Introduction
Breast cancer is the most common malignancy with a
high mortality in females worldwide. Tumor suppressor genes are
important for maintaining genome integrity and the cell cycle.
PTEN was the first phosphatase to be identified as a tumor
suppressor with diverse functions, including regulation of cell
cycle, apoptosis and metastasis (1–4).
Mutations or a reduced expression of the PTEN gene are
associated with a wide variety of human tumors (5). Germline mutations in PTEN are
known to cause Cowden syndrome (CS), which is characterized by a
high risk of breast cancer. In families with CS, ∼80% have
PTEN germline mutations and female CS patients have a 25–50%
lifetime risk of developing breast cancer (6). In sporadic breast carcinomas, the
frequency of PTEN loss is 30–40% and the somatic intragenic
PTEN mutation frequency is <5%. Besides genetic change,
aberrant DNA methylation is also responsible for the epigenetic
silencing of genes associated with tumor genesis and progression of
cancer. In the present study, the frequency of PTEN
mutations, the methylation of PTEN and its association with
the loss of PTEN expression were further investigated in
sporadic breast carcinoma in a Chinese population.
Material and methods
Patients and tissue samples
A total of 146 female Chinese patients, who were
diagnosed with breast cancer between 2003 and 2006, were included
in the present study. Clinical and pathological information,
including age, ethnicity, menopausal status, type of tumor, disease
stage, axillary lymph nodes, tumor size and biomarkers, were
collected. Paraffin blocks of tumor samples from all 146 patients
and fresh frozen tumor specimens from 45 patients were prepared. In
addition, 10 normal tissues adjacent to the tumors were also
collected. All patients were followed up until December 2009. This
study was approved by the ethics committee of the General Hospital
of Beijing Military Area. Written informed consent was obtained
from all patients.
Sample preparation
Tissue sections (thickness, 4–5 μm) were cut
from the paraffin blocks for the detection of PTEN
expression in the breast carcinoma or normal breast tissues.
Genomic DNA was isolated from frozen specimens using a NucleoSpin
Tissue kit (Clontech, Mountain View, CA, USA) according to the
manufacturer’s instructions. DNA samples were frozen at −70°C until
use.
Immunohistochemistry
Following deparaffinization and dehydration in
hydrogen peroxide, the sections were incubated at 37°C for 1 h with
anti-PTEN mouse monoclonal antibody (1:25 dilution). Corresponding
biotinylated anti-IgG was added and incubated for 30 min at 37°C.
Next, the sections were incubated with 3,3′-diaminobenzidine (DAB)
substrate chromogen solution and counterstained with hematoxylin.
Negative controls were incubated with phosphate-buffered saline
(PBS) instead of primary antibody. Known positive tissues were used
as positive controls. Immunohistochemical reactivity was graded
according to the percentage of positive tumor cells: −, 0; +,
<20; ++, 20–50; and +++, >50%. Grades − or + were considered
as low level expression and grades ++ or +++ were considered as
high level expression.
Analysis of PTEN gene mutation
Polymerase chain reaction-single strand conformation
polymorphism (PCR-SSCP) analysis was performed to analyze mutations
of the PTEN gene. The PTEN gene coding region was
amplified from genomic DNA by PCR. Due to the low sensitivity of
SSCP to detect sequences of >300 bp, exons 5, 8 and 9 were
amplified separately. The oligonucleotide primer pairs located in
exons 1–9 of the PTEN gene are listed in Table I. Following denaturation at 99°C for
10 min, the PCR products were chilled on ice, followed by
electrophoresis on a 8% polyacrylamide gel for 12 h at 40 W. The
gel was silver stained and the PCR products with aberrant bands or
mobility shift were retrieved and sequenced directly.
 | Table I.PCR primers designed for 9 PTEN exons
and MSP of PTEN |
Table I.
PCR primers designed for 9 PTEN exons
and MSP of PTEN
| Primer | Sense (5′-3′) | Antisense
(5′-3′) | bp |
|---|
| Exon | | | |
| 1 |
TTCTGCCATCTCTCTCCTCC |
ATCCGTCTACTCCCACGTTC | 194 |
| 2 |
GTTTGATTGCTGCATATTTCA |
TCTAAATGAAAACACAACATGAA | 201 |
| 3 |
AGCTCATTTTTGTTAATGGTGG |
CCTCACTCTAACAAGCAGATAACTTTC | 178 |
| 4 |
AAAGATTCAGGCAATGTTTGTTAG |
TGACAGTAAGATACAGTCTATCGGG | 200 |
| 5-1 |
TTTTTTCTTATTCTGAGGTTATC |
TCATTACACCAGTTCGTCC | 184 |
| 5-2 |
TCATGTTGCAGCAATTCAC |
GAAGAGGAAAGGAAAAACATC | 176 |
| 6 |
ATGGCTACGACCCAGTTACC |
AAGAAAACTGTTCCAATACATGG | 284 |
| 7 |
CAGTTAAAGGCATTTCCTGTG |
GCTTTTAATCTGTCCTTATTTTGG | 274 |
| 8-1 |
TTAACATAGGTGACAGATTTTC |
CACGCTCTATACTGCAAATG | 222 |
| 8-2 |
CATTCTTCATACCAGGACCAG |
TGGAGAAAAGTATCGGTTGG | 188 |
| 8-3 |
GCATTTGCAGTATAGAGCGTG |
TCAAGCAAGTTCTTCATCAGC | 217 |
| 9-1 |
AGATGAGTCATATTTGTGGG |
CTCTGGATCAGAGTCAGTGG | 185 |
| 9-2 |
AATCCAGAGGCTAGCAGTTC |
AAGGTCCATTTTCAGTTTATTC | 213 |
| MSP | | | |
| Methylated |
TTCGTTCGTCGTCGTATTT |
GCCGCTTAACTCTAAACCGCAACCG | 206 |
| Unmethylated |
GTGTTGGTGGAGGTAGTTGTTT |
ACCACTTAACTCTAAACCACAACCA | 162 |
Analysis of DNA methylation
Methylation of the PTEN promoter was assessed
by bisulfite treatment. This lead to chemical conversion of any
unmethylated cytosine to uracil, while the methylated cytosine
remained unmodified. As described previously (7), methylation specific PCR (MSP) using 2
primer pairs was designed to distinguish methylated DNA from
unmethylated DNA.
DNA modification by bisulfite
treatment
The bisulfite conversion was performed using 1
μg DNA. Briefly, the DNA was denatured by incubation with 10
μl NaOH (1 M) for 10 min at 37°C. The samples were then
treated with sodium bisulfite (3 M) and hydroquinone (10 mM) for 16
h at 55°C with salmon sperm DNA as a supporter. Modified DNA
samples were purified using the Wizard DNA Purification kit
(Promega, Madison, WI, USA) according to the manufacturer’s
instructions. NaOH (1 M) was added and incubated for 7 min at room
temperature to terminate the modification. The DNA was ethanol
precipitated and dissolved in double distilled water for PCR.
MSP
Two primer sets were used to amplify the promoter
region of the PTEN gene, which incorporated a number of CpG
sites, one specific for the methylated sequence (M, sense:
5′-TTCGTTCGTCGTCGTATTT-3′; antisense:
5′-GCCGCTTAACTCTAAACCGCAACCG-3′; PCR product, 206 bp) and the other
for the unmethylated sequence (U, sense:
5′-TGTTGGTGGAGGTAGTTGTTT-3′; antisense: 5′-ACCACT TA ACTCTA A
ACCACA ACCA-3′; PCR product, 162 bp) (7). The primers used in the present study
specifically detect the promoter sequence of the PTEN gene
rather than that of the PTEN pseudogene. The PCR volume (50
μl) included 200 ng modified DNA, 20 pmol of each primer,
1.5 mmol/l MgCl2, 5 μl PCR intensifier and 2.5 U
HotStart Taq. The PCR parameters consisted of 64°C for 10 cycles,
62°C for 15 cycles and 60°C for 10 cycles, for 60 sec at each
temperature. Each MSP was repeated at least 3 times.
Statistical analysis
All comparisons between categorical variables were
examined by the Fisher’s exact chi-squared test. Association
analysis was performed with the Spearman’s rank correlation.
Relapse-free survival was calculated using the Kaplan-Meier
survival estimates and the log-rank test from the date of diagnosis
until the last contact or relapse. A Cox regression analysis was
performed to estimate the relative risks (with 95% confidence
intervals). P<0.05 was considered to indicate a statistically
significant difference. Statistical analyses were conducted using
SPSS 13.0 for Windows (SPSS, Inc., Chicago, IL, USA).
Results
PTEN expression in breast cancer
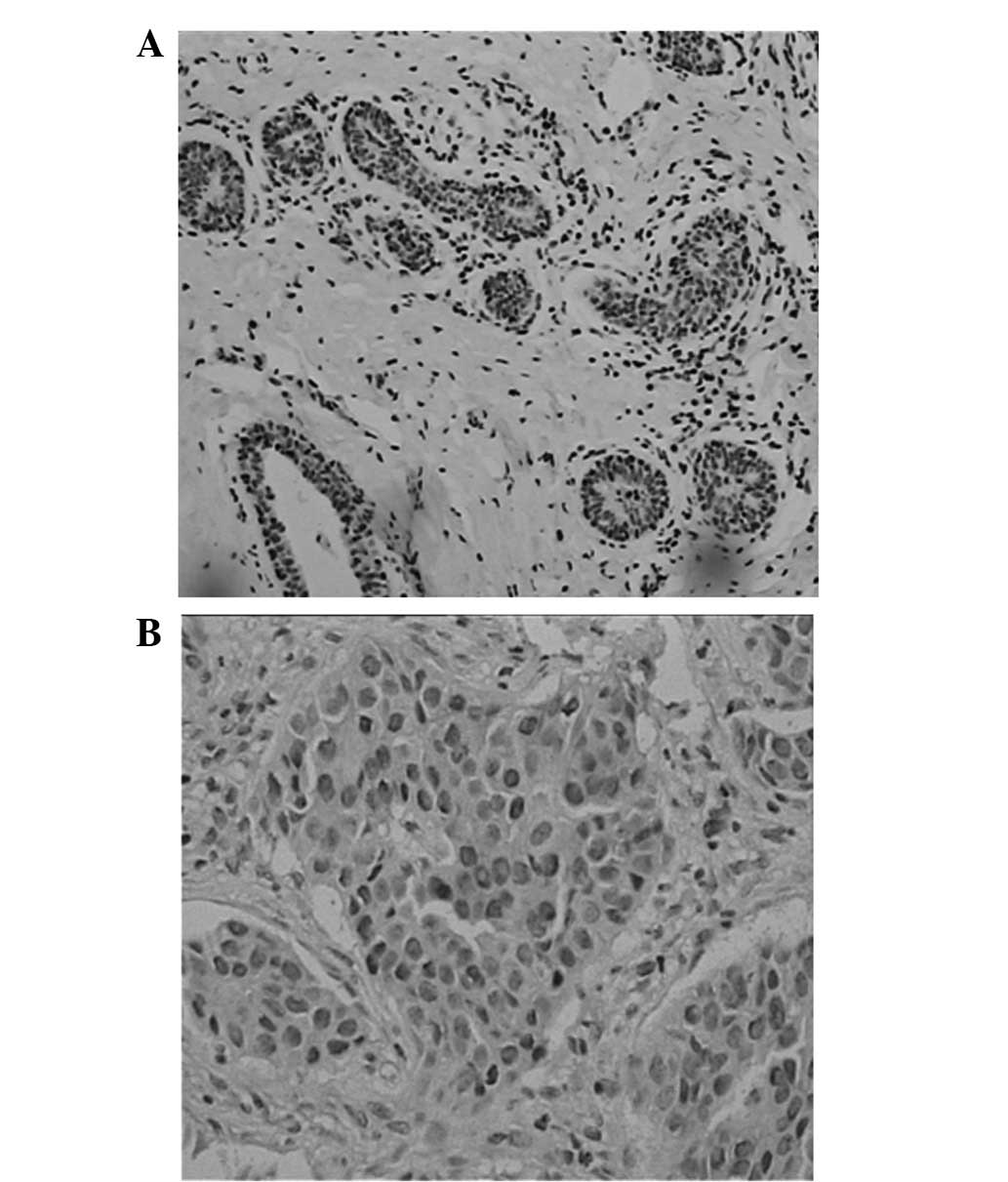
PTEN was markedly expressed in the cytoplasm and
nuclei of the breast cancer cells and in the normal duct epithelial
cells (Fig. 1). PTEN-positive cells
were diffusely distributed in the carcinoma. The positive
expression rate of PTEN was 57.5% (84/146) in the breast cancer
patients, but 100% in the normal breast tissues closely adjacent to
the carcinoma (Table II).
 | Table II.Correlation between PTEN expression
and breast cancer pathological characteristics. |
Table II.
Correlation between PTEN expression
and breast cancer pathological characteristics.
| Factors | n | PTEN
| P-value |
|---|
−
| +
| ++
| +++
|
|---|
| n | % | n | % | n | % | n | % |
|---|
| Age (years) | 146 | | | | | | | | | |
| <35 | | 14 | 56.0 | 5 | 20.0 | 5 | 20.0 | 1 | 4.0 | 0.150 |
| 35–55 | | 38 | 40.0 | 27 | 28.4 | 18 | 18.9 | 12 | 12.6 | |
| >55 | | 10 | 38.5 | 4 | 15.4 | 11 | 42.3 | 1 | 3.8 | |
| Menopause | 146 | | | | | | | | | |
| No | | 46 | 47.4 | 22 | 22.7 | 19 | 19.6 | 10 | 10.3 | 0.162 |
| Yes | | 16 | 32.7 | 14 | 28.6 | 15 | 30.6 | 4 | 8.2 | |
| T-stage | 146 | | | | | | | | | |
| T1 | | 8 | 22.2 | 11 | 30.6 | 10 | 27.8 | 7 | 19.4 | 0.002 |
| T2 | | 34 | 47.2 | 15 | 20.8 | 17 | 23.6 | 6 | 8.3 | |
| T3 | | 12 | 52.2 | 5 | 21.7 | 5 | 21.7 | 1 | 4.3 | |
| T4 | | 8 | 53.3 | 5 | 33.3 | 2 | 13.3 | 0 | 0.0 | |
| Lymph node
metastasis | 146 | | | | | | | | | |
| No | | 18 | 38.3 | 11 | 23.4 | 12 | 25.5 | 6 | 12.8 | 0.630 |
| Yes | | 45 | 45.5 | 25 | 25.3 | 22 | 22.2 | 7 | 7.1 | |
| Stage | 146 | | | | | | | | | |
| I | | 3 | 20.0 | 4 | 26.7 | 4 | 26.7 | 4 | 26.7 | 0.005 |
| II | | 25 | 39.1 | 16 | 25.0 | 16 | 25.0 | 7 | 10.9 | |
| III | | 28 | 47.5 | 15 | 25.4 | 13 | 22.0 | 3 | 5.1 | |
| IV | | 6 | 75.0 | 1 | 12.5 | 1 | 12.5 | 0 | 0.0 | |
The correlation between PTEN expression and the
clinicopathological parameters, including age, disease stage, lymph
node status, tumor grade, size and expression of ER, PR and
Her-2/neu, was analyzed. The results revealed a negative
correlation between PTEN expression and the tumor size or stage.
However, no correlation was observed between PTEN expression and
age, menopause or the presence of lymph node metastasis (Table II).
PTEN expression and tumor
immunophenotype
The correlation between PTEN expression and tumor
immunophenotype was also analyzed and the results demonstrated that
there was no correlation between PTEN expression and Her-2/neu
(P=0.865). However, there was a positive correlation between the
expression of PTEN and ER (P=0.023) or PR (P=0.038; Table III).
 | Table III.PTEN expression and tumor
immunophenotype (Spearmen’s rank correlation) |
Table III.
PTEN expression and tumor
immunophenotype (Spearmen’s rank correlation)
| Gene | PTEN
| χ2 | P-value |
|---|
−
| +
| ++
| +++
|
|---|
| n | % | n | % | n | % | n | % |
|---|
| ER | | | | | | | | | | |
| − | 21 | 42.9 | 16 | 32.7 | 12 | 24.5 | 0 | 0 | 2.303 | 0.023 |
| + | 32 | 48.5 | 15 | 22.7 | 12 | 18.2 | 7 | 10.6 | | |
| ++ | 7 | 35.0 | 3 | 15.0 | 6 | 30.0 | 4 | 20.0 | | |
| +++ | 2 | 18.2 | 2 | 18.2 | 4 | 36.4 | 3 | 27.3 | | |
| PR | | | | | | | | | | |
| − | 29 | 48.3 | 15 | 25.0 | 11 | 18.3 | 5 | 8.3 | 2.091 | 0.038 |
| + | 26 | 41.9 | 17 | 27.4 | 15 | 24.2 | 4 | 6.5 | | |
| ++ | 7 | 36.8 | 4 | 21.1 | 5 | 26.3 | 3 | 15.8 | | |
| +++ | 0 | 0.0 | 0 | 0.0 | 3 | 60.0 | 2 | 40.0 | | |
| HER-2 | | | | | | | | | | |
| − | 34 | 46.6 | 15 | 20.5 | 16 | 21.9 | 8 | 11.0 | 0.081 | 0.865 |
| + | 11 | 32.4 | 8 | 23.5 | 9 | 26.5 | 6 | 17.6 | | |
| ++ | 10 | 37.0 | 12 | 44.4 | 5 | 18.5 | 0 | 0.0 | | |
| +++ | 7 | 58.3 | 1 | 8.3 | 4 | 33.3 | 0 | 0.0 | | |
PTEN expression and relapse-free
survival
Breast cancer patients (98/101) were followed up for
40–266 months (median, 103 months; 3 patients were missed). A
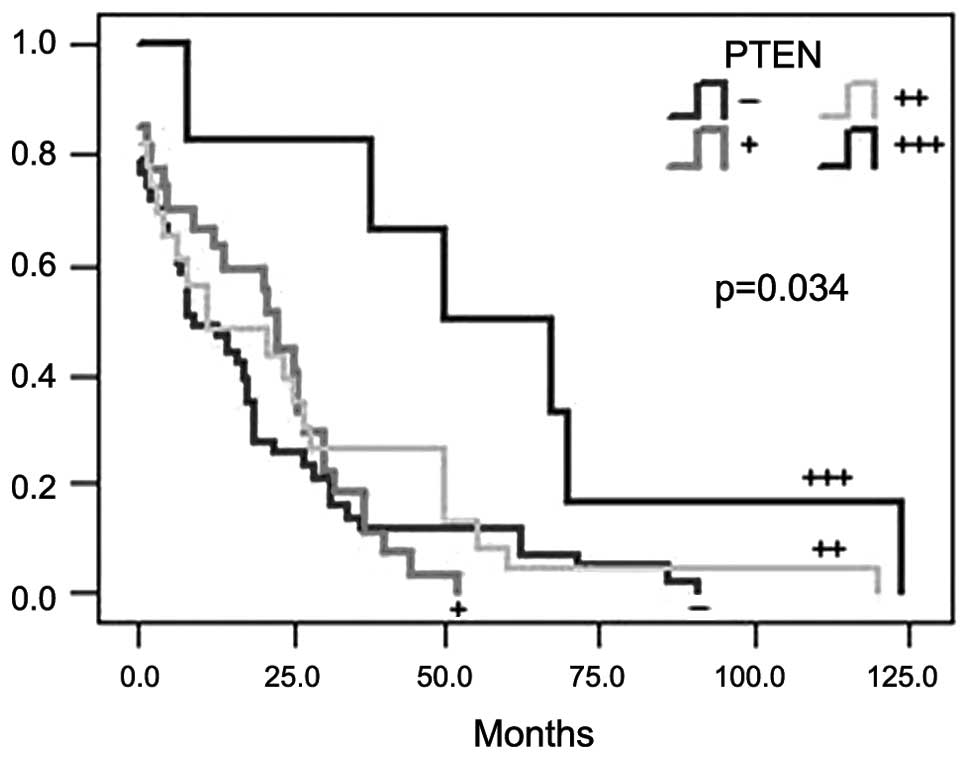
univariate analysis revealed that the high expression of PTEN was
significantly associated with a longer relapse-free survival
(χ2=7.965; P=0.034; Fig.
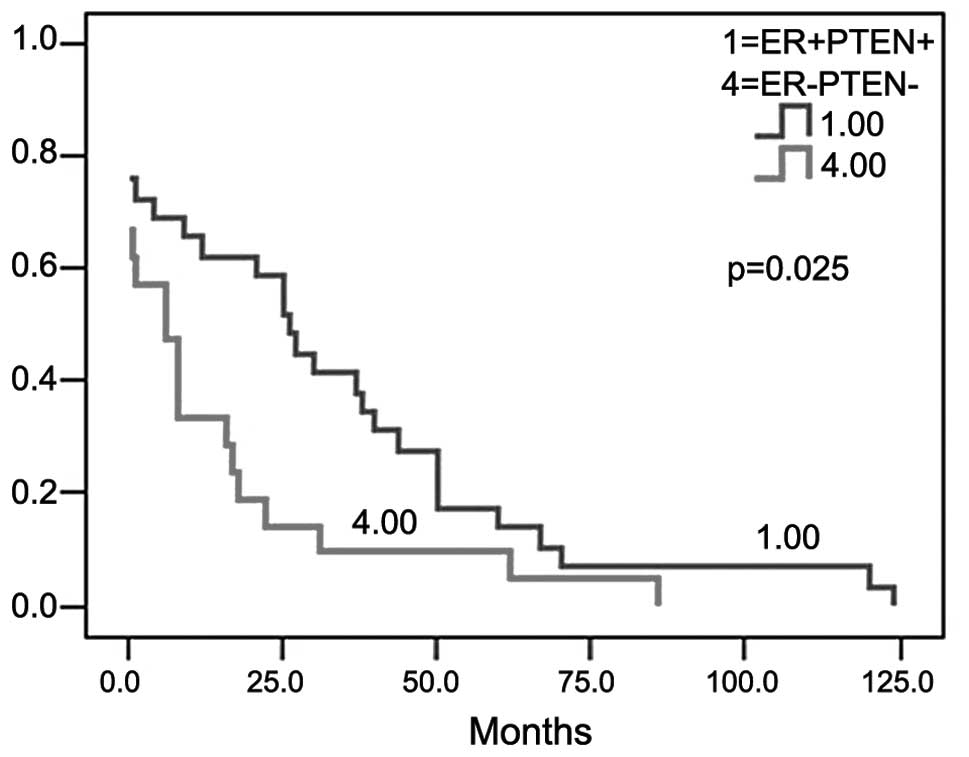
2). The longest relapse-free survival occurred in patients that
were ER- and PTEN-positive (χ2=5.044; P=0.025; Fig. 3). A Cox regression analysis
demonstrated that as the prognostic factors, including menopausal
status (r= 0.779; P=0.010), tumor size (r=0.836; P=0.000), lymph
nodes status (r=0.806; P=0.010), adjuvant therapy (r=−0.796;
P=0.016) and expression of ER (r=−0.793; P=0.018) and HER-2
(r=1.141; P= 0.002), were introduced into the equation, there was
no correlation between the expression of PTEN and relapse-free
survival (Table IV).
 | Table IV.Cox regression analysis estimated
prognostic value by hazard ratios and 95% CI for relapse-free
survival |
Table IV.
Cox regression analysis estimated
prognostic value by hazard ratios and 95% CI for relapse-free
survival
| Variable | B | SE | Wald | P-value | Exp (B) | 95% CI for Exp (B)
|
|---|
| Lower | Upper |
|---|
| Age | 0.190 | 0.348 | 0.298 | 0.585 | 1.209 | 0.611 | 2.393 |
| Menopause | 0.779 | 0.303 | 6.596 | 0.010 | 2.180 | 1.203 | 3.951 |
| Tumor size | 0.836 | 0.202 | 17.077 | 0.000 | 2.306 | 1.552 | 3.428 |
| Lymph node | 0.806 | 0.312 | 6.673 | 0.010 | 2.238 | 1.215 | 4.125 |
| Adjunct
therapy | −0.796 | 0.332 | 5.757 | 0.016 | 0.451 | 0.235 | 0.864 |
| ER | −0.793 | 0.334 | 5.636 | 0.018 | 0.453 | 0.235 | 0.871 |
| PR | 0.440 | 0.322 | 1.861 | 0.173 | 1.552 | 0.825 | 2.918 |
| HER | −1.141 | 0.373 | 9.328 | 0.002 | 0.320 | 0.154 | 0.665 |
| PTEN | −0.241 | 0.142 | 2.893 | 0.089 | 0.786 | 0.595 | 1.037 |
PTEN mutation in breast cancer
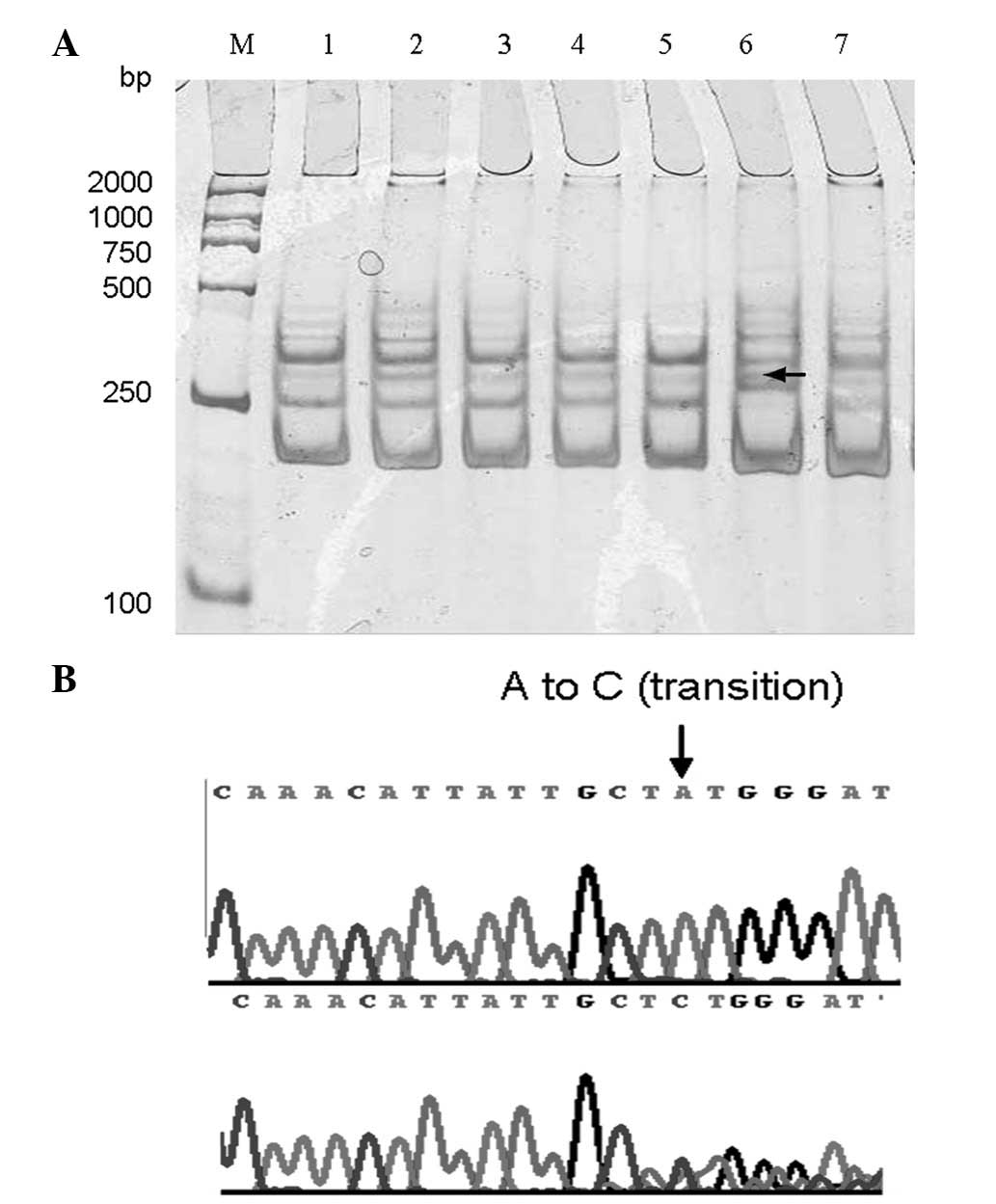
SSCP analysis identified no abnormal single-strand
conformation in the 10 normal tissues adjacent to the tumor. Among
the 45 fresh breast carcinoma samples, 3 band shifts were
identified by PCR-SSCP (Fig. 4).
However, only 1 mutation was confirmed in exon 2 by sequencing. The
PTEN mutation rate was 2.2% (1/45). Sequencing analysis
revealed that a codon 24 A→C missense mutation in exon 2 resulted
in a codogenic amino acid change from methionine to leucine
(Fig. 4).
PTEN promoter methylation in breast
cancer and its correlation with clinical manifestations
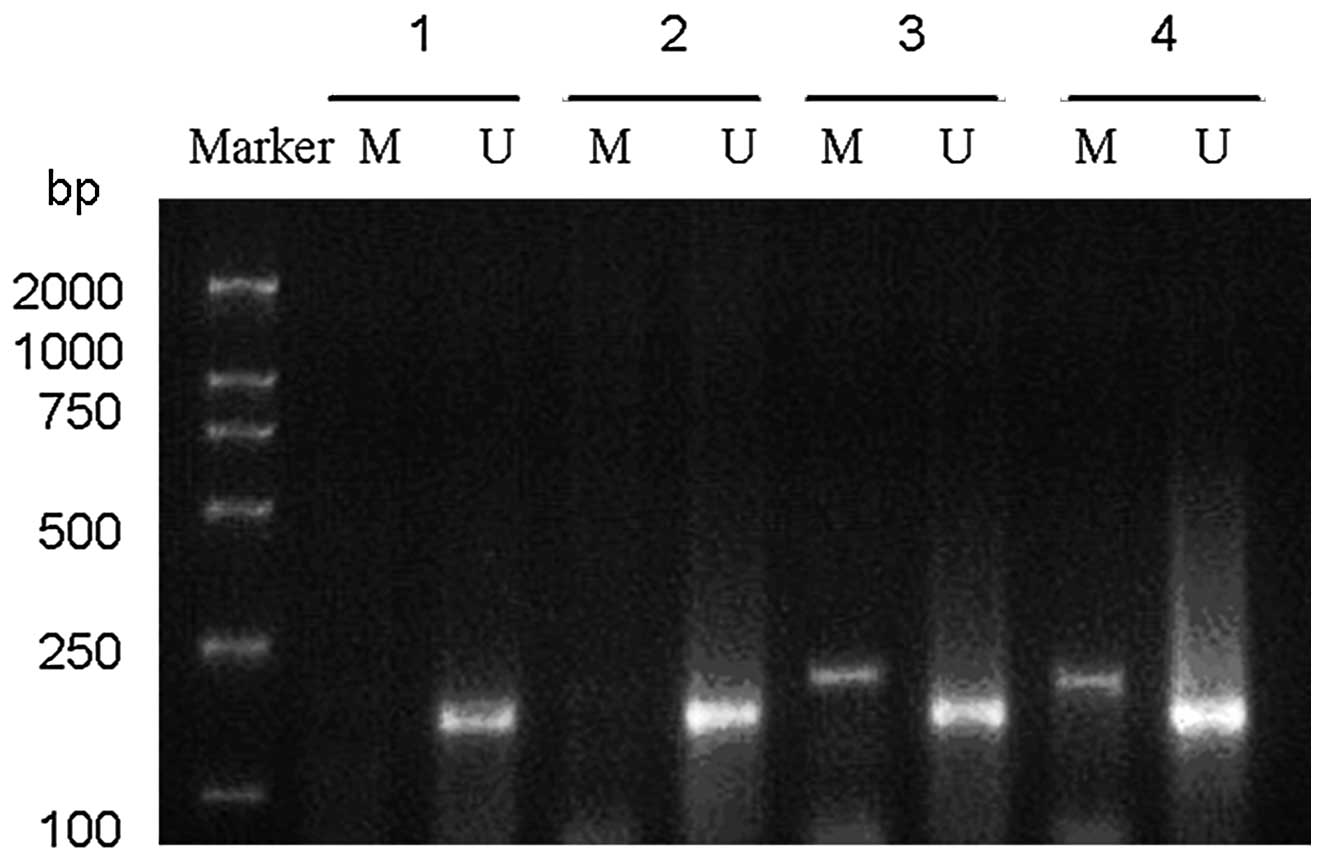
The results of the MSP analysis of PTEN
promoter methylation in the 45 breast cancer patients are
summarized in Table V and presented
in Fig. 5.
 | Table V.Clinicopathological features, PTEN
expression and promoter methylation in breast carcinoma. |
Table V.
Clinicopathological features, PTEN
expression and promoter methylation in breast carcinoma.
| Groups | Total cases
(n) | Methylation of PTEN
| χ2 | P-value |
|---|
| n | % |
|---|
| PTEN
expression | | | | | |
| − | 18 | 9 | 50.0 | 4.994 | 0.025 |
| + | 27 | 5 | 18.5 | | |
| Lymph node | | | | | |
| − | 18 | 5 | 27.8 | 0.733 | 0.392 |
| + | 27 | 9 | 33.3 | | |
| Stage | | | | | |
| I | 7 | 1 | 14.3 | 1.425 | 0.490 |
| II | 21 | 8 | 38.1 | | |
| III | 17 | 5 | 29.4 | | |
| T stage | | | | | |
| T1 | 15 | 3 | 20.0 | 2.118 | 0.347 |
| T2 | 24 | 10 | 41.7 | | |
| T3 | 6 | 1 | 16.7 | | |
| ER | | | | | |
| − | 25 | 6 | 24.0 | 0.249 | 1.327 |
| + | 20 | 8 | 40.0 | | |
| HER-2
expression | | | | | |
| Low | 33 | 11 | 33.3 | 0.285 | 0.593 |
| High | 12 | 3 | 25.0 | | |
The MSP analysis demonstrated that PTEN
promoter methylation was detected in 14 breast cancer cases with a
methylation rate of 31.1% (14/45; Table
V and Fig. 5). Among the cases
with PTEN promoter methylations, 64.3% (9/14) of the
patients lost PTEN expression. The occurrence of PTEN
promoter methylation in the patients with negative PTEN
expression was significantly higher that in the
PTEN-positive cases (χ2=4.994; P=0.025; Table V and Fig. 5).
Further clinicopathological analysis revealed that
among the 14 cases with positive PTEN promoter methylation,
7.1% (1/14) were stage I breast cancer, 64.3% (9/14) had axillary
lymph node metastasis, 42.9% (6/14) were ER-negative and 21.4%
(3/14) overexpressed HER-2. However, these cases were not found to
be significantly different compared with the PTEN
methylation negative group (P>0.05).
Discussion
PTEN was the first recognized tumor
suppressor with lipid phosphatase activity. A previous study
(3) demonstrated that PTEN
is mutated or inactivated in a number of malignant tumors,
including neuroglioma, endometrial, prostate, breast, thyroid and
liver cancer. Mutation rates range between 40 and 80% in prostatic
cancer, endometrial carcinoma and advanced neuroglioma, indicating
that the PTEN mutation is important for tumorigenesis and
cancer development (3). The
majority of PTEN mutations in tumors are localized to exons
5, 7 or 8 (5,7,8). In
CS, ∼40% of PTEN mutations are located in the exon 5
phosphatase-coding domain, leading to a reduction in its tumor
suppressor activity.
In breast cancer, a 10–40% loss of heterozygosity
(LOH) has been identified at the chromosome 10q23 region that
contains the PTEN gene (6,8,9). LOH
at the 10q23 region is functionally associated with the occurrence
of breast cancer and this region has also been found to
significantly correlate with tumor characteristics. In the study by
Garcia et al, the breast cancer patients with LOH were
younger, exhibiting a high incidence of lymph node metastases and a
high histological grade (10). The
loss of 10q23 is markedly associated with tumor progression
(9). LOH was not observed in pure
intraductal carcinomas, while it was observed in 40% (17/42) of the
invasive carcinomas. LOH at 10q23 was found to correlate with the
loss of ER (10).
Although LOH at 10q23 is frequent, the somatic
PTEN mutation rate is <5% (3) and the majority of PTEN gene
mutations occur in advanced and metastatic breast cancer. The
association between PTEN and breast cancer is controversial
and it has been hypothesized that another new gene is located in
the 10q23 region. Further studies have indicated that a reduced
PTEN expression rate occurs in 83% of breast cancer cases
(11). The frequency of reduced
PTEN expression in invasive cancer was higher than that in the
carcinoma in situ (12). It
was demonstrated that reduced PTEN expression was correlated with a
loss of ER expression, tumor size and lymph node metastasis.
Therefore, PTEN may represent an important prognostic factor.
In the present study, the expression rate of PTEN in
breast carcinoma (57.5%, 84/146) was found to be significantly
lower than that of normal breast tissues adjacent to tumors (100%,
10/10; P<0.05). In addition, PTEN expression was found to
significantly correlate with tumor size or stage, indicating that
the reduced expression of PTEN was associated with a larger tumor
size or advanced stages. In addition, PTEN expression was found to
correlate with ER or PR. A survival analysis revealed that the
2-year relapse-free survival rate of breast cancer patients with
PTEN expression grade +++ was higher than those with lower grades
of PTEN expression. The lowest rate of breast cancer relapse-free
survival occurred in ER- and PTEN-negative patients. Consistent
with studies by Perren et al (11) and Depowski et al (13), PTEN did not enter the equation of
the COX regression analysis in the present study. By contrast,
other studies have reported that PTEN is an independent prognostic
factor (14,15).
PCR-SSCP and sequencing analysis detected only 1
mutation in 45 breast cancer patients and this point mutation was a
missense mutation in exon 2. The patient with the PTEN
mutation had a high risk of recurrence, infiltrating duct
carcinoma, a tumor size of 1.2×2.0×2.0 cm and axillary lymph node
metastasis (4/10). In addition, this patient was at the
pathological stage of pT1N2M0, IIIA, ER -, PR - and Her2 ++. In the
present study, the loss of PTEN expression was observed in 42.5%
(62/146) of patients, while the mutation rate was only 2.2% (1/45),
indicating that mechanisms other than mutation caused the loss of
PTEN expression.
Shoman et al (15) previously demonstrated that reduced
PTEN expression was associated with the shorter relapse-free
survival of 100 tamoxifen-treated ER-positive breast cancer
patients. When stage I patients were analyzed separately, reduced
PTEN expression was a strong predictor of shorter relapse-free and
disease-specific survival. When patients were stratified by levels
of ER expression (≥50% vs. <50% positive cells), reduced PTEN
expression was associated with a less favorable outcome in each
patient group. Among the tumor patients with a normal expression of
PTEN, 30% relapsed and 25% succumbed to their condition. By
contrast, among the tumor patients with a reduced expression of
PTEN, 90% relapsed and 65% succumbed to their condition. However,
in this earlier study, ER-negative breast cancer cases and patients
not treated with tamoxifen were not included. This study and the
results of the present study indicate a correlation between PTEN
and ER, and the combination of PTEN and ER may predict an outcome
for breast cancer patients.
Estrogen-induced proliferation of mammary and
uterine epithelial cells is primarily mediated by ER via
estrogen-independent (AF-1) and estrogen-dependent (AF-2)
activation domains. These domains regulate gene transcription by
recruiting co-activators and interacting with the general
transcriptional machinery. In vitro studies have
demonstrated that phosphatidylinositol 3-kinase (PI3K) and AKT
activate ER in the absence of estrogen (16). PI3K was shown to increase the
activities of the AF-1 and AF-2 domains of ER, while AKT increased
the activity of the AF-1 domain only. PTEN and a catalytically
inactive AKT are able to downregulate PI3K-induced AF-1 activity,
indicating that PI3K activates ER via AKT-dependent and
-independent pathways. It has been demonstrated that the activation
of the PI3K/AKT survival pathways and the hormone-independent
activation of ER are associated with the inhibition of
tamoxifen-induced apoptotic regression. AKT protects breast cancer
cells from tamoxifen-induced apoptosis. The downregulation of PTEN
expression results in a loss of the inhibition of PI3K/AKT. By
contrast, the high expression of PTEN improves the response of
breast cancer to tamoxifen. Therefore, breast cancer patients with
a positive expression of PTEN and ER are associated with longer
survival (16).
In addition to gene mutation, epigenetic regulation,
including promoter hypermethylation, has been demonstrated to alter
tumor suppressor gene expression and contribute to tumorigenesis.
Thus, the methylation status of the PTEN promoter CpG island
was analyzed in the present study. As one of the most recurrent
gene alterations, DNA methylation significantly affects chromosomal
formation, gene expression and DNA replication (17,18).
CpG islands in the promoter or nearby regions are frequently
methylated, leading to the silencing of gene transcription.
Methylation of a growing number of tumor suppressor genes,
including p16, APC, MLH1 and BRCA1, has been revealed to be one of
the most frequent mechanisms of gene transcription inactivation and
loss of gene function. Soria et al (19) analyzed the methylation of the
PTEN promoter and the PTEN expression in 30 cases of
non-small cell lung cancer (NSCLC). The results indicated that
PTEN methylation occurred in 35% (7/20) of NSCLC cases and
69% (11/16) of NSCLC cell lines. In addition, PTEN
methylation was not detected in patients with positive PTEN
expression. Thus, the loss of PTEN expression was reported to
correlate with the methylation of its promoter. PTEN
methylation was considered as one of main causes of loss of PTEN
expression. In the present study, PTEN methylation was
detected in 31.1% (14/45) of breast cancer cases. Among these, 14
tissues with PTEN methylation, 64.3% (9/14), exhibited a
loss of PTEN expression. The PTEN promoter methylation rate
was 50% (9/18) in the PTEN-negative cases, which is statistically
different from the rate in the PTEN-positive cases (18.5%, 5/27;
P=0.025). Therefore, none of the normal breast tissues adjacent to
the tumor were found to exhibit methylated PTEN promoters.
These results are consistent with a previous study (20) demonstrating that PTEN
methylation occurred in 34% of cases of breast invasive ductal
cancer and that 60% of these were found to exhibit a loss of PTEN
expression. In addition, none of the breast cancer cell lines and
normal breast tissues were observed to have PTEN
methylation. These results indicated that the methylation of the
PTEN promoter may result in PTEN inactivation in
breast cancer.
Previous studies have demonstrated that the
PI3K/AKT/PTEN pathway is important for tumorigenesis. The PI3K
signaling pathway is associated with almost all aspects of tumor
biology, including cell transformation, growth, proliferation,
migration and apoptosis evasion and genomic instability,
angiogenesis, metastasis and cancer stem cell maintenance (21,22).
PTEN degrades the product PI3K by dephosphorylating
phosphatidylinositol 3,4,5-trisphosphate and phosphatidylinositol
3,4-bisphosphate at the 3’ position (23). The loss of function or reduced
expression of PTEN leads to the accumulation of critical messenger
lipids, which in turn increases AKT phosphorylation and activity,
leading to decreased apoptosis and/or increased mitogen signaling
(24–28). However, although the PI3K/AKT/PTEN
pathway contains a number of attractive therapeutic targets,
clinical trials of pathway-targeted drugs have not proved as
promising as expected. It is possible that a novel signaling
pathway playing a significant role in the PI3K/AKT/PTEN pathway has
yet to be identified or that present markers are not sufficient to
assess therapeutic response (29–30).
References
|
1.
|
Tamura M, Gu J, Matsumoto K, Aota S,
Parsons R and Yamada KM: Inhibition of cell migration, spreading,
and focal adhesions by tumor suppressor PTEN. Science.
280:1614–1617. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Waite KA and Eng C: Protean PTEN: form and
function. Am J Hum Genet. 70:829–844. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Lu Y, Lin YZ, LaPushin R, Cuevas B, Fang
X, Yu SX, Davies MA, Khan H, Furui T, Mao M, Zinner R, Hung MC,
Steck P, Siminovitch K and Mills GB: The PTEN/MMAC1/TEP tumor
suppressor gene decreases cell growth and induces apoptosis and
anoikis in breast cancer cells. Oncogene. 18:7034–7045. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Weng LP, Brown JL and Eng C: PTEN
coordinates (G1) arrest by down-regulating cyclin D1 via its
protein phosphatase activity and up-regulating p27 via its lipid
phosphatase activity in a breast cancer model. Hum Mol Genet.
10:599–604. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Tamguney T and Stokoe D: New insights into
PTEN. J Cell Sci. 120:4071–4079. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Marsh DJ, Coulon V, Lunetta KL,
Rocca-Serra P, Dahia PL, Zheng Z, Liaw D, Caron S, Duboué B, Lin
AY, Richardson AL, Bonnetblanc JM, Bressieux JM, Cabarrot-Moreau A,
Chompret A, Demange L, Eeles RA, Yahanda AM, Fearon ER, Fricker JP,
Gorlin RJ, Hodgson SV, Huson S, Lacombe D, Eng C, et al: Mutation
spectrum and genotype-phenotype analyses in Cowden disease and
Bannayan-Zonana syndrome, two hamartoma syndromes with germline
PTEN mutation. Hum Mol Genet. 7:507–515. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Baeza N, Weller M, Yonekawa Y, Kleihues P
and Ohgaki H: PTEN methylation and expression in glioblastomas.
Acta Neuropathol. 106:479–485. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Freihoff D, Kempe A, Beste B,
Wappenschmidt B, Kreyer E, Hayashi Y, Meindl A, Krebs D, Wiestler
OD, von Deimling A and Schmutzler RK: Exclusion of a major role for
the PTEN tumour-suppressor gene in breast carcinomas. Br J Cancer.
79:754–758. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Bose S, Wang SI, Terry MB, Hibshoosh H and
Parsons R: Allelic loss of chromosome 10q23 is associated with
tumor progression in breast carcinomas. Oncogene. 17:123–127. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Garcia JM, Silva JM, Dominguez G, Gonzalez
R, Navarro A, Carretero L, Provencio M, España P and Bonilla F:
Allelic loss of the PTEN region (10q23) in breast carcinomas of
poor pathophenotype. Breast Cancer Res Treat. 57:237–243. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Perren A, Weng LP, Boag AH, Ziebold U,
Thakore K, Dahia PL, Komminoth P, Lees JA, Mulligan LM, Mutter GL
and Eng C: Immunohistochemical evidence of loss of PTEN expression
in primary ductal adenocarcinomas of the breast. Am J Pathol.
155:1253–1260. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Bose S, Crane A, Hibshoosh H, Mansukhani
M, Sandweis L and Parsons R: Reduced expression of PTEN correlates
with breast cancer progression. Hum Pathol. 33:405–409. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Depowski PL, Rosenthal SI and Ross JS:
Loss of expression of the PTEN gene protein product is associated
with poor outcome in breast cancer. Mod Pathol. 14:672–676. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Lee JS, Kim HS, Kim YB, Lee MC, Park CS
and Min KW: Reduced PTEN expression is associated with poor outcome
and angiogenesis in invasive ductal carcinoma of the breast. Appl
Immunohistochem Mol Morphol. 12:205–210. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Shoman N, Klassen S, McFadden A, Bickis
MG, Torlakovic E and Chibbar R: Reduced PTEN expression predicts
relapse in patients with breast carcinoma treated by tamoxifen. Mod
Pathol. 18:250–259. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Campbell RA, Bhat-Nakshatri P, Patel NM,
Constantinidou D, Ali S and Nakshatri H: Phosphatidylinositol
3-kinase/AKT-mediated activation of estrogen receptor alpha: a new
model for anti-estrogen resistance. J Biol Chem. 276:9817–9824.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Keshet I, Lieman-Hurwitz J and Cedar H:
DNA methylation affects the formation of active chromatin. Cell.
44:535–543. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Antequera F: Structure, function and
evolution of CpG island promoters. Cell Mol Life Sci. 60:1647–1658.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Soria JC, Lee HY, Lee JI, Wang L, Issa JP,
Kemp BL, Liu DD, Kurie JM, Mao L and Khuri FR: Lack of PTEN
expression in non-small cell lung cancer could be related to
promoter methylation. Clin Cancer Res. 8:1178–1184. 2002.PubMed/NCBI
|
|
20.
|
Khan S, Kumagai T, Vora J, Bose N, Sehgal
I, Koeffler PH and Bose S: PTEN promoter is methylated in a
proportion of invasive breast cancers. Int J Cancer. 112:407–410.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Shaw RJ and Cantley LC: Ras, PI(3)K and
mTOR signalling controls tumour cell growth. Nature. 441:424–430.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Janzen V and Scadden DT: Stem cells: good,
bad and reformable. Nature. 441:418–419. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Simpson L and Parsons R: PTEN: life as a
tumor suppressor. Exp Cell Res. 264:29–41. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Dudek H, Datta SR, Franke TF, Birnbaum MJ,
Yao R, Cooper GM, Segal RA, Kaplan DR and Greenberg ME: Regulation
of neuronal survival by the serine-threonine protein kinase Akt.
Science. 275:661–665. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Ramaswamy S, Nakamura N, Vazquez F, Batt
DB, Perera S, Roberts TM and Sellers WR: Regulation of G1
progression by the PTEN tumor suppressor protein is linked to
inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc
Natl Acad Sci USA. 96:2110–2115. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Stahl JM, Sharma A, Cheung M, Zimmerman M,
Cheng JQ, Bosenberg MW, Kester M, Sandirasegarane L and Robertson
GP: Deregulated Akt3 activity promotes development of malignant
melanoma. Cancer Res. 64:7002–7010. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Curtin JA, Fridlyand J, Kageshita T, Patel
HN, Busam KJ, Kutzner H, Cho KH, Aiba S, Bröcker EB, LeBoit PE,
Pinkel D and Bastian BC: Distinct sets of genetic alterations in
melanoma. N Engl J Med. 353:2135–2147. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Lian Z and Di Cristofano A: Class reunion:
PTEN joins the nuclear crew. Oncogene. 24:7394–7400. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Miller TW, Pérez-Torres M, Narasanna A,
Guix M, Stål O, Pérez-Tenorio G, Gonzalez-Angulo AM, Hennessy BT,
Mills GB, Kennedy JP, Lindsley CW and Arteaga CL: Loss of
phosphatase and tensin homologue deleted on chromosome 10 engages
ErbB3 and insulin-like growth factor-I receptor signaling to
promote antiestrogen resistance in breast cancer. Cancer Res.
69:4192–4201. 2009. View Article : Google Scholar
|
|
30.
|
Saal LH, Johansson P, Holm K,
Gruvberger-Saal SK, She QB, Maurer M, Koujak S, Ferrando AA,
Malmström P, Memeo L, Isola J, Bendahl PO, Rosen N, Hibshoosh H,
Ringnér M, Borg A and Parsons R: Poor prognosis in carcinoma is
associated with a gene expression signature of aberrant PTEN tumor
suppressor pathway activity. Proc Natl Acad Sci USA. 104:7564–7569.
2007. View Article : Google Scholar : PubMed/NCBI
|