Introduction
Esophageal cancer ranks among the 10 most common
types of cancer in the world. The vast majority of the tumors are
squamous cell carcinomas. To date, surgical resection remains the
first treatment. However, nearly 95% of surgically resected
patients with advanced esophageal cancer succumb to recurrent or
metastatic disease within 5 years (1). Accordingly, it is necessary to
investigate the mechanism of tumorigenesis and metastasis of
esophageal squamous cell carcinoma (ESCC).
Previous studies have revealed that oncogenes and
tumor suppressor genes are implicated in tumorigenesis.
Inactivation, by loss or mutation, of tumor suppressor genes is
important in the genesis of many tumors. Tumor suppressor proteins
negatively regulate cell growth through a variety of mechanisms
controlling the cell cycle. The inhibitor of growth (ING) gene
family is newly recognised to be a part of this evolutionarily old
family of putative tumor suppressor genes. The currently identified
members of this family are the ING1, ING2 (ING1-L), ING3, ING4 and
ING5 genes. ING1, the first member of this family, was discovered
through a subtractive hybridization assay between normal mammary
epithelium and breast cancer cell lines and was shown to play an
essential role in neoplastic transformation (2–6). ING1
has been mapped to a locus on chromsome 13q33–34 and encodes four
isoforms, p47ING1a, p33ING1b,
p24ING1c and p27ING1d, which vary in mass
between 24 and 47 kDa (2,7). The p33ING1b protein is the
best characterized and most widely expressed in normal tissue
(8). Previous studies have shown
that p33ING1b is involved in the restriction of cell
growth and proliferation, apoptosis, tumor anchorage-independent
growth, cellular senescence, maintenance of genomic stability and
modulation of cell cycle checkpoints (9). A number of studies have been carried
out on altered p33ING1b in relation to tumors. Loss of
nuclear p33ING1b has been observed in melanoma, seminoma, papillary
thyroid carcinoma, oral squamous cell carcinoma, breast ductal
cancer and acute lymphoblastic leukemia (10,11).
To date, inactivation and/or decreased expression of
p33ING1b have been reported in various types of cancer,
including head and neck squamous cell, breast, lung, stomach, blood
and brain malignancies (7,12–16).
To the best of our knowledge, although there are a few studies of
p33ING1b in ESCC, little is known about its
clinicopathological significance in ESCC.
Particularly interesting new cysteine-histidine rich
protein (PINCH) is a newly discovered adapter protein, which
consists primarly of five LIM (double zinc finger) domains, and the
gene is located on chromosome 2q12.2. PINCH protein is able to
interact directly with integrin-linked kinase (ILK) and Nck-2
protein, and is associated with integrin signaling and the growth
factor signaling pathway (17–19).
It has been observed that PINCH expression is upregulated in
numerous types of malignancy, including oral and esophageal
squamous cell carcinoma, colorectal, pancreatic, skin, breast,
lung, prostate cancer and endometrioid endometrial carcinoma, as
well as gliomas (20–27). PINCH localizes to the peritumoral
stromal cells, particularly at the invasive edges of the tumor
(20). Furthermore, PINCH is an
independent prognostic factor in patients with colorectal cancer
(21). Our previous study on the
same series of cases used in the present study demonstrated that
PINCH expression was upregulated in ESCC compared with normal
esophageal squamous cells and the strong expression of PINCH was
correlated with lymph node metastasis (26). Recent studies have shown that the
genesis and metastasis of tumors are the result of the interaction
between tumor cells and tumor-associated stromal cells (28). Therefore, it is of significance to
explore whether there is a correlation between p33ING1b
expression in tumor cells and PINCH expression in the stromal cells
in human ESCC.
The aim of the present study was to investigate
p33ING1b expression in ESCC compared with normal
esophageal mucosa, and further to analyze the correlation between
p33ING1b expression in ESCC and clinicopathological
variables, including gender, age, tumor size, location, lymph node
status and the grade of differentiation, as well as PINCH
expression status.
Patients and methods
Patients
Formalin-fixed paraffin-embedded tissue samples were
obtained from 64 ESCC patients who underwent surgical resection at
the First Hospital of Hebei Medical University (Shijiazhuang,
Hebei, China), between 2000 and 2004. The study included 20 distant
normal mucosa specimens (all of which were matched with the primary
tumors) taken from the margin of distant resection. The primary
tumors were located in the upper, middle and lower sections of the
esophagus in 7, 36 and 21 cases, respectively, and 20 cases
involved lymph node metastasis. None of the patients had received
preoperative radiotherapy or chemotherapy. The patients’ gender,
age, tumor size, location, lymph node status and the grade of
differentiation were obtained from surgical and/or pathological
records at the hospital. The mean age of the patients was 59.5
years old (range 41–78 years). According to the WHO classification,
the tumor differentiation was graded as grade I (high
differentiation: 20 cases), grade II (moderate differentiation: 39
cases) and grade III (low differentiation: 5 cases). All
pathological slides, including normal specimens and tumors, were
confirmed by two pathologists (Z.L. Zhu and Z.M. Wang). The study
was approved by the ethical committee of the First Hospital of
Hebei Medical University, Shijiazhuang, Hebei, China. Written
informed consent was obtained from the patients.
Data of PINCH immunohistological staining in ESCC
were obtained from our previous study carried out at the Central
laboratory, The First Hospital of Hebei Medical University.
According to the intensity of PINCH staining in the
tumor-associated stromal cells, PINCH expression was graded as
negative group (none or <20% positive cells) and positive group
(≥20% positive cells) (26).
Immunohistological staining and
evaluation
Tissue sections (5 μm) from paraffin-embedded
tissue blocks were deparaffinised, hydrated and rinsed in distilled
H2O. In order to expose masked epitopes, the sections
were boiled in citrate buffer (pH 9.0) in a high pressure cooker
for 20 min, and then kept at room temperature for 30 min prior to
washing with phosphate-buffered saline (PBS, pH 7.4). The activity
of endogenous peroxidase was blocked with 3%
H2O2 in methanol for 10 min and then the
sections were washed three times in PBS. After blocking with 1.5%
horse serum in PBS for 10 min, the sections were incubated with a
goat polyclonal p33ING1 antibody raised against a peptide mapping
at the C-terminal of p33ING1 of human origin (C-19, sc-7566; Santa
Cruz Biotechnology, Inc., Santa Cruz, CA, USA) at 1:100 dilution at
4°C overnight. A biotinylated secondary antibody (Fuzhou Maixim
Biology Technology Co., Ltd., Fuzhou, Fujian, China) was then
applied for 30 min followed by incubation with an
avidin-biotin-peroxidase complex (Fuzhou Maxim Biotechnology Co.,
Ltd.) for 30 min. The sections were rinsed in PBS between the
incubation steps. The peroxidase reaction was developed using
diaminobenzidine (Beijing Zhongshan Biotechnology Co., Ltd,
Beijing, China) for 8 min. Following counterstaining with
hematoxylin, the sections were dehydrated and mounted. Sections of
ESCCs known to stain positively for p33ING1b were
included as negative (using PBS instead of the primary antibody)
and positive controls in all runs. There was no staining in the
negative controls, while the positive controls showed clear
staining.
p33ING1b immunohistological staining was
evaluated by two independent pathologists (Z.L. Zhu and Z.M. Wang)
in a blind fashion without knowledge of any clinicopathological
information. In normal squamous cells, only nuclear staining was
observed, while in tumors, cytoplasmic staining alone or staining
in the nucleus and cytoplasm were observed. According to the rate
of positive staining, we graded p33ING1b expression as
negative (no positive cells or <5% positive cells), weak (5–25%
positive cells), moderate (26–50% positive cells) and strong
positive (>50% positive cells). In statistical analysis, taking
into account similar clinicopathological features and facilitating
statistical analysis, we considered negative as the negative
staining group, and weak, moderate and strong positive as the
positive staining group. In order to avoid artificial effects,
cells on the margins of sections and in areas with poorly presented
morphology were not counted.
Statistical analysis
The statistical analyses were performed using SPSS
version 13.0 software. The Chi-square test was used to examine the
correlation between the frequencies of p33ING1b
expression in normal esophageal mucosa and ESCC, and the
correlation between p33ING1b expression in cancer and
clinicopathological variables or PINCH expression. All P-values
cited were two-sided and P<5% was considered to indicate a
statistically significant difference.
Results
p33ING1b expression in normal
mucosa and primary tumor
We examined p33ING1b protein expression
in normal esophageal mucosa and ESCC. In the 20 specimens of normal
mucosa, we found that the expression of p33ING1b was
only present in the nuclei of epithelial cells and there was no
cytoplasmic staining (Fig. 1A). Of
the specimens, 1 case was negative (5%) and 19 cases were positive
(95%), including 3 (15%) weak, 5 (25%) moderate and 11 (55%) strong
staining. However, in the primary cancers, none of the tumors
exhibited nuclear staining alone. There were 41 cases negative and
23 positive for p33ING1b, including 5 (8%) weak, 6 (9%)
moderate and 12 (19%) strong staining cases. Among the 23 positive
cases, 20 cases showed nuclear and cytoplasmic staining, mainly in
the cytoplasm (Fig. 1B) and 3 cases
had cytoplasmic staining alone.
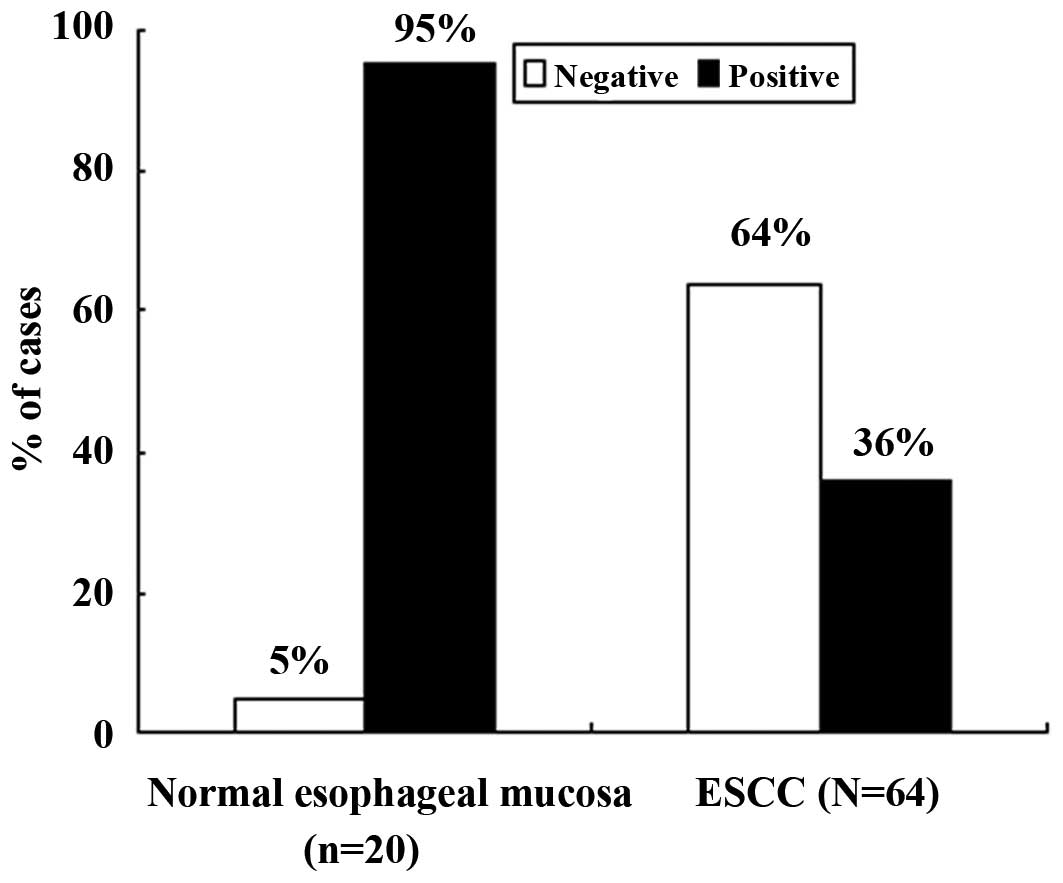
As shown in Fig. 2,
which presents the frequency of p33ING1b expression in
normal mucosa and ESCC, the rate of positive expression in the
normal mucosa specimens was 95% (19/20), which was significantly
higher than that in the ESCC specimens (36%, 23/64;
χ2=21.263, P<0.0001). We further compared nuclear and
cytoplasmic staining separately between the normal mucosa and ESCC
specimens; the results showed that the frequency of positive
p33ING1b expression in the nucleus (95% vs. 31%;
χ2=24.898, P=0.000) and in the cytoplasm (0 vs. 36%;
χ2=9.898, P=0.002) was significantly different.
Furthermore, we also observed the expression of
p33ING1b at the invasive margin and the inner part of
the tumor in all 64 ESCCs; there was no obvious difference between
the two sites.
p33ING1b protein expression in
relation to clinicopathological variables and PINCH expression in
ESCCs
Cytoplasmic staining of p33ING1b occurred
only in cancers and also dominated over nuclear staining, although
nuclear staining appeared in the majority of the cases with
cytoplasmic expression. For further statistical analysis,
regardless of nuclear staining, we investigated only
p33ING1b cytoplasmic staining (23 cases of positive
staining) in relation to clinicopathological variables (Table I).
 | Table ICorrelation of p33ING1b
protein expression with clinicopathological and biological
variables in patients with ESCC. |
Table I
Correlation of p33ING1b
protein expression with clinicopathological and biological
variables in patients with ESCC.
| | p33ING1b
expression
| | |
|---|
| Variables | N | Negative (%) | Positive (%) | χ2 | P-value |
|---|
| Gender | | | | | |
| Male | 50 | 32 (64) | 18 (36) | 0.000 | 0.984 |
| Female | 14 | 9 (64) | 5 (36) | | |
| Age (years) | | | | | |
| ≤50 | 19 | 13 (68) | 6 (32) | 0.223 | 0.637 |
| >50 | 45 | 28 (62) | 17 (38) | | |
| Tumor size
(cm) | | | | | |
| ≤3 | 26 | 17 (65) | 9 (35) | 0.033 | 0.855 |
| >3 | 38 | 24 (63) | 14 (37) | | |
| Location | | | | | |
| Upper | 7 | 5 (71) | 2 (29) | 0.208 | 0.901 |
| Middle | 36 | 23 (64) | 13 (36) | | |
| Lower | 21 | 13 (62) | 8 (38) | | |
| Lymph node
status | | | | | |
|
Non-metastasis | 44 | 34 (77) | 10 (23) | 10.673 | 0.001 |
| Metastasis | 20 | 7 (35) | 13 (65) | | |
| Grade | | | | | |
| I | 20 | 12 (60) | 8 (40) | 1.853 | 0.396 |
| II | 39 | 27 (69) | 12 (31) | | |
| III | 5 | 2 (40) | 3 (60) | | |
| PINCH | | | | | |
| Negative | 28 | 26 (93) | 2 (7) | 17.927 | <0.0001 |
| Positive | 36 | 15 (42) | 21 (58) | | |
As shown in Table I,
the cases with lymph node metastasis had a higher frequency of
p33ING1b positive expression than those without
metastasis in the lymph nodes (65% vs. 23%; χ2=10.673,
P=0.001). p33ING1b expression was not significantly
correlated with gender (P=0.984), age (P=0.637), tumor size
(P=0.855), tumor location (P=0.901) or grade of differentiation
(P=0.396).
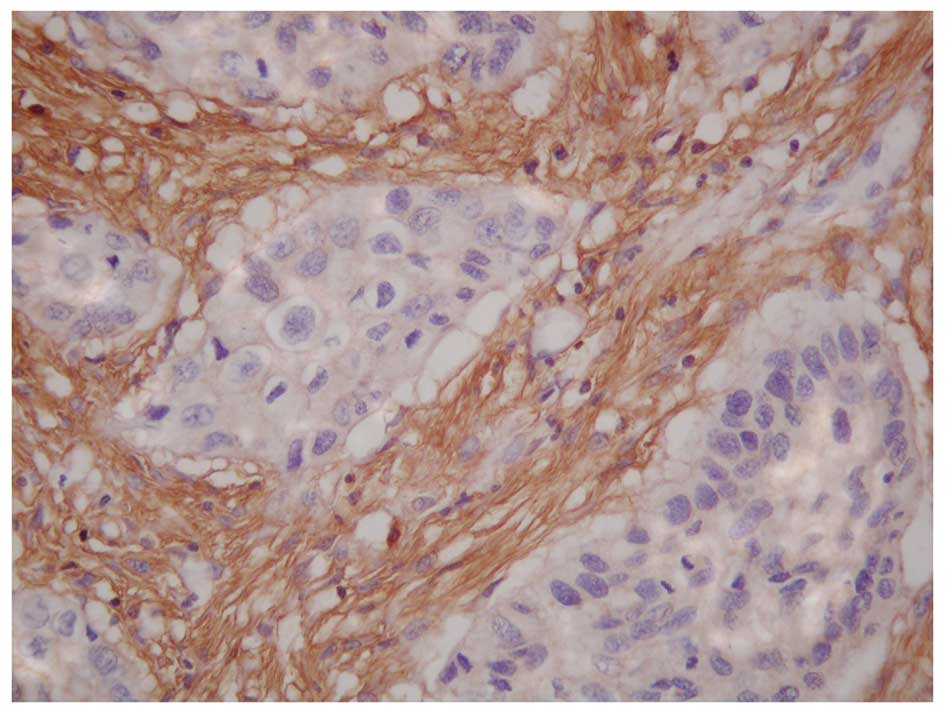
The results also revealed that p33ING1b
expression was positively related to the PINCH expression (Fig. 3) in all 64 ESCCs (Table I). Of the 36 cases with
PINCH-positive expression, 21 (58%) cases were p33ING1b
positive and 15 (42%) cases were p33ING1b negative.
However, in the 28 cases with PINCH-negative expression, there were
2 (7%) cases of p33ING1b positive and 26 (93%) cases of
p33ING1b negative (χ2=17.927, P=0.000).
Moreover, we found that the cases positive for both proteins had
the highest frequency of lymph node metastasis (13/20, 65%), the
cases negative for both proteins had the lowest frequency of
metastasis (2/20, 10%) and cases positive for either protein had a
moderate frequency (5/20, 25%; χ2=14.550, P=0.001).
Discussion
Studies have shown that the evolution and
development of ESCC results from multiple stepwise alterations of
cellular and molecular pathways in the squamous cells (1). Genetic changes may cause some
individuals to be more sensitive to these environmental factors,
although lifestyle factors account for the majority of ESCCs. The
activation of oncogenes and inactivation of tumor suppressor genes
(TSGs) are implicated in tumorigenesis. Tumor suppressor genes are
often referred to as ‘gatekeepers’ as they are able to prevent
tumor genesis and development by direct control of the cell cycle.
The ING gene family is a newly discovered TSG class. The currently
identified members of this family are the ING1, ING2, ING3, ING4
and ING5 genes. ING1 is the first member of the ING family, has
been mapped to a locus on chromsome 13q33–34 and encodes four
isoforms, p47ING1a, p33ING1b,
p24ING1c and p27ING1d. Currently,
p33ING1b is the most widely studied in malignancies and
is a focus of medical studies (2–7).
Nouman et al studied 76 melanocytic lesions by
immunohistochemistry for the expression of p33ING1b and
identified that there was a loss of nuclear p33ING1b
expression in invasive malignant melanoma compared with normal
cutaneous melanocytes or the melanocytes of benign melanocytic
naevi, and enhancement of cytoplasmic p33ING1b
expression in invasive malignant melanoma (29). In another study, Hoque et al
examined the mRNA expression of p33ING1b by reverse
transcription-PCR of 28 oral squamous cell cancers and found 2 (7%)
tumors with loss of p33ING1 expression (30). Thereafter, our research group
explored 49 oral squamous cell carcinoma specimens for
p33ING1b expression by immunohistochemistry and found
that 37 (76%) of the primary tumors were negative for
p33ING1b expression although the majority (90%) of
normal mucosa specimens showed p33ING1b-positive
expression in the nucleus (11).
Recently, Luo et al also identified that p33ING1b expression
was lost in the nucleus in 115 of 217 cases of human non-small cell
lung cancer (31). In the present
study, we used immunohistological staining and observed that, in 20
cases of normal mucosa, p33ING1b expression was only
present in the nuclei of the epithelial cells and 19 (95%) cases
were positive for p33ING1b (including 11 cases of strong
staining). By contrast, in 64 primary tumor samples, none of the
cancers showed nuclear staining alone and 41 (64%) cases had
negative p33ING1b expression; this was significant
difference (95 vs. 36%; χ2=21.263, P=0.000).
Results from previous studies have shown that
p33ING1b, as a candidate type II TSG, is involved in a
variety of processes, including DNA repair, cell cycle control,
senescence, apoptosis and chromatin remodeling, which are critical
points for genomic integrity and stability (9). p33ING1b gene and TP53
products are interrelated and the optimum functioning of both is
required for efficient cell growth suppression. Moreover, the tumor
suppression of TP53 and the transactivation activity of WAF1 are
partially dependent upon the fidelity and activity of
p33ING1b. Thus, the loss of p33ING1b function
may have similar consequences to loss of TP53 function and may
contribute to tumorgenesis by augmenting genomic instability and
refractivity to pro-apoptotic stimuli (9,32). The
observation by other groups of loss of p33ING1b
expression in tumors and our results in the present study, indicate
that the loss of p33ING1b nuclear expression in tumors
may be a key point in tumorigenesis.
Notably, in the present study, we also observed that
23 (36%) tumor samples had cytoplasmic expression of
p33ING1b, including 20 cases with nuclear and
cytoplasmic staining and 3 cases with cytoplasmic staining alone.
From these results, a doubt may be raised as to whether the
p33ING1b cytoplasmic expression was specific or
background staining. In order to clarify this issue, we re-observed
the staining results of all sections and confirmed the specificity
of the cytoplasmic staining of p33ING1b for the
following reasons: firstly, the negative controls did not show any
cytoplasmic staining; and secondly, there was no cytoplasmic
staining in the normal epithelial cells. Furthermore, this evidence
has been confirmed in certain tumors, including melanoma (10), brain tumor (15), breast cancer (7), oral squamous cell carcinoma (11) and acute lymphoblastic leukemia
(14), where p33ING1b
was also found to localize mainly in the cytoplasm. In addition, we
also identified that the cases with lymph node metastasis had a
higher frequency of positive p33ING1b expression in the
cytoplasm than those without metastasis (65% vs. 23%;
χ2=10.673, P=0.001). This result suggests a role for
p33ING1b cytoplasmic expression in promoting metastasis
of the ESCCs. Therefore, from the results of the present study and
other studies, the p33ING1b cellular compartment shift
from the nucleus to the cytoplasm may cause loss of normal cellular
function and play a central role in tumorigenesis and
progression.
However, the mechanism behind this shift of
p33ING1b protein from the nucleus to the cytoplasm is
not fully understood. Riabowol’s research group has reported that
p33ING1b particularly binds to members of the 14-3-3
family through phosphorylation at serine residue 199 (33). Studies revealed that 14-3-3 family
members primarily reside in the cytoplasm and are associated with
phosphorylated ligands involved in numerous cellular processes,
including regulation of the cell cycle and DNA damage checkpoints.
Binding to 14-3-3 causes tethering of significant amounts of
p33ING1b in the cytoplasm (33,34).
Moreover, other studies have demonstrated that cytoplasmic
p33ING1b may be imported into the nucleus through
interactions between its intrinsin nuclear location signal and
karyopherins α2 and β1. In the nucleus, lamin A binds and targets
ING1 and regulates its levels and biological function (35,36).
Therefore, 14-3-3, karyopherins α2 and β1, and lamin A are involved
in the cytoplasmic accumulation of p33ING1b in tumors.
However, the function of cytoplasmic p33ING1b is unclear
and requires further study.
There have been a few studies on the correlation
between p33ING1b expression and clinicopathological
variables. Li et al found that high expression of
cytoplasmic p33ING1b was significantly correlated with poor
differentiation, T staging, lymph node metastasis and TNM staging
in head and neck squamous cell carcinoma (37). In the present study, we also
observed that high cytoplasmic expression of p33ING1b
was significantly correlated with lymph node metastasis, but no
significant correlation was found between cytoplasmic expression of
p33ING1b and other clinicopathological variables,
including gender, age, tumor size, tumor location and the grade of
differentiation.
In the present study, we also found that the
cytoplasmic expression of p33ING1b had a positive
correlation with PINCH expression in the primary tumors. More
importantly, we further observed that cases positive for both
proteins had the highest frequency of lymph node metastasis (65%),
cases negative for both proteins had the lowest frequency of
metastasis (10%) and cases positive for either protein had a
moderate frequency (25%). The results suggest that
p33ING1b and PINCH cooperate in the metastasis of ESCC.
Taken together with the results of our previous study of
p33ING1b expression in oral squamous cell carcinoma
(11), we propose that, during
tumor development and metastasis, p33ING1b in the tumor
cells interacts with PINCH by a signaling pathway in the
associated-tumor stroma, particularly at the site of cell adhesion.
PINCH may be a marker for stroma manifesting the ability to
facilitate metastasis in human ESCC. If so, p33ING1b and
PINCH may be considered as novel biomarkers for the target of
therapy. Thus, it is necessary to further study this issue in a
large number of samples to verify this result.
The results suggest that p33ING1b
cellular compartment shift from the nucleus to the cytoplasm causes
a loss of normal cellular function and may play a central role in
the tumorigenesis and metastasis in human ESCC, particularly in
combination with PINCH expression.
Acknowledgements
This study was supported by the
Science and Technology Research and Development Program of Hebei,
China, 2011, No.11276103D-40.
References
|
1
|
Wu ZB and Yang GH: Chinese Surgical
Pathology. People’s Health Press; Beijing: pp. 619–627. 2002
|
|
2
|
Garkavtsev I, Kazarov A, Gudkov A and
Riabowol K: Suppression of the novel growth inhibitor
p33ING1 promotes neoplastic transformation. Nat Genet.
14:415–420. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nagashima M, Shiseki M, Miura K, et al:
DNA damage-inducible gene p33ING2 negatively regulates cell
proliferation through acetylation of p53. Proc Natl Acad Sci USA.
98:9671–9676. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shimada Y, Saito A, Suzuki M, Takahashi E
and Horie M: Cloning of a novel gene (ING1L) homologous to ING1, a
candidate tumor suppressor. Cytogenet Cell Genet. 83:232–235. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nagashima M, Shiseki M, Pedeux RM, et al:
A novel PHD-finger motif protein, p47ING3, modulates p53-mediated
transcription, cell cycle control, and apoptosis. Oncogene.
22:343–350. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shiseki M, Nagashima M, Pedeux RM, et al:
p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity.
Cancer Res. 63:2373–2378. 2003.PubMed/NCBI
|
|
7
|
Nouman GS, Anderson JJ, Crosier S,
Shrimankar J, Lunec J and Angus B: Downregulation of nuclear
expression of the p33ING1b inhibitor of growth protein
in invasive carcinoma of the breast. J Clin Pathol. 56:507–511.
2003.PubMed/NCBI
|
|
8
|
Saito A, Furukawa T, Fukushige S, Koyama
S, Hoshi M, Hayashi Y and Horii A: p24/ING1-ALT1 and p47/ING1-ALT2,
distinct alternative transcripts of p33/ING1. J Hum Genet.
45:177–181. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nouman GS, Anderson JJ, Lunec J and Angus
B: The role of the tumor suppressor p33 ING1b in human neoplasia. J
Clin Pathol. 56:491–496. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nouman GS, Angus B, Lunec J, Crosier S,
Lodge A and Anderson JJ: Comparative assessment expression of the
inhibitor of growth 1 gene (ING1) in normal and neoplastic tissue.
Hybridoma Hybridomics. 21:1–10. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang JT, Wang DW, Li QX, et al: Nuclear
to cytoplasmic shift of p33ING1b protein from normal
oral mucosa to oral squamous cell carcinoma in relation to
clinicopathological variables. J Cancer Res Clin. 134:421–426.
2008.PubMed/NCBI
|
|
12
|
Oki E, Maehara Y, Tokunaga E, Kakeji Y and
Sugimachi K: Reduced expression of p33ING1 and the
relation with p53 expression in human gastric cancer. Cancer Lett.
147:157–162. 1999.
|
|
13
|
Kameyama K, Huang CL, Liu D, et al:
Reduced ING1b gene expression plays an important role in
carcinogenesis of non-small cell lung cancer patients. Clin Cancer
Res. 9:4926–4934. 2003.PubMed/NCBI
|
|
14
|
Nouman GS, Anderson JJ, Wood KM, Lunec J,
Hall AG, Reid MM and Angus B: Loss of nuclear expression of the
p33ING1b inhibitor of growth protein in childhood acute
lymphoblastic leukaemia. J Clin Pathol. 55:596–601. 2002.PubMed/NCBI
|
|
15
|
Vieyra D, Senger DL, Toyama T, et al:
Altered subcellular localization and low frequency of mutation of
ING1 in human brain tumors. Clin Cancer Res. 9:5952–5961.
2003.PubMed/NCBI
|
|
16
|
Li X, Nishida T, Noguchi A, et al:
Decreased nuclear expression and increased cytoplasmic expression
of ING5 may be linked to tumorigenesis and progression in human
head and neck squamous cell carcinoma. J Cancer Res Clin.
136:1573–1583. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rearden A: A new LIM protein containing an
autopitope homologous to ‘senescent cell antigen’. Biochem Biophys
Res Commun. 201:1124–1134. 1994.
|
|
18
|
Wu C: PINCH, N(i)ck and the ILK: network
wiring at cell-matrix adhesions. Trends Cell Biol. 15:460–466.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tu Y, Li F, Goicoechea S and Wu C: The
LIM-only protein PINCH directly interacts with integrin-linked
kinase and is recruited to integrin-rich sites in spreading cells.
Mol Cell Biol. 19:2425–2434. 1999.PubMed/NCBI
|
|
20
|
Wang-Rodriquez J, Dreilinger AD, Alsharabi
GM and Rearden A: The signaling adapter protein PINCH is
up-regulated in the stroma of common cancer, notably at invasive
edges. Cancer. 95:1387–1395. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao J, Arbman G, Readen A and Sun XF:
Expression of PINCH protein is an independent prognostic factor in
colorectal cancer patients. Neoplasia. 6:796–801. 2004. View Article : Google Scholar
|
|
22
|
Zhao ZR, Zhang ZY, Cui DS, Li J, Zhang HJ,
Wang MW and Sun XF: Particularly interesting new cysteine-histidine
rich protein expression in colorectal adenocaccinomas. World J
Gastroenterol. 12:298–301. 2006.PubMed/NCBI
|
|
23
|
Wang MW, Gu P, Zhang ZY, Zhu ZL, Li YH,
Zhao HM and Sun XF: Expression of PINCH protein in gliomas and its
clinicopathological significance. Oncology. 72:343–346. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang JT, Li QX, Wang DW, et al:
Upregulation of PINCH in the stroma of oral squamous cell carcinoma
predicts nodal metastasis. Oncol Rep. 14:1519–1522. 2005.PubMed/NCBI
|
|
25
|
Yan BY, Wang DW, Zhu ZL, et al:
Overexpression of MAC30 in the cytoplasm of oral squamous cell
carcinoma predicts nodal metastasis and poor differentiation.
Chemotherapy. 56:424–428. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu Z, Yang Y, Zhang Y, et al: PINCH
expression and its significance in esophageal squamous cell
carcinoma. Dis Markers. 25:75–80. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang HZ, Li XH, Zhang X, et al: PINCH
protein expression in normal endometrium, atypical endometrial
hyperplasia and endometrioid endometrial carcinoma. Chemotherapy.
56:291–297. 2010. View Article : Google Scholar
|
|
28
|
Hwang RF, Moore T, Arumugam T, et al:
Cancer-associated stromal fibroblasts promote pancreatic tumor
progression. Cancer Res. 68:918–926. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nouman GS, Anderson JJ, Mathers ME,
Leonard N, Crosier S, Lunec J and Angus B: Nuclear to cytoplasmic
compartment shift of the p33ING1b tumor suppressor protein is
associated with malignacy in melanocytic lesions. Histopathology.
40:360–366. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hoque MO, Kawamata H, Nakashiro K, et al:
Dysfunction of the p53 tumor suppressor pathway in head and neck
cancer. Int J Oncol. 21:119–126. 2002.PubMed/NCBI
|
|
31
|
Luo ZG, Tang H, Li B, Zhu Z, Ni CR and Zhu
MH: Genetic alterations of tumor suppressor ING1 in human non-small
cell lung cancer. Oncol Rep. 25:1073–1081. 2011.PubMed/NCBI
|
|
32
|
Garkavtsev I, Grigorian IA, Ossovskaya VS,
Chernov MV, Chumakov PM and Gudkov AV: The candidate tumor
suppressor p33ING1 cooperates with p53 in cell growth
control. Nature. 391:295–298. 1998. View
Article : Google Scholar
|
|
33
|
Gong W, Russel M, Suzuki K and Riabowol K:
Subcellular targeting of p33ING1b by phosphorylation-dependent
14-3-3 binding regulates p21WAF1 expression. Mol Cell Biol.
26:2947–2954. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hermeking H and Benzinger A: 14-3-3
proteins in cell cycle regulation. Semin Cancer Biol. 16:183–192.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Russell MW, Soliman MA, Schriemer D and
Riabowol K: ING1 protein targeting to the nucleus by karyopherins
is necessary for activation of p21. Biochem Bioph Res Commun.
374:490–495. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Han X, Feng X, Rattner JB, et al:
Tethering by lamin A stabilizes and targets the ING1 tumor
suppressor. Nat Cell Biol. 10:1333–1340. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li XH, Kikuchi K and Takanol Y: ING genes
work as tumor suppressor genes in the carcinogenesis of head and
neck squamous cell carcinoma. J Oncol. 2011:9636142011.PubMed/NCBI
|