Introduction
Hepatocellular carcinoma (HCC) is one of the most
common and aggressive types of cancer worldwide. Its highly
invasive and metastatic phenotypes are the key reasons for
treatment failure and a poor prognosis. Despite advances in
diagnostic and therapeutic techniques, a high failure rate and a
low median survival rate are currently observed in patients
following multimodality therapies with recurrent, intractable HCC.
To improve patient survival, it is important to elucidate the
regulatory mechanisms that control the tumor-initiating and
metastatic properties of HCC.
Cancer stem cells (CSCs), also defined as
tumor-initiating stem-like cells (TISCs), are a subpopulation of
neoplastic cells that possess characteristics of normal stem cells,
most notably the ability to self-renew and to differentiate into
heterogeneous non-tumorigenic cancer cells that comprise the bulk
of the tumor. There has been abundant evidence that supports the
existence of cancer stem cells in HCC (1). Under the CSC hypothesis, factors
contributing to a poor prognosis, including resistance to
conventional therapies, tumor recurrence and metastasis, may be
associated with a small subset of highly tumorigenic stem-like
cells (2). However, despite growing
evidence for the existence of tumor-initiating cells, the stem cell
properties of tumor-initiating cells remain largely unresolved
(3,4). Studies have suggested that CSCs have
common molecular signatures that are similar to those of
pluripotent embryonic stem cells (5,6). The
core transcription factors of this stem cell-like signature,
including Oct4, Sox2, Klf4, c-Myc, Nanog and Lin28, have been used
to successfully reprogram differentiated somatic cells into
pluripotent stem cells (7,8). Studies have also revealed that the
expression of stem cell-related genes may help to distinguish
tumors associated with distinct clinical outcomes in certain solid
types of cancer, such as medulloblastoma and colorectal cancer
(9,10). However, thus far, a possible
correlation between pluripotent stem cell genes and the prognosis
has not been established in HCC. Our study aimed to evaluate the
expression profiles of the pluripotent stem cell genes (Oct4, Sox2,
Klf4, c-Myc, Nanog and Lin28) using quantitative reverse
transcription (qRT)-polymerase chain reaction (PCR), and to
determine its possible prognostic significance in HCC.
Materials and methods
Patients and sample collection
A total of 57 pairs of HCC tissues and adjacent
non-tumor liver tissues were obtained from patients who underwent
curative liver resection consecutively in a single group at the
Department of Hepatic Surgery, Zhongshan Hospital, Fudan University
from January, 2006 to December, 2007. Tissue samples were frozen in
liquid nitrogen immediately following resection and were stored at
−80°C until RNA extraction was performed. Patient selection was
according to the following inclusion criteria: i) confirmed
pathological diagnosis of HCC; ii) without anticancer treatments
and distant metastases prior to surgery; iii) undergoing curative
liver resection for HCC, defined as macroscopically complete
removal of the tumor as described previously by Sun et
al(11); and iv) availability
of fresh tissue specimens and their complete clinical data. The
patients included 48 males (84.2%) and 9 females (15.8%), with a
median age of 49 years (range, 33–67 years). Fifty-four patients
(94.7%) were positive for hepatitis B virus. Tumor size ranged from
1.5 to 14.0 cm, with a median size of 6.0 cm. All tumors were
histologically diagnosed as HCC with Edmondson grade I in 2 cases,
grade II in 47 cases and grade III in 8 cases. The tumor stages
were classified according to the 7th edition tumor-node-metastasis
(TNM) classification of the International Union Against Cancer.
Twenty-four cases were classified as stage I, 25 as stage II and 8
as stage III. The study was performed in accordance with the
ethical standards of the Declaration of Helsinki and was approved
by the Ethics Committee of Zhongshan Hospital, Fudan University.
Informed consent was obtained from all patients participating in
the present study.
Real-time quantitative PCR analysis
Total RNA was extracted from HCC tissues and
adjacent non-tumor liver tissues using TRIzol (Invitrogen Life
Technologies; Carlsbad, CA, USA) according to the manufacturer’s
instructions. Total RNA (2 μg) was reverse transcribed using
the Primescript RT reagent kit (Takara Bio, Inc.; Tokyo, Japan).
Expression levels of Oct4 (NM_203289), Sox2 (NM_003106) and Klf4
(NM_004235), as well as the c-Myc (NM_002467), Nanog (NM_002467)
and Lin28 (NM_024674) genes were evaluated by quantitative
real-time PCR. Real-time PCR for quantification was performed using
SYBR Premix Ex Taq (Takara Bio, Inc.). The reactions were performed
in triplicate. The expression levels of target genes were
normalized to the expression level of GAPDH (NM_002046), a
housekeeping gene control. Primer sequences of target genes and
GAPDH were as follows: forward: 5’-GAGAAGGATGTGGTCCGAGTGTG-3’ and
reverse: 5’-GGCAGATGGTCGTTTGGCTGAATA-3’ for human Oct4; forward:
5’-CGCCCCCAGCAGACTTCACA-3’ and reverse:
5’-CTCCTCTTTTGCACCCCTCCCATTT-3’ for human Sox2; forward:
5’-AGAGGAGCCCAAGCCAAAGAG-3’ and reverse:
5’-CGAATTTCCATCCACAGCCGTC-3’ for human Klf4; forward:
5’-CAGAGTGCATCGACCCCTCG-3’ and reverse: 5’-TTCCTCCTCAGAGTCGCTGC-3’
for human c-Myc; forward: 5’-TGAACCTCAGCTACAAACAGGTG-3’ and
reverse: 5’-AACTGCATGCAGGACTGCAGAG-3’ for human Nanog; forward:
5’-CTCCGTGTCCAACCAGCAG-3’ and reverse:
5’-CACGTTGAACCACTTACAGATGC-3’ for human Lin28; forward:
5’-AGCCACATCGCTCAGACA-3’ and reverse: 5’-GCCCAATACGACCAAATCC-3’ for
human GAPDH. The relative genomic expression was calculated by
2−ΔΔCt, as previously described by Schmittgen and Livak
(12).
Follow-up
Patients were followed up until December, 2011 and
the median follow-up period was 22 months (range, 5–58 months).
Follow-up procedures were performed according to a uniform
guideline previously established in our institute (13). Briefly, patients were monitored by
abdominal ultrasonography, serum α-fetoprotein (AFP) levels and
chest radiography with an interval of 2–4 months following
discharge. When a recurrence was suspected, computed tomography
(CT) scanning or magnetic resonance imaging (MRI) was performed
immediately. Treatment modalities following relapse were
administered according to a uniform guideline by Sun et
al(14). Overall survival (OS)
was defined as the time interval between the date of surgery and
the date of mortality or the last observation time. For surviving
patients, the data were censored at the last follow-up.
Recurrence-free survival (RFS) was defined as the time interval
between the date of surgery and the date of diagnosis of any type
of tumor relapse (intrahepatic recurrence and extrahepatic
metastasis).
X-tile analysis
As there are no established cut-off points available
for pluripotent stem cell gene expression in HCC, X-tile analysis
(X-tile software, version 3.6.1; Yale University School of
Medicine; New Haven, CT, USA) was performed to generate an optimal
cut-off point for categorization of the stem cell gene expression
levels. The X-tile program split the cohort randomly into a matched
training and validation set as a method for selecting an optimal
cut-off point. It calculated a P-value for every possible division
of the cohort expression data. A two-dimensional graph with its
corresponding survival curves was plotted, where each colored pixel
was proportional to its χ2 value. The program
automatically calculated the maximum χ2 value, which
served as a cut-off point that predicted the prognosis (15).
Statistical analysis
Numerical data are presented as mean ± standard
deviation or as median (range). Quantitative data were compared
using a Wilcoxon signed rank sum test. Categorical data were
analyzed by the χ2 or Fisher exact tests as appropriate.
A Spearman’s correlation test was applied to analyze the
correlations. OS and RFS curves were generated using the
Kaplan-Meier method, and the differences between curves were
assessed by the log-rank test. Independent prognostic factors were
estimated by the Cox proportional hazards stepwise regression
model. The Statistical Package for the Social Sciences (SPSS) 13.0
for Windows (SPSS, Inc.; Chicago, IL, USA) was the software used
for assessment. A two-tailed P<0.05 was considered to indicate a
statistically significant difference.
Results
Pluripotent stem cell gene expression in
HCC tissues
The expression of six pluripotent genes was
evaluated in 57 HCC patients through qPCR analysis. The results
revealed that the expression levels of Oct4, Sox2, Klf4, c-Myc and
Nanog in HCC specimens were signifcantly higher than those in the
corresponding adjacent non-tumor tissues (Fig. 1). Analysing all possible
combinations of transcription factors demonstrated that a
significant correlation was only achieved between Oct4 and Nanog
(Spearman’s correlation coefficient, 0.44; P<0.001), suggesting
a possible functional link between Oct4 and Nanog in HCC cases.
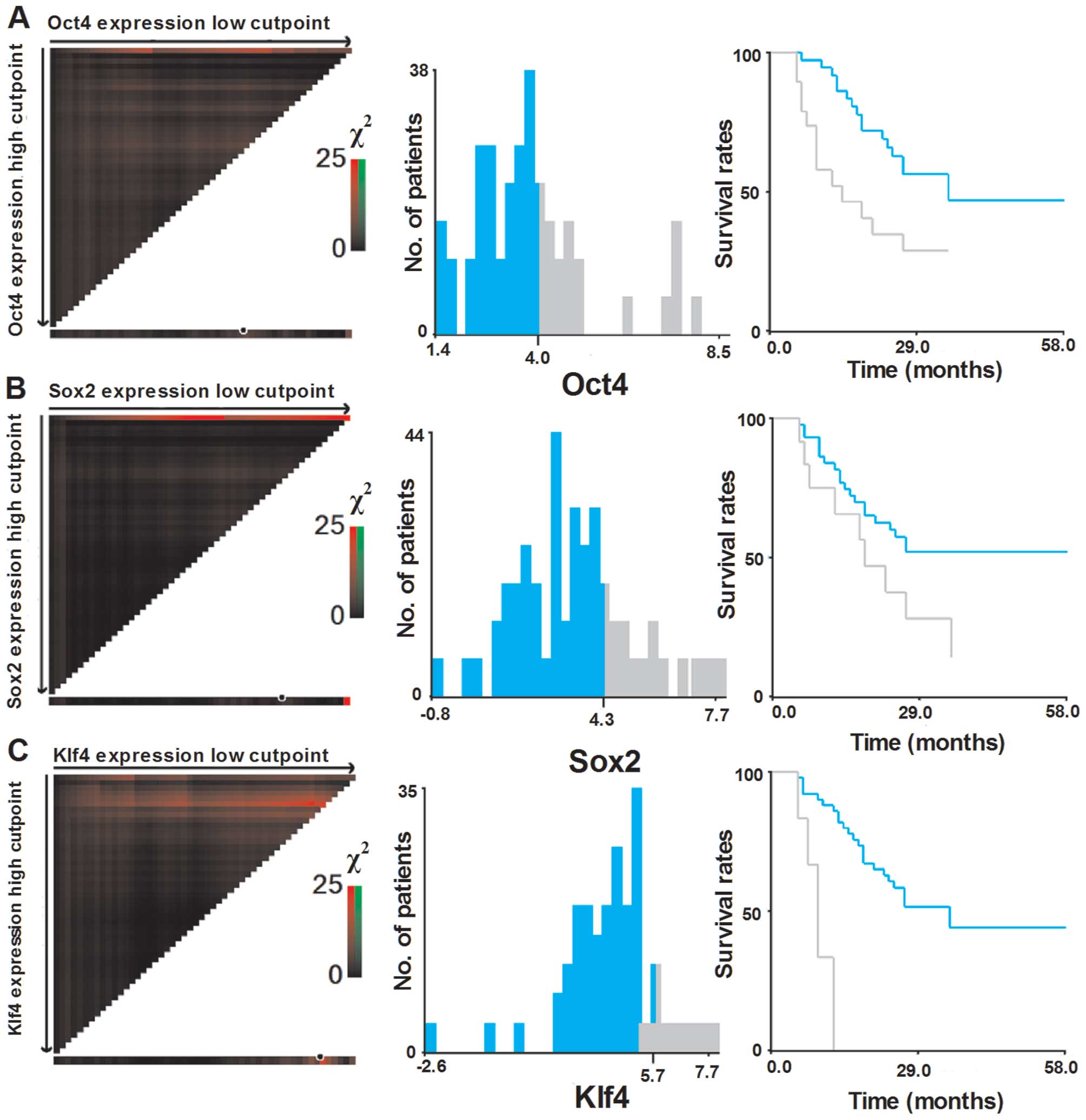
Selection of optimal cut-off values
To assess the statistical significance and to avoid
the problems of multiple cut-off point selection, the X-tile
program was employed to determine the cut-off values for the
expression of six pluripotent genes. According to the X-tile
program, the patient cohort was divided into low expression and
high expression populations based on a cut-off point of 4.0 for
Oct4 (Pmin=0.02), 4.3 for Sox2 (Pmin=0.05),
5.7 for Klf4 (Pmin<0.001), 4.7 for Nanog
(Pmin=0.01) 7.3 for c-Myc (Pmin=0.26) and 4.2
for Lin28 (Pmin=0.38) (Figs.
2 and 3).
Correlation of pluripotent stem cell gene
expression with clinicopathologic features
As shown in Table I,
the expression levels of the Sox2 and Lin28 genes were
significantly correlated with tumor size. The mean Sox2 and Lin28
expression levels in the 28 cases with a large tumor size (>5
cm) were 3.79±0.29 and 4.66±0.40, respectively; whereas in the 29
cases with a small tumor size, the levels were 2.79±0.29 and
3.55±0.31 (P=0.02 and 0.03), respectively. With respect to vascular
invasion, marked increases in Klf4 (P=0.02) and c-Myc (P=0.01)
levels were detected in HCC patients with vascular invasion. With
regard to tumor differentiation, a significant correlation was
identified between Klf4 expression and poor tumor differentiation
(P=0.03). However, no statistical difference was identified in the
expression of pluripotent genes when compared with other
clinicopathological factors, including age, gender, hepatitis B
surface antigen (HBsAg), AFP, liver cirrhosis, tumor number, tumor
encapsulation and TNM stage.
 | Table ICorrelation between pluripotent stem
cell gene expression and clinicopathological variables in
hepatocellular carcinoma (HCC) patients. |
Table I
Correlation between pluripotent stem
cell gene expression and clinicopathological variables in
hepatocellular carcinoma (HCC) patients.
| Oct4 | Sox2 | Klf4 | c-Myc | Nanog | Lin28 |
|---|
|
|
|
|
|
|
|
|---|
| Variables | High n=19 | Low n=38 | P-value | High n=13 | Lo n=44 | P-value | High n=7 | Low n=50 | P-value | High n=21 | Low n=36 | P-value | High n=13 | Low n=44 | P-value | High n=28 | Low n=29 | P-value |
|---|
| Gender | | | | | | | | | | | | | | | | | | |
| Male | 16 | 32 | 1.0 | 9 | 39 | 0.09 | 4 | 44 | 0.04 | 19 | 29 | 0.32 | 10 | 38 | 0.41 | 21 | 27 | 0.06 |
| Female | 3 | 6 | | 4 | 5 | | 3 | 6 | | 2 | 7 | | 3 | 6 | | 7 | 2 | |
| Age (years) | | | | | | | | | | | | | | | | | | |
| ≤50 | 8 | 23 | 0.19 | 3 | 28 | 0.01 | 3 | 28 | 0.51 | 13 | 18 | 0.38 | 5 | 26 | 0.19 | 12 | 19 | 0.09 |
| >50 | 11 | 15 | | 10 | 16 | | 4 | 22 | | 8 | 18 | | 8 | 18 | | 16 | 10 | |
| HBsAg | | | | | | | | | | | | | | | | | | |
| Positive | 1 | 2 | 1.0 | 1 | 2 | 0.66 | 0 | 3 | 0.51 | 0 | 3 | 0.17 | 0 | 3 | 0.33 | 2 | 1 | 0.53 |
| Negative | 18 | 36 | | 12 | 42 | | 7 | 47 | | 21 | 33 | | 13 | 41 | | 26 | 28 | |
| Liver
cirrhosis | | | | | | | | | | | | | | | | | | |
| Presence | 2 | 4 | 1.0 | 3 | 3 | 0.09 | 2 | 4 | 0.10 | 1 | 5 | 0.28 | 1 | 5 | 0.71 | 3 | 3 | 0.96 |
| Absence | 17 | 34 | | 10 | 41 | | 5 | 46 | | 20 | 31 | | 12 | 39 | | 25 | 26 | |
| AFP
(μg/l) | | | | | | | | | | | | | | | | | | |
| Positive
>20 | 6 | 12 | 1.0 | 2 | 16 | 0.15 | 1 | 17 | 0.29 | 7 | 11 | 0.83 | 4 | 14 | 0.94 | 9 | 9 | 0.93 |
| Negative
<20 | 13 | 26 | | 11 | 28 | | 6 | 33 | | 14 | 25 | | 9 | 30 | | 20 | 19 | |
| Tumor number | | | | | | | | | | | | | | | | | | |
| Single | 12 | 30 | 0.20 | 8 | 34 | 0.26 | 5 | 37 | 0.89 | 14 | 28 | 0.36 | 10 | 32 | 0.76 | 21 | 21 | 0.83 |
| Multiple | 7 | 8 | | 5 | 10 | | 2 | 13 | | 7 | 8 | | 3 | 12 | | 7 | 8 | |
| Tumor sizea (cm) | | | | | | | | | | | | | | | | | | |
| <5 | 10 | 18 | 0.71 | 4 | 24 | 0.02 | 4 | 24 | 0.65 | 11 | 17 | 0.71 | 7 | 21 | 0.70 | 10 | 15 | 0.03 |
| >5 | 9 | 20 | | 9 | 20 | | 3 | 26 | | 10 | 19 | | 6 | 23 | | 18 | 11 | |
| Vascular
invasion | | | | | | | | | | | | | | | | | | |
| Yes | 6 | 10 | 0.17 | 5 | 11 | 0.34 | 4 | 12 | 0.02 | 10 | 6 | 0.01 | 6 | 10 | 0.10 | 7 | 9 | 0.60 |
| No | 13 | 28 | | 8 | 33 | | 3 | 38 | | 11 | 30 | | 7 | 34 | | 21 | 20 | |
| Tumor
encapsulation | | | | | | | | | | | | | | | | | | |
| Complete | 11 | 16 | 0.26 | 5 | 22 | 0.34 | 5 | 22 | 0.17 | 13 | 14 | 0.09 | 9 | 18 | 0.07 | 13 | 14 | 0.90 |
| Incomplete | 8 | 22 | | 8 | 22 | | 2 | 28 | | 8 | 22 | | 4 | 26 | | 15 | 15 | |
|
Differentiation | | | | | | | | | | | | | | | | | | |
| I–II | 14 | 35 | 0.12 | 11 | 38 | 0.87 | 5 | 44 | 0.03 | 18 | 31 | 0.97 | 10 | 39 | 0.26 | 24 | 25 | 0.96 |
| III–IV | 5 | 3 | | 2 | 6 | | 2 | 6 | | 3 | 5 | | 3 | 5 | | 4 | 4 | |
| TNM stage | | | | | | | | | | | | | | | | | | |
| I | 8 | 16 | 0.85 | 4 | 20 | 0.62 | 3 | 21 | 0.49 | 9 | 15 | 0.21 | 5 | 19 | 0.56 | 10 | 14 | 0.341 |
| II | 9 | 16 | | 7 | 18 | | 4 | 21 | | 7 | 18 | | 5 | 20 | | 15 | 10 | |
| III | 2 | 6 | | 2 | 6 | | 0 | 8 | | 5 | 3 | | 3 | 5 | | 3 | 5 | |
Involvement of pluripotent stem cell gene
expression in the prognosis of HCC
The OS and RFS rates for the whole study population
were 76.9 and 66.9% at 1 year, 42.4 and 36.7% at 3 years, as well
as 42.4 and 19.7% at 5 years, respectively. Upon univariate
analysis, Oct4, Klf4 and Nanog mRNA expression were correlated with
an unfavorable OS (Fig. 4A–C). High
Klf4 expression was also correlated with poor RFS compared with low
Klf4 expression (median, 9.0 vs. 15.5 months; P=0.009; Fig. 5C). The multivariate analysis
revealed that Klf4 expression was independently correlated with OS
(HR, 8.61; 95% CI, 2.7–27.5; P<0.001) and RFS (HR, 3.96; 95% CI,
1.3–11.6; P=0.01). Additionally, vascular invasion and complete
tumor encapsulation were also independent predictors for OS
(P=0.006 and 0.04, respectively). Moreover, tumor number and TNM
stage were observed to be independently correlated with RFS (P=0.04
and P=0.03, respectively).
Discussion
Although the number of HCC cases was limited in our
study, the results were intriguing. We demonstrated that positive
expression of Klf4 was significantly correlated with vascular
invasion, poor tumor differentiation, short post-operative
recurrence time and a poor prognosis.
Klf4, previously known as gut-enriched Krüppel-like
factor (GKLF), is highly expressed in the post-mitotic cells of the
gut and skin. Klf4 was first identified as a tumor suppressor,
owing to frequent loss of Klf4 expression in gastric, colon,
esophageal, bladder and lung cancer, as well as in pancreatic
ductal carcinoma (16–18). However, subsequent studies proposed
the expression of Klf4 was upregulated in oral squamous carcinoma,
primary breast ductal carcinoma and human skin squamous cell
carcinoma, and correlated with carcinogenesis and tumor progression
(19–21). These studies suggested that Klf4 may
function as either a tumor suppressor or an onco-gene, depending on
the tumor type. To date, the role of Klf4 in HCC carcinogenesis and
progression remains unclear. Our study indicated that the
expression of Klf4 mRNA was significantly correlated with vascular
invasion and poor tumor differentiation. Notably, positive
expression of Klf4 mRNA was also correlated with tumor relapse and
a poor prognosis in patients with HCC. It is difficult to explain
the effects of Klf4 on HCC invasion and progression, and there is
no direct evidence to indicate this clearly in vivo and
in vitro. Further study is required to determine the precise
role of KLF4 in HCC and in other human cancer types.
Sox2, along with Oct4 and Nanog, plays a crucial
role in the maintenance of embryonic stem cell pluripotency
(22). Previous studies have
demonstrated that Sox2 is also involved in promoting tumorigenesis,
proliferation, and dedifferentiation of human lung squamous cell
carcinoma (23) and breast cancer
(24). Reduced Sox2 levels in
glioblastoma tumor-initiating cells can cause proliferation to
cease and a loss of tumorigenicity (25), while overexpression of Sox2 in
pancreatic cancer is correlated with an invasive and metastatic
phenotype (26). To elucidate the
role of Sox2 in HCC, we investigated the correlation between Sox2
mRNA expression and clinicopathological variables associated with
tumor progression. The results suggested that Sox2 mRNA is
upregulated in HCC tissue, compared with in the adjacent non-tumor
tissue and is correlated with a large tumor size. The survival
analysis further indicated that patients with a high level of Sox2
had a short survival time (25 months) compared with those with a
low level of Sox2 (17 months); however, the difference was not
significant (P=0.05). These results suggested that the upregulation
of Sox2 may play an important oncogenic role in HCC and represent
an acquired malignant proliferative phenotypic feature of tumor
cells.
Oct4, a transcription factor in the POU protein
family, expressed in both embryonic and adult stem cells, has been
proposed to be associated with the pluripotency, proliferative
potential and self-renewal properties of embryonic stem cells
(ESCs) and germ cells (27). Nanog,
a downstream target of Oct4 that contributes to cell fate
determination of the pluripotent inner cell mass during embryonic
development, is also specifically expressed in human embryonic
pluripotent stem cells (28). The
expression of Oct4 and Nanog is usually confined to pluripotent
cells of the developing embryo. Studies have suggested that Oct4
and Nanog may participate in the tumorigenicity and tumor
progression of various types of cancer, including breast cancer,
glioma, human endometrial adenocarcinoma, gastric cancer and
colorectal cancer (29–33). Coexpression of Oct4 and Nanog has
also been identified to be correlated with pancreatic
carcinogenesis as well as with a poor prognosis of oral squamous
cell carcinoma patients (34,35).
In the present study, we found that Oct4 and Nanog expression
levels were elevated in HCC tissue compared with the adjacent
non-tumor liver tissue. Moreover, Oct4 mRNA expression was
positively correlated with Nanog mRNA expression (Spearman’s
correlation coefficient, 0.44; P<0.001). Patients with a
positive expression of Oct4 or Nanog had a relatively low survival
rate (Fig. 3A and B). These results
indicated that Oct4 and Nanog, the two master transcription
factors, which are correlated with stem cell self-renewal and
differentiation, may also be correlated with HCC carcinogenesis and
the unfavorable prognosis of HCC patients following surgery.
The Myc transcription factor is one of the most
important somatically mutated oncogenes in human cancer. There is
increasing evidence to support the role of the c-Myc protooncogene
in tumor onset and progression. The Myc protein is able to confer a
selective advantage in cancer cells by promoting proliferation,
cell survival, differentiation blockade, genetic instability and
angiogenesis, all of which may indirectly contribute to metastasis
(36–38). Upregulating c-Myc expression drives
cell growth and vasculogenesis, reduces cell adhesion and promotes
metastasis (39). c-Myc also
induces the production of the vascular endothelial growth factor
(VEGF) by tumor cells, leading to tumor vasculo genesis in
pancreatic islet cancer and non-small cell lung cancer (38,40).
In accordance with these studies, our data also demonstrated that
an abnormal expression of c-Myc mRNA in HCC tissue was correlated
with vascular invasion, which suggested an important role of c-Myc
in HCC vasculogenesis.
The expression of Lin28 has been demonstrated to
promote oncogenesis and participate in the tumor progression of
cancer. Dangi-Garimella et al(41) proposed that upregulated Lin28
expression accelerated metastasis in breast cancer, as Lin28
repressed let-7 processing that in turn inhibited HMGA2, a
chromatin remodeling protein that activated pro-invasive and
pro-metastatic genes. Saiki et al(10) and Hamano et al(42) demonstrated that Lin28 expression was
closely correlated with cancer progression in colorectal and
oesophageal cancer. However, the current study did not yield
definitive findings with respect to the potential role of Lin28
genes in HCC carcinogenesis, aggressiveness and a poor prognosis.
The expression of Lin28 mRNA was slightly higher in HCC tissue than
in the adjacent tumor tissue (P=0.18). No correlation was observed
between Lin28 expression and HCC patient survival, post-operative
recurrence or prognosis.
In summary, the results of our comprehensive
analysis of six pluripotent stem cell genes in 57 HCC cases suggest
that upregulated expression of pluripotency genes in HCC cells may
be important in HCC carcinogenesis. Altered expression of Klf4,
Sox2, Oct4, Nanog and c-Myc genes indicates the abundance of cancer
stem-like cells (CSLCs) in primary tumors that may be key resources
for tumor progression and the poor prognosis of HCC. As there are
no relevant therapeutic strategies to specifically target cells
with a CSC-like phenotype in HCC, further investigations to
establish the appropriate adjuvant strategies are required.
Acknowledgements
This study was supported by a grant
from the Natural Science Foundation of China (No. 81201901) and the
National Clinical Key Special Subject of China.
References
|
1
|
Yang ZF, Ngai P, Ho DW, Yu WC, Ng MN, Lau
CK, Li ML, Tam KH, Lam CT, Poon RT and Fan ST: Identification of
local and circulating cancer stem cells in human liver cancer.
Hepatology. 47:919–928. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pardal R, Clarke MF and Morrison SJ:
Applying the principles of stem-cell biology to cancer. Nat Rev
Cancer. 3:895–902. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rossi DJ and Weissman IL: Pten,
tumorigenesis, and stem cell self-renewal. Cell. 125:229–231. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Widschwendter M, Fiegl H, Egle D,
Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M,
Young J, Jacobs I and Laird PW: Epigenetic stem cell signature in
cancer. Nat Genet. 39:157–158. 2007. View
Article : Google Scholar
|
|
5
|
Ben-Porath I, Thomson MW, Carey VJ, Ge R,
Bell GW, Regev A and Weinberg RA: An embryonic stem cell-like gene
expression signature in poorly differentiated aggressive human
tumors. Nat Genet. 40:499–507. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wong DJ, Liu H, Ridky TW, Cassarino D,
Segal E and Chang HY: Module map of stem cell genes guides creation
of epithelial cancer stem cells. Cell Stem Cell. 2:333–344. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Takahashi K and Yamanaka S: Induction of
pluripotent stem cells from mouse embryonic and adult firoblast
cultures by defined factors. Cell. 126:663–676. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu J, Vodyanik MA, Smuga-Otto K,
Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA,
Ruotti V, Stewart R, Slukvin II and Thomson JA: Induced pluripotent
stem cell lines derived from human somatic cells. Science.
318:1917–1920. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rodini CO, Suzuki DE, Saba-Silva N,
Cappellano A, de Souza JE, Cavalheiro S, Toledo SR and Okamoto OK:
Expression analysis of stem cell-related genes reveal OCT4 as a
predictor of poor clinical outcome in medulloblastoma. J
Neurooncol. 106:71–79. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Saiki Y, Ishimaru S, Mimori K, Takatsuno
Y, Nagahara M, Ishii H, Yamada K and Mori M: Comprehensive analysis
of the clinical significance of inducing pluripotent
stemness-related gene expression in colorectal cancer cells. Ann
Surg Oncol. 16:2638–2644. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun HC, Zhang W, Qin LX, Zhang BH, Ye QH,
Wang L, Ren N, Zhuang PY, Zhu XD, Fan J and Tang ZY: Positive serum
hepatitis B e antigen is associated with higher risk of early
recurrence and poorer survival in patients after curative resection
of hepatitis B-related hepatocellular carcinoma. J Hepatol.
47:684–690. 2007. View Article : Google Scholar
|
|
12
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang XR, Xu Y, Shi GM, Fan J, Zhou J, Ji
Y, Sun HC, Qiu SJ, Yu B, Gao Q, He YZ, et al: Cytokeratin 10 and
cytokeratin 19: predictive markers for poor prognosis in
hepatocellular carcinoma patients after curative resection. Clin
Cancer Res. 14:3850–3859. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun HC, Zhuang PY, Qin LX, Ye QH, Wang L,
Ren N, Zhang JB, Qian YB, Lu L, Fan J and Tang ZY: Incidence and
prognostic values of lymph node metastasis in operable
hepatocellular carcinoma and evaluation of routine complete
lymphadenectomy. J Surg Oncol. 96:37–45. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Raeside DE: Monte Carlo principles and
applications. Phys Med Biol. 21:181–197. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Evans PM and Liu C: Roles of Krüpel-like
factor 4 in normal homeostasis, cancer and stem cells. Acta Biochim
Biophys Sin. 40:554–564. 2008.
|
|
17
|
Zhou Y, Hofstetter WL, He Y, Hu W, Pataer
A, Wang L, Wang J, Zhou Y, Yu L, Fang B and Swisher SG: KLF4
inhibition of lung cancer cell invasion by suppression of SPARC
expression. Cancer Biol Ther. 9:507–513. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zammarchi F, Morelli M, Menicagli M, Di
Cristofano C, Zavaglia K, Paolucci A, Campani D, Aretini P, Boggi
U, Mosca F, Cavazzana A, et al: KLF4 is a novel candidate tumor
suppressor gene in pancreatic ductal carcinoma. Am J Pathol.
178:361–372. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pandya AY, Talley LI, Frost AR, Fitzgerald
TJ, Trivedi V, Chakravarthy M, Chhieng DC, Grizzle WE, Engler JA,
Krontiras H, Bland KI, LoBuglio AF, Lobo-Ruppert SM and Ruppert JM:
Nuclear localization of KLF4 is associated with an aggressive
phenotype in early-stage breast cancer. Clin Cancer Res.
10:2709–2719. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Foster KW, Liu Z, Nail CD, Li X,
Fitzgerald TJ, Bailey SK, Frost AR, Louro ID, Townes TM, Paterson
AJ, Kudlow JE, et al: Induction of KLF4 in basal keratinocytes
blocks the proliferation-differentiation switch and initiates
squamous epithelial dysplasia. Oncogene. 24:1491–1500. 2005.
View Article : Google Scholar
|
|
21
|
Chen YJ, Wu CY, Chang CC, Ma CJ, Li MC and
Chen CM: Nuclear Krüppel-like factor 4 expression is associated
with human skin squamous cell carcinoma progression and metastasis.
Cancer Biol Ther. 7:777–782. 2008.
|
|
22
|
Fong H, Hohenstein KA and Donovan PJ:
Regulation of self-renewal and pluripotency by Sox2 in human
embryonic stem cells. Stem Cells. 26:1931–1938. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hussenet T, Dali S, Exinger J, Monga B,
Jost B, Dembelé D, Martinet N, Thibault C, Huelsken J, Brambilla E
and du Manoir S: Sox2 is an oncogene activated by recurrent 3q26.3
amplifications in human lung squamous cell carcinomas. PLoS One.
5:e89602010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen Y, Shi L, Zhang L, Li R, Liang J, Yu
W, Sun L, Yang X, Wang Y, Zhang Y and Shang Y: The molecular
mechanism governing the oncogenic potential of Sox2 in breast
cancer. J Biol Chem. 283:17969–17978. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gangemi RM, Griffero F, Marubbi D, Perera
M, Capra MC, Malatesta P, Ravetti GL, Zona GL, Daga A and Corte G:
Sox2 silencing in glioblastoma tumor-initiating cells causes stop
of proliferation and loss of tumorigenicity. Stem Cells. 27:40–48.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sanada Y, Yoshida K, Ohara M, Oeda M,
Konishi K and Tsutani Y: Histopathologic evaluation of stepwise
progression of pancreatic carcinoma with immunohistochemical
analysis of gastric epithelial transcription factor Sox2:
comparison of expression patterns between invasive components and
cancerous or nonneoplastic intraductal components. Pancreas.
32:164–170. 2006.
|
|
27
|
Nichols J, Zevnik B, Anastassiadis K, Niwa
H, Klewe-Nebenius D, Chambers I, Schöler H and Smith A: Formation
of pluripotent stem cells in the mammalian embryo depends on the
POU transcription factor Oct4. Cell. 95:379–391. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chambers I, Colby D, Robertson M, Nichols
J, Lee S, Tweedie S and Smith A: Functional expression cloning of
Nanog, a pluripotency sustaining factor in embryonic stem cells.
Cell. 113:643–655. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chang CC: Recent translational research:
stem cells as the roots of breast cancer. Breast Cancer Res.
8:1032006. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Du Z, Jia D, Liu S, Wang F, Li G, Zhang Y,
Cao X, Ling EA and Hao A: Oct4 is expressed in human gliomas and
promotes colony formation in glioma cells. Glia. 57:724–733. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou X, Zhou YP, Huang GR, Gong BL, Yang
B, Zhang DX, Hu P and Xu SR: Expression of the stem cell marker,
Nanog, in human endometrial adenocarcinoma. Int J Gynecol Pathol.
30:262–270. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lin T, Ding YQ and Li JM: Overexpression
of Nanog protein is associated with poor prognosis in gastric
adenocarcinoma. Med Oncol. 29:878–885. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Meng HM, Zheng P, Wang XY, Liu C, Sui HM,
Wu SJ, Zhou J, Ding YQ and Li JM: Overexpression of Nanog predicts
tumor progression and poor prognosis in colorectal cancer. Cancer
Biol Ther. 9:295–302. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wen J, Park JY, Park KH, Chung HW, Bang S,
Park SW and Song SY: Oct4 and Nanog expression is associated with
early stages of pancreatic carcinogenesis. Pancreas. 39:622–626.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chiou SH, Yu CC, Huang CY, Lin SC, Liu CJ,
Tsai TH, Chou SH, Chien CS, Ku HH and Lo JF: Positive correlations
of Oct-4 and Nanog in oral cancer stem-like cells and high-grade
oral squamous cell carcinoma. Clin Cancer Res. 14:4085–4095. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Grandori C, Cowley SM, James LP and
Eisenman RN: The Myc/Max/Mad network and the transcriptional
control of cell behavior. Annu Rev Cell Dev Biol. 16:653–699. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ma L, Young J, Prabhala H, Pan E, Mestdagh
P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S,
Westermann F, et al: miR-9, a MYC/MYCN-activated microRNA,
regulates E-cadherin and cancer metastasis. Nat Cell Biol.
12:247–256. 2010.PubMed/NCBI
|
|
38
|
Rapp UR, Korn C, Ceteci F, Karreman C,
Luetkenhaus K, Serafin V, Zanucco E, Castro I and Potapenko T: MYC
is a metastasis gene for non-small-cell lung cancer. PLoS One.
4:e60292009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dominguez-Sola D, Ying CY, Grandori C,
Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J and
Dalla-Favera R: Non-transcriptional control of DNA replication by
c-Myc. Nature. 448:445–451. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Soucek L, Lawlor ER, Soto D, Shchors K,
Swigart LB and Evan GI: Mast cells are required for angiogenesis
and macroscopic expansion of Myc-induced pancreatic islet tumors.
Nat Med. 13:1211–1218. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dangi-Garimella S, Yun J, Eves EM, Newman
M, Erkeland SJ, Hammond SM, Minn AJ and Rosner MR: Raf kinase
inhibitory protein suppresses a metastasis signalling cascade
involving LIN28 and let-7. EMBO J. 28:347–358. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hamano R, Miyata H, Yamasaki M, Sugimura
K, Tanaka K, Kurokawa Y, Nakajima K, Takiguchi S, Fujiwara Y, Mori
M and Doki Y: High expression of Lin28 is associated with tumour
aggressiveness and poor prognosis of patients in oesophagus cancer.
Br J Cancer. 106:1415–1423. 2012. View Article : Google Scholar : PubMed/NCBI
|