17 (13). Results of the western blot analysis
showed whether STAT3 in the DLD-1 cells was phosphorylated by
IL-23; however, only a minimal amount of phosphorylated STAT3 was
detected (data not shown).
Another possible mechanism for the IL-23-induced
proliferative and invasive activity of DLD-1 cells was suggested by
our results. The TGF-β1 production by DLD-1 cells was significantly
increased by IL-23 stimulation (Fig.
4), whereas IL-23 did not affect the TGF-β production in the
MIP101 and KM12c cells. These results correlated with those of the
cell proliferation and invasion assay (Fig. 3). Therefore, an autocrine mechanism
via TGF-β production might underlie the IL-23-induced proliferative
and invasive activity observed in the DLD-1 cells.
During the early stages of tumor development, the
TGF-β/Smad pathway acts as a tumor suppressor (15,16).
However, TGF-β also promotes the invasion or the proliferation of
certain tumor cells (15–18). Epithelial-mesenchymal transition
(EMT) is known to promote carcinoma progression through a variety
of mechanisms such as endowing cells with migratory and invasive
properties (19). TGF-β signaling
is important in inducing the EMT (20). It is suggested that IL-23 stimulates
DLD-1 cells to produce TGF-β and therefore promote malignancy via
induction of the EMT. Moreover, Forkhead box P3 (FOXP3) production
is elevated in colorectal carcinoma and the IL-23/IL-23R signaling
pathway may affect the FOXP3-associated expression-repression and
proliferation-suppression observed in tumor cells (14). The gene transcription of FOXP3 is
regulated by TGF-β signaling, including the NFAT and Smad3
pathways, in regulatory T cells (21,22).
Considering that IL-23 stimulated DLD-1 cells to produce TGF-β in
the present study, our findings suggest that the IL-23/IL-23R
signaling pathway may affect FOXP3 expression via TGF-β
signaling.
In conclusion, this is the first study to
demonstrate the direct effects of IL-23 on colorectal carcinoma
cells and a correlation between IL-23/IL-23R and TGF-β in carcinoma
cells. IL-23 is a potential target for colon cancer immunotherapy.
However, the heterogeneity in the IL-23R expression and the effects
of IL-23 on the colorectal carcinoma cells should be considered.
Further characterization of IL-23R-positive colorectal carcinoma
cells is required before IL-23-associated biotherapy becomes
available in the clinic.
References
|
1
|
Oppmann B, Lesley R, Blom B, et al: Novel
p19 protein engages IL-12p40 to form a cytokine, IL-23, with
biological activities similar as well as distinct from IL-12.
Immunity. 13:715–725. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yen D, Cheung J, Scheerens H, et al: IL-23
is essential for T cell-mediated colitis and promotes inflammation
via IL-17 and IL-6. J Clin Invest. 116:1310–1316. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sarra M, Pallone F, Macdonald TT, et al:
IL-23/IL-17 axis in IBD. Inflamm Bowel Dis. 16:1808–1813. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Monteleone I, Pallone F and Monteleone G:
Interleukin-23 and Th17 cells in the control of gut inflammation.
Mediators Inflamm. 2009:2976452009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hue S, Ahern P, Buonocore S, et al:
Interleukin-23 drives innate and T cell-mediated intestinal
inflammation. J Exp Med. 203:2473–2483. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Abraham C and Cho J: Interleukin-23/Th17
pathways and inflammatory bowel disease. Inflamm Bowel Dis.
15:1090–1100. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cua DJ, Sherlock J, Chen Y, et al:
Interleukin-23 rather than interleukin-12 is the critical cytokine
for autoimmune inflammation of the brain. Nature. 421:744–748.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Parham C, Chirica M, Timans J, et al: A
receptor for the heterodimeric cytokine IL-23 is composed of
IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J
Immunol. 168:5699–5708. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cheung PF, Wong CK and Lam CW: Molecular
mechanisms of cytokine and chemokine release from eosinophils
activated by IL-17A, IL-17F, and IL-23: implication for Th17
lymphocytes-mediated allergic inflammation. J Immunol.
180:5625–5635. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fukuda M, Ehara M, Suzuki S, et al: IL-23
promotes growth and proliferation in human squamous cell carcinoma
of the oral cavity. Int J Oncol. 36:1355–1365. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Langowski JL, Zhang X, Wu L, et al: IL-23
promotes tumour incidence and growth. Nature. 442:461–465. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kortylewski M, Xin H, Kujawski M, et al:
Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the
tumor microenvironment. Cancer Cell. 15:114–123. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu H, Pardoll D and Jove R: STATs in
cancer inflammation and immunity: a leading role for STAT3. Nat Rev
Cancer. 9:798–809. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lan F, Zhang L, Wu J, et al: IL-23/IL-23R:
potential mediator of intestinal tumor progression from adenomatous
polyps to colorectal carcinoma. Int J Colorectal Dis. 26:1511–1518.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Meulmeester E and Ten Dijke P: The dynamic
roles of TGF-β in cancer. J Pathol. 223:205–218. 2011.
|
|
16
|
Ten Dijke P, Goumans MJ, Itoh F, et al:
Regulation of cell proliferation by Smad proteins. J Cell Physiol.
191:1–16. 2002.PubMed/NCBI
|
|
17
|
Tian M and Schiemann WP: The TGF-β paradox
in human cancer: an update. Future Oncol. 5:259–271. 2009.
|
|
18
|
Bruna A, Darken RS, Rojo F, et al: High
TGFβ-Smad activity confers poor prognosis in glioma patients and
promotes cell proliferation depending on the methylation of the
PDGF-B gene. Cancer Cell. 11:147–160. 2007.
|
|
19
|
Thiery JP, Acloque H, Huang RY, et al:
Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Micalizzi DS, Farabaugh SM and Ford HL:
Epithelial-mesenchymal transition in cancer: parallels between
normal development and tumor progression. J Mammary Gland Biol
Neoplasia. 15:117–134. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Su J and Liu YC: Foxp3 positive regulatory
T cells: a functional regulation by the E3 ubiquitin ligase Itch.
Semin Immunopathol. 32:149–156. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu L, Kitani A and Strober W: Molecular
mechanisms regulating TGF-β-induced Foxp3 expression. Mucosal
Immunol. 3:230–238. 2010.
|