Introduction
Gelatinases, also named type IV collagenases,
include gelatinase A (MMP-2) and gelatinase B (MMP-9) (1). It is known that gelatinases are
important in tumor progression and are overexpressed in different
types of tumor cells, thus representing important tumor-associated
antigens (2,3). Solid tumors are usually composed
mainly of tumor cells and partly of stromal cells; the latter
consist of endothelial cells, neutrophil cells and hemopoietic
progenitor cells, which comprise the significant tumor
microenvironment and are important in tumor development and drug
therapy (4). It has been
demonstrated that gelatinases are also overexpressed in the tumor
stromal cells (4,5). Therefore, it may be promising to
develop new therapeutic drugs using gelatinases as a potential
target, which could kill the tumor cells as well as imbalance the
tumor microenvironment homeostasis.
Lidamycin is an extremely potent antitumor
antibiotic, which is composed of a highly active enediyne
chromophore and a protecting apoprotein (6). The chromophore and the apoprotein may
be disconnected and reconstituted under certain conditions
(6). Taking advantage of the
specific targeting capability of antibody fragments, different
types of fusion protein, which were composed of small antibody
fragments and the lidamycin apoprotein, were created. Following
stimulation with enediyne, the fusion protein demonstrated
antitumor efficiency in vitro and in vivo(6–10).
It is known that the heavy-chain
complementarity-determining region-3 (CDR3) domain of scFv is
important in antigen binding (11).
Qiu et al demonstrated that the fusion of several different
CDR3 domains led to a good targeting efficiency (12). Our previous studies also
demonstrated that fusion proteins containing the LDP and
oligopeptides specific for tumor antigens exhibited potent
antitumor activities (7,13), which suggested that a fusion protein
containing LDP and tumor specific oligopeptides was a promising
agent for development. We suggested that the combination of the
enediyne-energized fusion protein with its analog led to augmented
antitumor efficiency in vivo(10); however, the high-dose intravenous
administration of fusion protein may facilitate an increase in
immunogenicity. The possibility that a human anti-mouse antibody
(HAMA) response may be induced by the antibody of murine origin,
and that the down-sized antibody may more easily penetrate the core
of the solid tumor with lower immunogenicity were considered. In
the present study, fusion proteins containing the lidamycin
apoprotein and one or two heavy-chain CDR3 domains of
anti-gelatinases scFv were created, and their biomedical
characterization was investigated.
Materials and methods
Construcion of recombiant plasmids
pET-CDR3-LDP and pET-CDR3-LDP-CDR3
The sequence of the heavy chain CDR3 domain of
antigelatinase scFv (GenBank No. FJ037775) was
TGTGCTAGAGGGGACTACTATAGGCGCTACTTTGAC. To construct the CDR3-LDP
fusion protein, (GGGGS)2 was inserted betweeen the LDP and CDR3
domains as a linker (Fig. 1). Two
primers was designed and the sequences used were as follows: P1:
5′-GGAATTCCATATGTGTGCTAGA
GGGGACTACTATAGGCGCTACTTTGACGGTGGAGGT GGTTCAGGTGGA-3′ (the
NdeI site is underlined) and P2: 5′-CCGCTCGAGGCCGAAGGTCAGAGCCACGTG-3′
(the XhoI site is underlined). Using the plasmid
pET-Ec-ldp-Hr (7) as a template,
and primers P1 and P2, PCR amplification was performed. The
obtained fragment was NdeI/XhoI digested and was
inserted into a pET30a(+) expression vector to generate the
recombinant plasmid pET-CDR3-LDP. DNA sequencing analysis was
conducted to verify that the gene sequence was correct (Invitrogen
Life Technologies, Carlsbad, CA, USA).
To construct the pET-CDR3-LDP-CDR3 recombinant
plasmid, three different primer were designed and the sequences
used were as follows: P3: 5′-CCTTGCC
GAAGATCCTCCACCTCCAGATCCTCCCCCGCCGCCG AAGGTCAGACCAC-3′; P4:
5′-CCGCTCGAGATCGAAAT ATCGTCTGATAATCTCCCCTTGCCGAAGATCCTCC-3′; P5:
5′-GGAATTCCATATGTGTGCT-3′. Using the pETEc-ldp-Hr as a template,
and primers P1 and P3, PCR amplification was conducted. The
amplified product was used as the next template, and P4 and P5 as
the primers in the second PCR amplification. The final product was
NdeI/XhoI digested and inserted into to the pET30a(+)
plasmid to generate the recombinant plasmid pET-cdr3-ldp-cdr3.
Additionally, the sequence analysis of pET-cdr3-ldp-cdr3 was
verified (Invitrogen Life Technologies).
Expression and purification of CDR3-LDP
and CDR3-LDP- CDR3
The sequence-verified plasmids pET-CDR3-LDP and
pET-CDR3-LDP-CDR3 were transformed into the E.coli BL21
(DE3) expression strain (Novagen/MerckKGaA, Darmstadt, Germany) to
produce the recombinant protein. Expression, purification of
CDR3-LDP and CDR3-LDP-CDR3 fusion protein was carried out according
to the manufacturer’s protocol (Novagen). The purified protein was
analyzed by SDS-PAGE and the protein concentration was determined
by the BCA kit (Pierce Biotechnology, Inc., Rockford, IL, USA).
Binding with gelatinases
Gelatinases were coated in a 96-well plate
overnight, and a serial dilution of purified fusion proteins
CDR3-LDP and CDR3-LDP-CDR3 was added. The detailed procedure was
described previously (14), and the
final affintiy constant was determined by Graphpad Prism 5 software
(San Diego, CA, USA).
Binding activities of fusion protein
CDR3-LDP with tumor cells
Binding with tumor cells was determined by ELISA
assay. Human Bel-7402 and HepG2 hepatoma cell lines were seeded in
96-well plate at a density of 1×104cells/well and
cultivated overnight at 37°C. The following procedure was performed
according to that of our previous study (14).
To further identify the binding affinity of fusion
protein to target tumor cells, we used a fluorescence-activated
cell sorting (FACS)-based analysis assay. Protein bovine serum
albumin (BSA), LDP and CDR3-LDP were FITC labeled for 16 h in a
carbonate buffer solution (100 mmol/l NaHCO3; 10 mmol/l
Na2CO3, pH 9.0) at 4°C. Labeled protein was
separated from unbound FITC using the Sephadex G-25 column (GE
Healthcare, Waukesha, WI, USA). Each FITC-labeled protein, BSA, LDP
and CDR3-LDP, were incubated with 5×105 Bel-7402 and
HepG2 cells in a 100-μl volume of FACS buffer (PBS with 2% fetal
bovine serum) for 2 h at room temperature. Following three washes
with 500 μl of FACS buffer, cells were analyzed with a BD
FACSCalibur (BD Biosciences San Jose, CA, USA).
Additionally, the binding specificity of CDR3-LDP
with cancer cells was assessed by immunofluorescence. HepG2 cells
(1×105) were grown on coverslides overnight, fixed with
ice-cold 70% methanol, blocked with 5% BSA, then incubated with
CDR3-LDP fusion protein (100 μg/ml) for 2 h at 37°C. After washing
with PBS, cells were incubated with mouse anti-His tag monoclonal
antibody (dilution 1:200; Novagen) for 1 h, followed with
FITC-conjugated goat anti-mouse antibody (dilution 1:500; Zhongshan
Golden Bridge Biotechnology, Beijing, China). The images were
observed under a fluorescence microscope and collected by
fluorescence microscopy (Nikon TE 2000u, Tokyo, Japan).
Preparation of enediyne-energized fusion
protein CDR3-LDP-AE
To establish the potent antitumor activity of fusion
protein CDR3-LDP, assembly of the fusion protein with the enediyne
chromophore was performed. The detailed procedures and the HPLC
analysis were all performed according to our previous study
(10).
MTT assay
The MTT assay was used for measuring in vitro
cytotoxicity of stimulated CDR3-LDP-AE fusion protein as described
previously (10). Cells were seeded
at 3,000 cells/well in 96-well plates and incubated in 37°C for
overnight. Subsequently, cells were exposed to different
concentrations of lidamycin and stimulated CDR3-LDP-AE fusion
protein for 48 h. MTT (Sigma, St. Louis, MO, USA) solution (5
mg/ml, 20 μl) was added to each well and incubated for a further 4
h at 37°C. The supernatant was removed and 150 μl DMSO was added to
each well. The absorbance at 570 nm was measured using an ELISA
reader (Thermo Fisher Scientific). Growth inhibition was calculated
as a percentage of the nontreated controls.
In vivo antitumor activity
The in vivo experiment was performed with
7-week-old female Kunming mice (KM), which were purchased from the
Institute of Animal Research, Chinese Academy of Medical Science.
The study protocols were according to the regulations of the Good
Laboratory Practice for non-clinical laboratory studies of drugs
issued by the National Scientific and Technologic Committee of
People’s Republic of China.
Hepatoma 22 (H22) cells suspended in sterile saline
were inoculated subcutaneously (at day 0) in the right axilla of
mice, at a density of 2.0×106 cells/0.2 ml/mouse. The
mice were divided into seven groups, with 10 mice in each group. At
24 h of H22 cell transplantation (day 1), CDR3-LDP-AE was
administered at doses of 0.25 and 0.5 mg/kg of body weight.
lidamycin and CDR3-LDP fusion proteins were administered at doses
of 0.06 and 10 mg/kg, respectively. The combination of CDR3-LDP (10
mg/kg) fusion protein with CDR3-LDP-AE (0.25 and 0.5 mg/kg) was
also administered. All treatments were administered by intravenous
injection into the tail vein in 200 ml of sterile saline solution.
The experiment was terminated at day 11, and the tumors were
excised and weighed. The mean tumor weight was calculated and the
results were expressed as the mean ± standard deviation. The tumor
inhibition rate was calculated by: 1 - Tumor weight (treated) /
tumor weight (control) × 100.
Statistical methods
Results of quantitative data in this study were
presented as the mean ± standard deviation. Significant differences
between two values were determined using the Student’s t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression and purification of CDR3-LDP
and CDR3-LDP-CDR3 fusion proteins
As shown in Fig. 1,
the recombinant plasmids pET-CDR3-LDP and pET-CDR3-LDPCDR3 were
constructed. The plasmid were transferred to E.coli BL21
(DE3) for expression. Following cultivation for 4 h at 30°C and
induction by 0.2 mM IPTG for an additional 3 h, the cultures were
collected and washed with Tris-HCl buffer. Cultures were then
treated with ultrasound to break down the cells. The supernatant
was collected and loaded onto an Ni-affinity column for
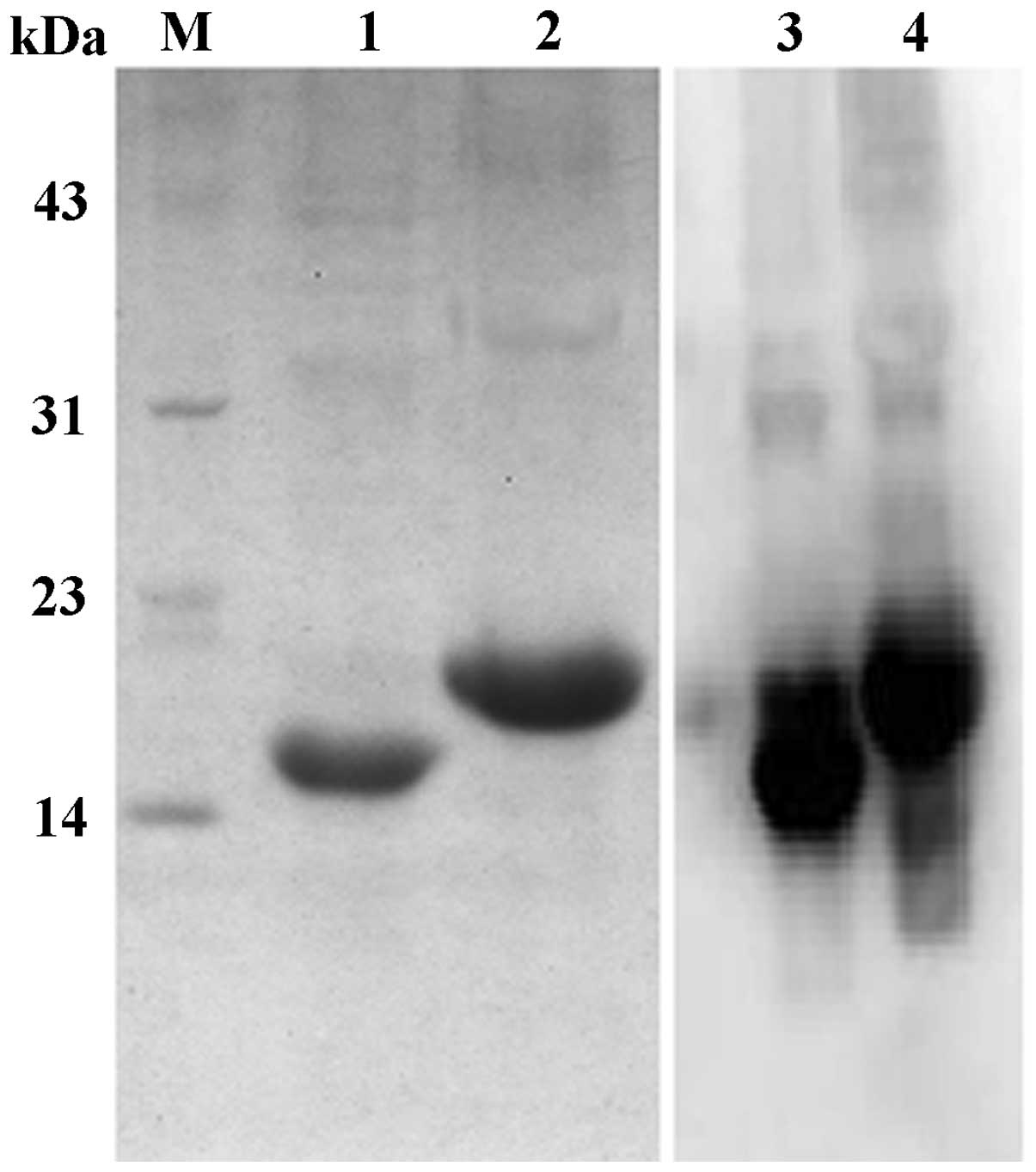
purification. SDS-PAGE was used to analyze the purified CDR3-LDP
and CDR3-LDP-CDR3 fusion proteins. As shown in Fig. 2, the CDR3-LDP fusion protein
migrated at a molecular weight of 15000 Da, while that of
CDR3-LDP-CDR3 was 18,000 Da. These findings were in accordance with
the theoretical prediction. Western blot analysis using anti-His
tag antibody confirmed that the (His)6 tag was
introduced into the fusion protein successfully.
Affinity assay
ELISA was used to determine the binding of the
CDR3-LDP and CDR3-LDP-CDR3 fusion proteins with antigen
gelatinases. As shown in Fig. 3, no
difference in affinity activity between fusion proteins CDR3-LDP
and CDR3-LDP-CDR3 with gelatinases was observed; their affinity
constant Kd values were 5.78×10−6 and
6.563×10−6 M, respectively. By contrast, the binding
affinity of the CDR3-LDP fusion protein with gelatinases was
relatively higher compared with that of the CDR3-LDP-CDR3 protein.
The reason for this may be that the two CDR3 domains did not have a
synergistic effect on binding with gelatinases, and the fused
(His)6 tag may also have had a negative impact on
antigen binding, thus resulting in that the binding activity of
CDR3-LDP-CDR3 did not exhibit a higher affinity compared with that
of CDR3-LDP. As the volu-metric productivity of the CDR3-LDP-CDR3
fusion protein expressed in E.coli in a soluble form was
lower compared with that of CDR3-LDP and did not demonstrate
enhanced binding activity with gelatinases, the CDR3-LDP fusion
protein was chosen for the following investigation.
Immunofluorescence assay
Immunofluorescence was used to investigate the
binding specifity of the CDR3-LDP fusion protein with HepG2 cells
in vitro. As shown in Fig.
4, the CDR3-LDP fusion protein was able to bind well with HepG2
cells, indicating their gelatinases were abundantly expressed
around the cell membrane.
Flow cytometry analysis of CDR3-LDP with
tumor cells
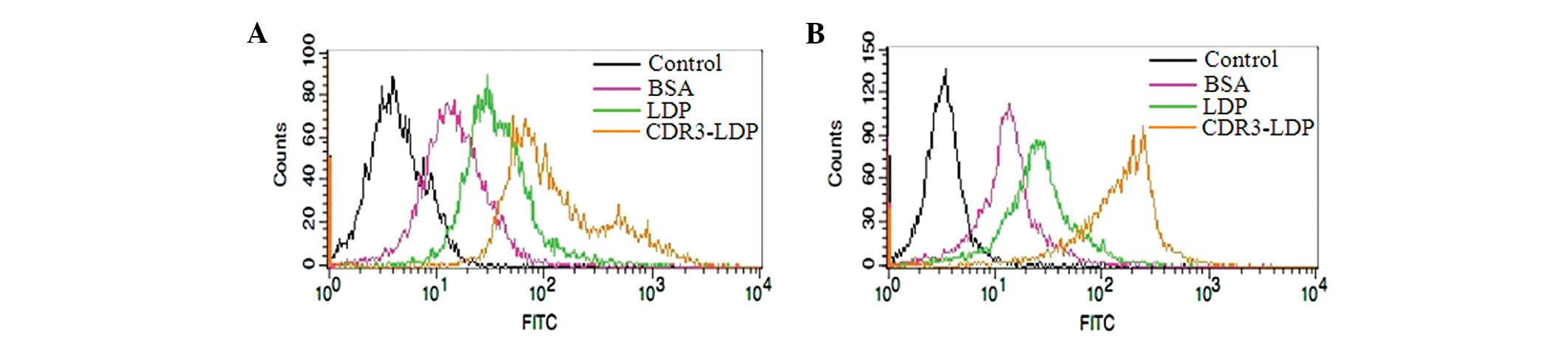
Flow cytometry was conducted to further testify that
the attachment of the CDR3-LDP domain was capable of improving the
binding of LDP with tumor cells. As shown in Fig. 5, compared with the FITC-labeled BSA
and LDP protein, the FITC-labeled CDR3-LDP was able to enhance the
binding intensity with tumor cells; the attachment of a single CDR3
domain increased the binding of LDP with tumor cells.
The assembly of CDR3-LDP fusion protein
with lidamycin AE
To establish the potent antitumor activity of
CDR3-LDP, the assembly procedure was performed. As shown in
Fig. 6A, the RP-HPLC chromatogram
indicated that the purity of purified CDR3-LDP was >90%, which
satisfied the requirements of the experiment. Following AE
assembly, the enediyne-energized CDR3-LDP-AE fusion protein was
obtained. As shown in Fig. 6B, an
additional peak was evident in the chromatogram and the retention
time was ∼7.3 min, which was similar to that of our previous study
(10).
MTT assay
The MTT assay was used to determined
enediyneenergized CDR3-LDP-AE fusion protein cytotoxicity to
Bel-7402 and HepG2 cells. As shown in Fig. 7, CDR3-LDP demonstrated extremely
potent cytotoxicity to tumor cells; the IC50 values for Bel-7402
and HepG2 cells were 1.05×10−11 and 6.6×10−14
M, respectively. By contrast, the IC50 values of lidamycin to
Bel-7402 and HepG2 cells were 5.6×10−10 and
4.2×10−11 M, respectively. The enhancement in
cytotoxicity of CDR3-LDP-AE may be related to the increased binding
capability with tumor cells compared with that of lidamycin.
In vivo animal experiment
To evaluate the antitumor efficiency of CDR3-LDP-AE
in vivo and to further demonstrate that the combined therapy
of fusion protein with enediyne-energized fusion protein augmented
the antitumor effect in vivo as observed in our previous
study (10), the H22 cell
xenografts in mice were used to evaluate the antitumor efficiency
of fusion proteins CDR3-LDP and CDR3-LDP-AE, both alone and in
combination.
As shown in Table I,
10 mg/kg CDR3-LDP had an inhibition rate of 50%, which indicated
that the fusion protein alone exerted certain antitumor effects.
Following assembly with AE, 0.25 and 0.5 mg/kg enediyne-energized
CDR3-LDP-AE fusion protein demonstrated tumor inhibition rates of
78.8 and 87.1%, respectively (P<0.05 compared with those of
CDR3-LDP and lidamycin). This indicated that the assembly of AE
improved the antitumor efficacy of the CDR3-LDP fusion protein, and
also suggested that the attachment of CDR3 domain increased the
accumulation of lidamycin at the tumor site. The combination of
CDR3-LDP (10 mg/kg) and CDR3-LDP-AE (0.25 and 0.5 mg/kg) further
augmented the tumor inhibitory efficiency; the tumor inhibition
rates were increased to 85.2 and 92.7%, respectively (P<0.05
compared with CDR3-LDP-AE). The increase in antitumor efficacy was
not accompanied with a loss of body weight, which indicated that
the combined therapy was effective and further improved the
antitumor efficacy.
 | Table IThe tumor growth inhibition effects of
fusion protein CDR3-LDP, CDR3-LDP-AE and their combination on
hepatoma 22 cells in mice. |
Table I
The tumor growth inhibition effects of
fusion protein CDR3-LDP, CDR3-LDP-AE and their combination on
hepatoma 22 cells in mice.
| Groups | Dosage (mg/kg) | Mouse number
(begin/end) | BWC (g) | Tumor weight (g) | Inhibition ratio
(%) |
|---|
| Control | | 10/10 | +5.72 | 1.401±0.234 | - |
| LDM | 0.06 | 10/10 | +4.09 | 0.513±0.281 | 63.4 |
| CDR3-LDP | 10.0 | 10/10 | +4.69 | 0.613±0.523 | 56.2 |
| CDR3-LDP-AE | 0.25 | 10/10 | +2.43 | 0.297±0.137 | 78.8ab |
| 0.5 | 10/10 | −0.47 | 0.180±0.091 | 87.1ab |
| CDR3-LDP | 10+0.25 | 10/10 | +2.15 | 0.208±0.067 | 85.2c |
| + CDR3-LDP-AE | 10+0.5 | 10/10 | −0.16 | 0.103±0.037 | 92.7c |
Discussion
Lidamycin is a potent antitumor antibiotic, which
consisted of a protecting apoprotein and a highly active enediyne
chromophore. The apoprotein and the chromophore may be detached and
reassembled under certain conditions (6,15).
Taking advantage of the targeting property of antibodies or
antibody fragments, types of chimeric fusion proteins, composed of
antibody fragments and apoprotein, were created and then
reassembled with the detached chromophore (6–9). In
the present study, two fusion proteins CDR3-LDP and CDR3-LDP-CDR3,
composed of a heavy-chain CDR3 domain fused with the lidamycin
apoprotein, were designed. The aim of the study was to develop a
targeting fusion protein with a lowered immunogenicity. As
observed, both CDR3-LDP and CDR3-LDP-CDR3 were easily expressed in
E.coli, and mainly existed in the soluble form. However, the
volumetric productivity of CDR3-LDP-CDR3 was relatively lower
compared with that of CDR3-LDP, and was easy to deposit, which may
have been due to the addition of a further CDR3 domain and thus the
increase in hydrophobicity. The results of the ELISA suggested that
there was no significant difference in the binding activity between
CDR3-LDP and CDR3-LDP-CDR3 fusion proteins. The addition of a
further CDR3 domain failed to increase the binding activities.
Therefore, for the development of this type of lidamycin fusion
protein, it is necessary to consider the negative influences of
steric hindrance or the existence of His tag on the affinity
property of oligopeptide. However, the ELISA is relatively accurate
approach to determine the affinity activity, which may be
determined by surface plasmon resonance (SPR) or other more
accurate equipment (16,17). As the CDR3-LDP-CDR3 fusion protein
did not demonstrate improved binding activity, the CDR3-LDP fusion
protein was chosen to perform the following investigations.
The results of ELISA and immunofluorescence
demonstrated that the CDR3-LDP fusion protein could bind well with
the tumor cells, and the results of FACS further confirmed that the
CDR3 domain increased the binding of LDP with tumor cells.
Following AE assembly, the enediyne-energized CDR3-LDP-AE fusion
protein displayed more potent cytotoxicity activity to tumor cells
compared with lidamycin, this may have been due to the increased
binding capability of CDR3-LDP-AE with tumor cells, as the protein
was fused with a tumor-binding oligopeptide CDR3 domain. Notably,
in the mouse model for the transplantation of H22 cells, 10 mg/kg
CDR3-LDP fusion protein alone had moderate antitumor efficiency,
and this was similar to the in vivo result of apoprotein
produced by genetic engineering (18). The lidamycin apoprotein does not
merely act as a carrier of chromophore; its antitumor mechanism
requires further elucidation. Following the assembly of the
chromophore with CDR3-LDP, the assembly product CDR3-LDP-AE
demonstrated enhanced antitumor efficiency. Additionally, the
combination of a large concentration of fusion protein CDR3-LDP and
a small concentration of enediyne-energized CDR3-LDP-AE fusion
protein further augmented the antitumor effect, indicating that the
combined therapeutic strategy was effective and has further
potential for the development of fusion protein containing
lidamycin apoprotein. This strategy would enhance the antitumor
effect without being accompanied with an increase in side effects.
As the molecular weight of CDR3-LDP was ∼15000 Da, the
immunogenicity of CDR3-LDP may be greatly lower compared with that
of dFv-LDP-AE as demonstrated previously (10). However, the binding activity of
CDR3-LDP was ∼100-fold lower compared with that of dFv-LDP.
Although the higher affinity does not negate superior tumor
targeting capability (12), it has
been demonstrated that the higher the affinity, the lower the tumor
penetrating ability (19,20). Therefore, in the development of a
lidamycin fusion protein, our long-term aim was to design a
tumor-targeting peptide or antibody fragment, and select the fusion
protein with lower immunogenicity, and stronger tumor targeting and
penetrating properties for further development. Our efforts should
also be directed toward the development of a novel administrative
approach or the humanization of antibodies.
Acknowledgements
This study was supported by the
National Natural Science Foundation of China (Grant Nos. 81201765
and 40901125).
References
|
1
|
Björklund M and Koivunen E:
Gelatinase-mediated migration and invasion of cancer cells. Biochim
Biophys Acta. 1755:37–69. 2005.PubMed/NCBI
|
|
2
|
Kallakury BV, Karikehalli S, Haholu A,
Sheehan CE, Azumi N and Ross JS: Increased expression of matrix
metalloproteinases 2 and 9 and tissue inhibitors of
metalloproteinases 1 and 2 correlate with poor prognostic variables
in renal cell carcinoma. Clin Cancer Res. 7:3113–3119. 2001.
|
|
3
|
Svagzdys S, Lesauskaite V, Pangonyte D,
Saladzinskas Z, Tamelis A and Pavalkis D: Matrix
metalloproteinase-9 is prognostic marker to predict survival of
patients who underwent surgery due to rectal carcinoma. Tohoku J
Exp Med. 223:67–73. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kessenbrock K, Plaks V and Werb Z: Matrix
metalloproteinases: regulators of the tumor microenvironment. Cell.
141:52–67. 2010. View Article : Google Scholar
|
|
5
|
Escaff S, Fernández JM, González LO, et
al: Comparative study of stromal metalloproteases expression in
patients with benign hyperplasia and prostate cancer. J Cancer Res
Clin Oncol. 137:551–555. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shao RG and Zhen YS: Enediyne anticancer
antibiotic lidamycin: chemistry, biology and pharmacology.
Anticancer Agents Med Chem. 8:121–131. 2008.
|
|
7
|
Guo XF, Zhu XF, Shang Y, Zhang SH and Zhen
YS: A bispecific enediyne-energized fusion protein containing
ligand-based and antibody-based oligopeptides against epidermal
growth factor receptor and human epidermal growth factor receptor 2
shows potent antitumor activity. Clin Cancer Res. 16:2085–2094.
2010. View Article : Google Scholar
|
|
8
|
Xin C, Ye S, Ming Y, et al: Efficient
inhibition of B-cell lymphoma xenografts with a novel recombinant
fusion protein: anti-CD20Fab-LDM. Gene Ther. 17:1234–1243. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhong GS, Wu MN, Guo XF, Zhang SH, Miao QF
and Zhen YS: Antitumor activities of dFv-LDP-AE: An
enediyne-energized fusion protein targeting tumor-associated
antigen gelatinases. Oncol Rep. 28:1193–1199. 2012.
|
|
10
|
Zhong G, Zhang S, Li Y, Liu X, Gao R, Miao
Q and Zhen Y: A tandem scFv-based fusion protein and its
enediyne-energized analogue show intensified therapeutic efficacy
against lung carcinoma xenograft in athymic mice. Cancer Lett.
295:124–133. 2010. View Article : Google Scholar
|
|
11
|
Lowe DC, Gerhardt S, Ward A, et al:
Engineering a high-affinity anti-IL-15 antibody: crystal structure
reveals an α-helix in VH CDR3 as key component of paratope. J Mol
Biol. 406:160–175. 2011.PubMed/NCBI
|
|
12
|
Qiu XQ, Wang H, Cai B, Wang LL and Yue ST:
Small antibody mimetics comprising two complementarity-determining
regions and a framework region for tumor targeting. Nat Biotechnol.
25:921–929. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo XF, Zhong GS, Miao QF and Zhen YS:
Construction of energized fusion protein consisting of epidermal
growth factor receptor oligopeptide ligand and lidamycin and its
antitumor activity. Ai Zheng. 28:561–568. 2009.(In Chinese).
|
|
14
|
Miao QF, Shang B, Ouyang Z, Liu X and Zhen
Y: Generation and antitumor effects of an engineered and energized
fusion protein VL-LDP-AE composed of single-domain antibody and
lidamycin. Sci China C Life Sci. 50:447–456. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shao RG and Zhen YS: Relationship between
the molecular composition of C1027, a new macromolecular antibiotic
with enediyne chromophore, and its antitumor activity. Yao Xue Xue
Bao. 30:336–342. 1995.(In Chinese).
|
|
16
|
Ashish B and Murthy GS: Analysis of human
chorionic gonadotropin-monoclonal antibody interaction in BIAcore.
J Biosci. 29:57–66. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Quinn JG and O’Kennedy R: Biosensor-based
estimation of kinetic and equilibrium constants. Anal Biochem.
290:36–46. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ru Q, Shang BY, Miao QF, Li L, Wu SY, Gao
RJ and Zhen YS: A cell penetrating peptide-integrated and
enediyne-energized fusion protein shows potent antitumor activity.
Eur J Pharm Sci. 47:781–789. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Adams GP, Schier R, McCall AM, et al: High
affinity restricts the localization and tumor penetration of
single-chain Fv antibody molecules. Cancer Res. 61:4750–4755.
2001.PubMed/NCBI
|
|
20
|
Carter PJ: Potent antibody therapeutics by
design. Nat Rev Immunol. 6:343–357. 2006. View Article : Google Scholar : PubMed/NCBI
|