Introduction
Epithelial ovarian cancer (EOC) has a high mortality
rate (1); it is the leading cause
of death among gynecological tumors, and the fourth leading cause
of cancer-related mortality among women in the United States
(2). Due its nonspecific symptoms
and lack of effective screening methods (3), approximately two-thirds of cases are
diagnosed in stages III and IV, with a five-year survival rate of
10–20% (4,5). Approximately 90% of ovarian tumors
originate from epithelial cells (6,7). The
mortality rate has not changed in the last two decades (8).
A group of enzymes known as the DNA mismatch repair
(MMR) system is responsible for repairing mutations. Hereditary
nonpolyposis colorectal cancer (HNPCC) is the third leading cause
of hereditary ovarian cancer, and is caused by mutations in genes
of the MMR system. One of the consequences of deficient MMR is
microsatellite instability (9),
which carries somatic mutations in tumor suppressor genes,
oncogenes, apoptosis and detoxification genes, and is involved in
both the initiation and progression of tumors (10).
HNPCC has been studied using a panel of five
National Cancer Institute (NCI) markers, which includes two
mononucleotides (BAT25 and BAT26) and three dinucleotides (D2S123,
D5S346 and D17S250) (11). MSI is
identified when the alleles detected in the microsatellite DNA of
tumor samples are not present in normal tissue samples from the
same individual (12). It is also
believed that genetic changes may occur in response to constant
ovulation (13,14).
The identification of MMR system mutations by
microsatellite instability (MSI) in women with EOC may help us to
understand tumor biology and its pathogenesis (11,15,16).
Despite the evidence of the involvement of the MMR system in the
complex process of ovarian carcinogenesis, the actual function of
MSI and the optimal panel of markers for EOC are not well
established (9,17). This study uses the NCI markers with
the aim of evaluating the expression of MSI in patients with
ovarian serous cystadenocarcinoma, compared with ovarian serous
cystadenoma and normal ovaries.
Materials and methods
Patients
A total of 37 patients were prospectively evaluated
in three different groups, as follows: ovarian serous
cystadenocarcinoma (n=13), ovarian serous cystadenoma (n=10) and
normal ovaries (n=14), from February 2008 to July 2010. The study
was approved by the ethics committee of UNA University Center
(protocol 0005.0.391.000-10) and all patients signed informed
consent forms.
All patients underwent clinical and gynecological
examination and transvaginal ultrasound, prior to the study.
Surgical staging was performed in patients with ovarian serous
cystadenocarcinoma, according to the International Federation of
Gynecology and Obstetrics (FIGO). Normal ovarian tissue was
obtained from patients undergoing oophorectomy, during total
abdominal hysterectomy for treatment of benign gynecological
disease. Histological evaluation was performed by a pathologist.
None of the patients had received prior treatment with chemotherapy
and/or radiotherapy, or acute infectious peritoneal process.
Polymorphisms and microsatellite
instability
Peripheral blood samples were collected prior to the
induction of anesthesia in tubes containing EDTA (Becton Dickinson,
Franklin Lakes, NJ, USA). Ovarian tissue samples were collected
intraoperatively from the solid portion of the tumor without
necrosis, and immediately frozen in liquid nitrogen. DNA was
extracted with 1 m1 TRIzol® reagent (Invitrogen,
Carlsbad, CA, USA), using 50–100 mg frozen ovarian tissue or 500
μl blood. The gDNA was quantified using the NanoVue
spectrophotometer Pathlength Fluid Calibration kit (GE Healthcare,
Little Chalfont, Buckinghamshire, UK) at wavelengths of 260 and 280
nm.
The MSI was evaluated using the primers described in
Table I, in two different PCR
reactions (blood and ovarian tissue). We used
GoTaq®-Green Master mix 1X (Promega, Sao Paulo, SP,
Brazil), 1 μM of each primer, and 10 ng DNA from each
sample. Tubes were incubated at 95°C for 2 min to denature the
sample. Cycles of PCR amplification were performed as follows:
denaturation at 94°C for 30 sec, annealing at 52, 55 or 56°C for 45
sec, extension at 72°C for 30 sec, and a final extension at 72°C
for 5 min (Table I). A 15-μl
sample of the PCR products was analyzed by 7.5% polyacrylamide gel
electrophoresis at 100 volts. The gels were then incubated in
freshly prepared silver nitrate solution (0.2%). PCR was performed
with negative and positive controls.
 | Table IDescription of National Cancer
Institute primers for PCR. |
Table I
Description of National Cancer
Institute primers for PCR.
| Markers | Primers | AT (°C) | Product size
(bp) |
|---|
| BAT25 | Forward: TCG CCT CCA
AGA ATG TAA GT | | |
| Reverse: TCT GGA TTT
TAA CTA TGG CTC | 56 | 110–130 |
| BAT26 | Forward: TGA CTA CTT
TTG ACT TCA GCC | | |
| Reverse: AAC CAT TCA
ACA TTT TTA ACC C | 56 | 100–120 |
| D2S123 | Forward: AAA CAG GAT
GCC TGC CTT TA | | |
| Reverse: GGA CTT TCC
ACC TAT GGG AC | 55 | 200–230 |
| D5S346 | Forward: AGC AGA TAA
GAC AGT ATT ACT AGT T | | |
| Reverse: ACT CAC TCT
AGT GAT AAA TCG GG | 55 | 100–130 |
| D17S250 | Forward: GGA AGA ATC
AAA TAG ACA AT | | |
| Reverse: GCT GGC CAT
ATA TAT ATT TAA ACC | 52 | 140–170 |
The identification of polymorphisms and analysis of
genomic instability were performed by comparing amplified alleles
in samples of ovarian tissue and peripheral blood. Presence of MSI
was confirmed when monomorphic or polymorphic variants identified
in microsatellite DNA in ovarian tissue samples were not present in
the peripheral blood sample from the same individual. The level of
MSI was classified as high (MSI-H) when two or more of the markers
tested demonstrated instability, low (MSI-L) when one of the
markers tested demonstrated instability, or stable (MSS) when no
instability was detected. All analyses were reviewed by two authors
independently.
Real-time PCR
cDNA was generated from 2 mg total RNA using
Illustra Ready-to-Go RT-PCR beads (GE Healthcare) in a total volume
of 50 μl, according to the manufacturer’s instructions. PCR
primers were used as described in previous publications:
MLH1: forward, 5′-CTGAAGGCACTTCCGTT GAG-3′ and reverse,
5′-TGGCCGCTGGATAACTTC-3′; MSH2: forward,
5′-GAGGCTCTCCTCATCCAGATTG-3′ and reverse,
5′-GGCCTGGAATCTCCTCTATCAC-3′; TATA: forward,
5′-TGCACAGGAGCCAAGAGTGAA-3′ and reverse, 5′-CACATCACAGCTCCCCACCA-3′
(18). qRT-PCR was performed using
10 μl duplicate reactions with 1X Brilliant II
SYBR®-Green qPCR Master mix (Agilent Technologies, La
Jolla, CA, USA), 0.2 μl Rox (1:500), 0.25–0.30 μM of
the primers, and 40 ng/μl cDNA (RNA equivalent) for each
experiment. The Agilent MX 3005P detection system (Stratagene) was
used. The reference loci TATA binding protein (TBP) was used
as the normalization gene. PCR amplification was performed as
follows: 95°C for 10 min; 40 cycles of 95°C for 30 sec, annealing
at 60°C for 60 sec and extension at 72°C for 60 sec. The
optimization of the RT-qPCR reaction was performed according to the
manufacturer’s instructions. No template controls were included in
the assay for any gene. A melting curve was constructed for each
primer pair to confirm the product specificity.
Statistical analysis was performed with SPSS 18.0
(SPSS Inc., Chicago, IL, USA). The Chi-square and Fisher’s exact
tests were used to establish the differences between the groups.
Gene expression levels from qPCR were compared using the
Kruskal-Wallis test. P<0.05 was considered to indicate a
statistically significant result.
Results
The FIGO stage was I/II in three patients (23.1%)
and III/IV in 10 patients (76.9%) in the serous cystadenocarcinoma
group. There were no differences between the groups regarding age
(P=0.254) or parity (P=0.994), but there was a difference with
regard to menopausal status (P=0.013; Table II).
 | Table IIComparison between serous
cystadenocarcinoma, serous cystadenoma and normal ovary. |
Table II
Comparison between serous
cystadenocarcinoma, serous cystadenoma and normal ovary.
| Variablea |
Cystadenocarcinoma | Cystadenoma | Normal ovary | P-value |
|---|
| Number of
patients | 13 | 10 | 14 | |
| Age, years (mean ±
SD) | 58.8±12.2 | 52.3±16.4 | 51.2±8.7 | 0.254 |
| Menopause, n | 10 | 5 | 14 | 0.013 |
| Parity, mean ±
SD | 2.23±1.87 | 2.2±2.86 | 2.14±1.91 | 0.994 |
| NCI markers, n
(%) |
| BAT25 | 4 (30.8) | 3 (30) | 7 (50) | 0.492 |
| BAT26 | 10 (76.9) | 5 (50) | 10 (71.4) | 0.363 |
| D2S123 | 7 (53.8) | 4 (40) | 10 (71.4) | 0.298 |
| D5S346 | 9 (69.2) | 6 (60) | 9 (64.3) | 0.898 |
| D17S250 | 9 (69.2) | 3 (30) | 9 (64.3) | 0.131 |
Polymorphisms were found using at least one marker
in 32 women (86.4%), and were observed with D2S123 (83.7%), D17S250
(81.1%), D5S346 (72.9%), BAT25 (21.6%) and BAT26 (16.2%) markers.
Polymorphisms were similar between MSS samples for D2S123, while
the polymorphism observed for D5S346 differed between the MSI
samples of ovarian tissue and peripheral blood. Fig. 1 shows the results of MSI analysis in
patients with cystadenocarcinoma, cystadenoma and normal ovaries,
respectively.
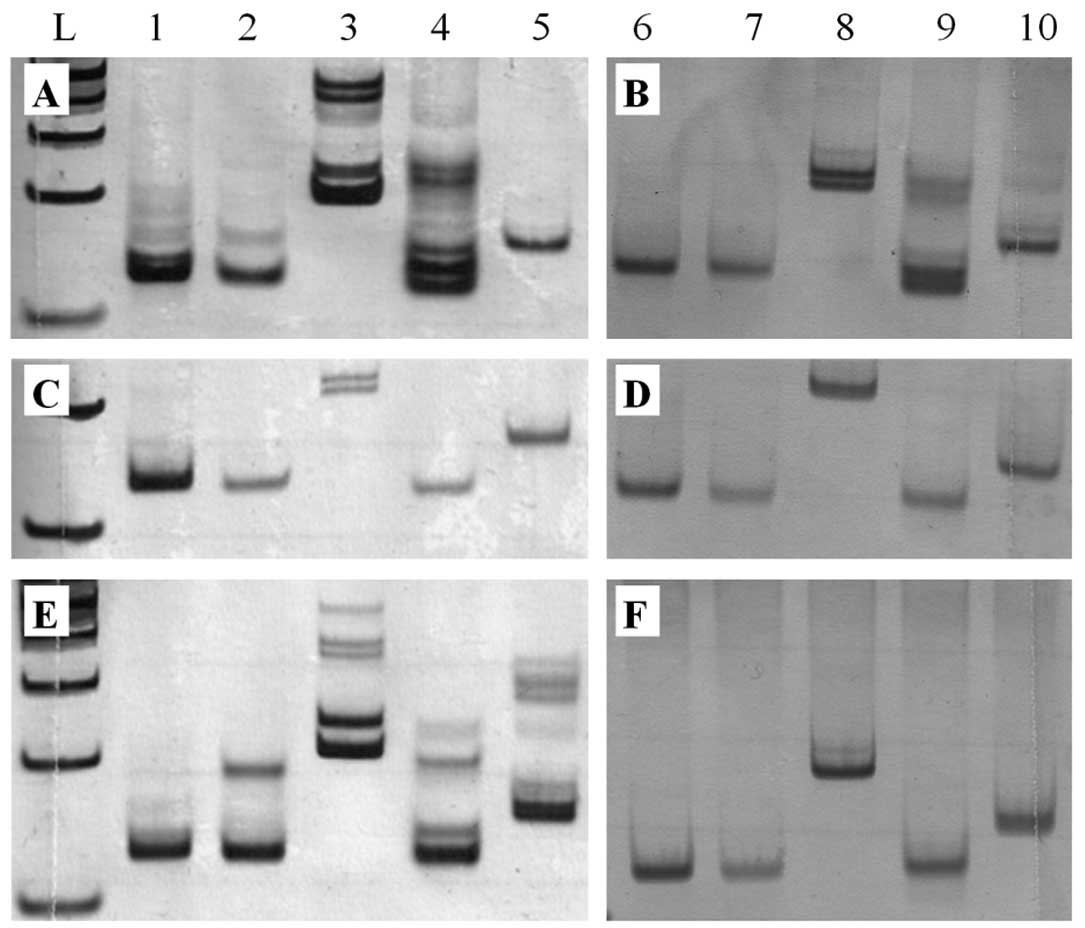 | Figure 1Polyacrylamide gel electrophoresis
(7.5%) of NCI markers (A, C, E) and peripheral blood samples (B, D,
F) of patients. (A, B) Cystadenocarcinoma, (C, D) cystadenoma, and
(E, F) normal ovarian tissue. Columns 1 and 6, BAT25; 2 and 7,
BAT26; 3 and 8, D2S123; 4 and 9, D5S346; and 5 and 10, D17S250.
BAT25, BAT26, D2S123 (A), D2S123 (C) and BAT26, D2S123, D5S346,
D17S250 (E) polymorphic alleles are present in the ovarian tissues
and absent in the peripheral blood samples (B, D, F), characterized
as MSI-H, MSI-L and MSI-H, respectively. L, 100-bp DNA ladder. |
MSI was identified in 25 cases (67.6%) with BAT26,
24 cases (64.9%) with D5S346, 21 cases (56.8%) with D2S123 and
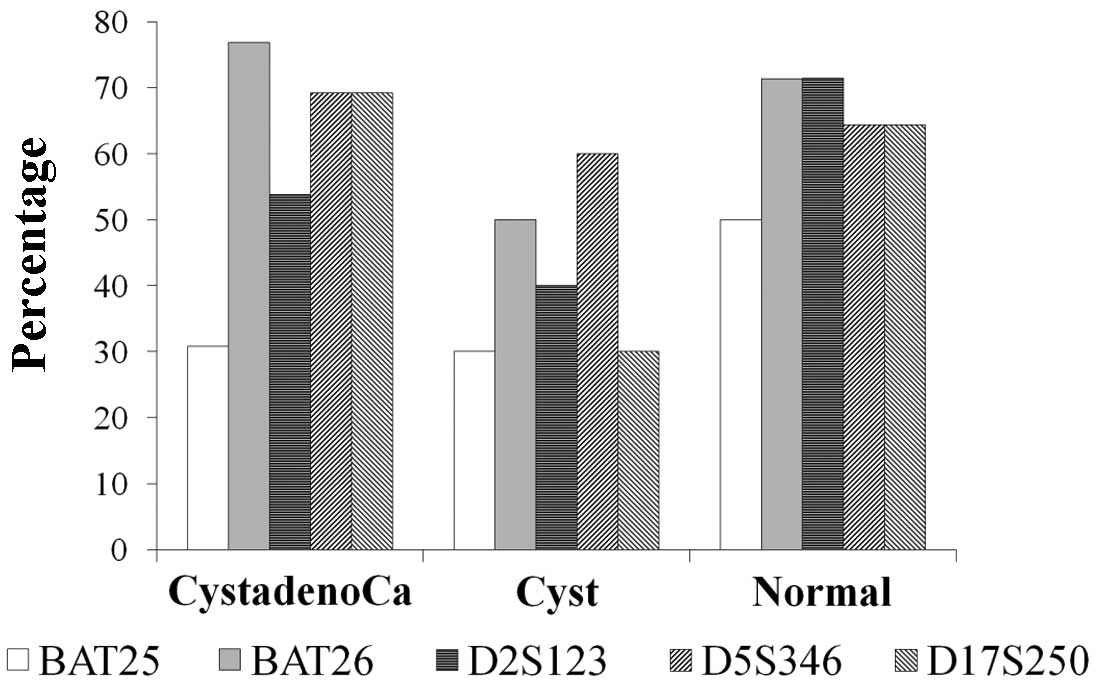
D17S250, and 14 cases (37.8%) with BAT25. In the cystadenocarcinoma
group, BAT25, BAT26, D2S123, D5S346 and D17S250 markers were
positive in 30.8, 76.9, 53.8, 69.2 and 69.2% of patients,
respectively. The same markers were positive for 30, 50, 40, 60 and
30% in the cystadenoma group, and 50, 71.4, 71.4, 64.3 and 63.3% of
the normal ovary group, respectively. There were no differences
between the specific NCI markers among the three studied groups
(Fig. 2, Table II).
MSI-H was present in 84.6, 60 and 78.6% of the
cystadenocarcinoma, cystadenoma and normal patients, respectively.
Although there was a lower incidence of MSI-H in the cystadenoma
group, the difference was not statistically significant. MSI-L was
detected in 0, 30 and 7.1%, and MSS was identified in 15.4, 10 and
14.3% of the cystadenocarcinoma, cystadenoma and normal patients,
respectively (Fig. 3).
MLH1 and MSH2 gene expression by qPCR
revealed no statistically significant difference among the three
studied groups (P=0.089 and P=0.122, respectively; Fig. 4).
Discussion
Despite advances in EOC therapy, mortality and
morbidity have not changed in recent decades (8). The MMR system is a well-defined
molecular pathway of carcinogenesis in hereditary and sporadic
tumors (9).
Several techniques have been used to evaluate the
MMR system, and, in the present study, we assessed MMR deficiencies
through the analysis of MSI in patients with EOC compared with
benign and normal ovarian tissue, which is a technique frequently
used by other researchers. A variety of markers used to identify
MSI in EOC have been described in the literature, but the optimal
markers are not yet well defined.
In our study, MSI was observed in 84.6% of serous
cystadenocarcinoma patients, and all of them had MSI-H. In 2001,
Sood et al were the first to use the NCI markers to
determine MSI in patients with EOC (11). These authors reported an MSI
frequency of 19%, of which 11% had MSI-H, and 8% had MSI-L. In
2006, Lu et al used the same NCI markers and identified MSI
in 53% of patients, of which 20% had MSI-H (19). In 2008, Yoon et al reported
an MSI frequency of 8%, of which 4% had MSI-H (20). The sample size may explain the
differences found in the frequency of MSI between the present study
and those in the literature. The highest frequency of MSI was found
with the BAT26 marker (67.6%) followed by the D5S346 marker
(64.9%). Sood et al reported that BAT25 was the most
frequent (11%), followed by D5S346 (10%).
An important feature taken into account in the study
of Sood et al was the polymorphic variation in the
amplification of alleles of NCI markers. Polymorphism
identification can prevent a polymorphic marker from being
characterized as unstable, which would undermine the results. In
the present study, polymorphism was also considered for the
determination of MSI. Among the 37 women studied, 32 (86.4%)
revealed polymorphism in the microsatellite analysis. The highest
frequency of polymorphism was observed in the D2S123 (83%) and
D17S250 markers (81%).
To assist in the identification of polymorphisms and
MSI we compared DNA leukocytes with the DNA of ovarian tissue. The
present study used peripheral blood samples, similar to Sood et
al in 2001, while in 2008 Yoon et al utilized samples
from paraffinized gynecological tissue for normal DNA extraction
(11,20).
Data in the literature suggests that women with
malignant ovarian tumors associated with a deficiency of the MMR
system have a higher survival rate, possibly related to less
aggressive tumor behavior (21,22).
In addition, MMR deficiency may be a predictor of tumor resistance
to chemotherapy (15,23). However, a systematic review
involving 22 studies found that the association between clinical
and/or epidemiological factors with MSI or MMR system deficiencies
in EOC has not been adequately studied (24). In this study, there was no
statistically significant association of MSI with clinical data in
the different comparison groups. The menopausal status was the only
statistically significant difference between groups, but this
factor was not associated with MSI (P=0.542).
In the present study, MSI of EOC was compared with
cystadenoma and normal ovarian tissue. To the best of our
knowledge, no other studies have used identical comparison groups.
The frequency of MSI in both benign epithelial ovarian neoplasms
and normal ovaries was high, as well as in EOC, with no
statistically significant difference between groups. This suggests
that MSI may arise as a consequence of the ovulatory process, and
not solely as a feature of malignant ovarian tumor development.
Repeated injuries in ovarian epithelium, due to an incessant
ovulatory process, would result in genetic alterations that
compromise the MMR system, culminating in MSI.
Additionally, to better assess the DNA mismatch
repair system, we studied MLH1 and MSH2 gene
expression using qPCR. Our results did not demonstrate any
difference between groups when comparing normal, cystadenoma and
cystadenocarcinoma samples.
Ovulation requires intense cell replication to
repair and restore epithelial ovarian microtrauma and may induce
permanent genetic changes that accumulate in cellular DNA, causing
a malfunction of the cell, which predisposes it to epithelial
ovarian mutagenesis (13,14). The presence of MSI as a consequence
of the ovulatory process reinforces the importance of certain
clinical risk factors, including early menarche, late menopause and
infertility, while factors that decrease the number of ovulatory
cycles, such as pregnancy, lactation and contraceptive use, reduce
the risk of ovarian cancer throughout life (25).
The results revealed a high frequency of MSI in
normal ovarian tissue, benign and malignant tumors of the ovary,
with no difference in the expression of the MMR system genes,
suggesting that MSI may be inherent to the ovulatory process. In
conclusion, MSI does not appear to play a role in ovarian
carcinogenesis.
References
|
1
|
Silva-Filho AL, Carmo GA, Athayde GR,
Assis ME, Almeida RC, Leal RH, Lamaita RM, Santos-Júnior JL and
Castro e Silva JG: Safe fertility-preserving management in
gynecological malignancies. Arch Gynecol Obstet. 275:321–330. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar
|
|
3
|
Dorigo O and Berek JS: Personalizing CA125
levels for ovarian cancer screening. Cancer Prev Res (Phila).
4:1356–1359. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brewer MA, Johnson K, Follen M, Gershenson
D and Bast R Jr: Prevention of ovarian cancer: intraepithelial
neoplasia. Clin Cancer Res. 9:20–30. 2003.PubMed/NCBI
|
|
5
|
Roett MA and Evans P: Ovarian cancer: an
overview. Am Fam Physician. 80:609–616. 2009.
|
|
6
|
Feeley KM and Wells M: Precursor lesions
of ovarian epithelial malignancy. Histopathology. 38:87–95. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dubeau L: The cell of origin of ovarian
epithelial tumours. Lancet Oncol. 9:1191–1197. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Berek JS, Chalas E, Edelson M, Moore DH,
Burke WM, Cliby WA and Berchuck A; Society of Gynecologic
Oncologists Clinical Practice Committee. Prophylactic and
risk-reducing bilateral salpingo-oophorectomy: recommendations
based on risk of ovarian cancer. Obstet Gynecol. 116:733–743. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pal T, Permuth-Wey J and Sellers TA: A
review of the clinical relevance of mismatch-repair deficiency in
ovarian cancer. Cancer. 113:733–742. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sidransky D: Emerging molecular markers of
cancer. Nat Rev Cancer. 2:210–219. 2002. View Article : Google Scholar
|
|
11
|
Sood AK, Holmes R, Hendrix MJ and Buller
RE: Application of the National Cancer Institute international
criteria for determination of microsatellite instability in ovarian
cancer. Cancer Res. 61:4371–4374. 2001.PubMed/NCBI
|
|
12
|
Singer G, Kallinowski T, Hartmann A,
Dietmaier W, Wild PJ, Schraml P, Sauter G, Mihatsch MJ and Moch H:
Different types of microsatellite instability in ovarian carcinoma.
Int J Cancer. 112:643–646. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fathalla MF: Incessant ovulation-a factor
in ovarian neoplasia? Lancet. 2:1631971. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hennessy BT, Coleman RL and Markman M:
Ovarian cancer. Lancet. 374:1371–1382. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Massey A, Offman J, Macpherson P and
Karran P: DNA mismatch repair and acquired cisplatin resistance in
E. coli and human ovarian carcinoma cells. DNA Repair
(Amst). 2:73–89. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Crijnen TE, Janssen-Heijnen ML, Gelderblom
H, Morreau J, Nooij MA, Kenter GG and Vasen HF: Survival of
patients with ovarian cancer due to a mismatch repair defect. Fam
Cancer. 4:301–305. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lawes DA, SenGupta S and Boulos PB: The
clinical importance and prognostic implications of microsatellite
instability in sporadic cancer. Eur J Surg Oncol. 29:201–212. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vaughn CP, Lyon E and Samowitz WZ:
Confirmation of single exon deletions in MLH1 and MSH2 using
quantitative polymerase chain reaction. J Mol Diagn. 10:355–360.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu Y, Liu XS, Wang YX, Song HY and Zhong
N: Study of microsatellite instability in epithelial ovarian
tumors. Beijing Da Xue Xue Bao. 38:62–65. 2006.PubMed/NCBI
|
|
20
|
Yoon BS, Kim YT, Kim JH, Kim SW, Nam EJ,
Cho NH, Kim JW and Kim S: Clinical significance of microsatellite
instability in sporadic epithelial ovarian tumors. Yonsei Med J.
49:272–278. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gryfe R, Kim H, Hsieh ET, Aronson MD,
Holowaty EJ, Bull SB, Redston M and Gallinger S: Tumor
microsatellite instability and clinical outcome in young patients
with colorectal cancer. N Engl J Med. 342:69–77. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ribic CM, Sargent DJ, Moore MJ, Thibodeau
SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R,
Shepherd LE, Tu D, Redston M and Gallinger S: Tumor
microsatellite-instability status as a predictor of benefit from
fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J
Med. 349:247–257. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sakamoto-Hojo ET and Balajee AS: Targeting
poly (ADP) ribose polymerase I (PARP-1) and PARP-1 interacting
proteins for cancer treatment. Anticancer Agents Med Chem.
8:402–416. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Murphy MA and Wentzensen N: Frequency of
mismatch repair deficiency in ovarian cancer: a systematic review.
International Journal of Cancer. 129:1914–1922. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guppy AE, Nathan PD and Rustin GJ:
Epithelial ovarian cancer: a review of current management. Clin
Oncol (R Coll Radiol). 17:399–411. 2005. View Article : Google Scholar : PubMed/NCBI
|