Introduction
Telomeres are nuclear protein complexes located at
the ends of chromosomes, which shorten with cell division. This
shortens the telomere and, after 50–70 such divisions (a number
known as the Hayflick limit, after its discoverer), a chromosome
can grow no shorter and the cell it is in can divide no more. Thus,
the cell begins the process of aging, followed by death (1). Telomerase is composed of human
Telomerase RNA (hTR), Telomerase-associated protein 1 (TP1) and
human Telomerase reverse transcriptase (hTERT). Telomerase is
capable of extending or stabilizing the shortened telomeres in the
process of cell division by using the subunit hTERT and hTR as a
template for synthesizing the telomeric repeat sequence to the ends
of chromosomes. Telomerase is important in cell immortalization,
and in the occurrence and development of malignant tumors. Positive
telomerase expression has been found in 90% of tumor cells, while
negative telomerase expression has been identified in the majority
of normal human cells (2). Numerous
studies (3,4) have indicated that hTR and hTP1 are
widely expressed in both tumor and normal tissue. However, hTERT,
which is the determined part of telomerase activity, has only been
found in the majority of tumors, germ cells and proliferative stem
cells (along with its encoded mRNA), and has not been detected in
normal tissues (5). Based on these
findings, it was concluded that hTERT is important in
tumor-specific telomerase activiation. Therefore, how to apply data
concerning hTERT activity to the diagnosis and treatment of tumors
is the current issue in hTERT research.
In vivo bioluminescence imaging technology is
a novel type of sensitive optical imaging system. In the present
study, cells, proteins or DNA labelled with bioluminescence
technology were directly monitored using sensitive optical
detection equipment. The movement of cells, protein expression and
the genetic behavior of living organisms were monitored in
vivo. Bioluminescence technology is extremely sensitive, with
∼102 labelled cells having been observed in vivo
in previous studies that have used this type of technology
(6). Compared with the traditional
imaging techniques, such as computed tomography (CT) and magnetic
resonance imaging (MRI), bioluminescence technology is simple,
intuitive, rapid, highly sensitive and inexpensive. Additionally,
it is a safe technique that does not require the use of radioactive
substances.
The hTERT tumor-specific bioluminescence eukaryotic
expression vector constructed in the present study was generated
with regard to the bioluminescence imaging system, in order that
its expression could be detected in cells and animals. Stable
expression of luciferase in the HeLa-luc cell lines was screened
for in this study, and the constructed vector was inoculated in
nude mice to observe the tumor growth in vivo.
Materials and methods
Ethics
The present study was approved by the Ethics Review
Committee of Zhongnan Hospital of Wuhan University, China.
Cell culture
Human cervical cancer, HeLa; human breast cancer,
MCF-7; human kidney epithelial, 293T and human embryonic lung
fibroblast, MRC-5 cell lines were purchased from the Cell Bank of
the Chinese Academy of Sciences (Yunnan). Human osteosarcoma cell
lines U2OS and Saos were a gift from the Microscopy Orthopedic
Laboratory, Research Center of Wuhan University, China.
MCF-7 cell lines were cultured in minimum essential
medium (MEM; HyClone Laboratories Inc., Logan, UT, USA) mixed with
10% insulin and 20% fetal bovine serum (FBS). MRC-5 and Saos cell
lines were cultured in Roswell Park Memorial Institute (RPMl)-1640
medium (HyClone Laboratories Inc.) mixed with 20% FBS. The
remainder of the cell lines were cultured in RPMl-1640 medium mixed
with 10% FBS. All the cells were cultured in a humidified incubator
at 5% CO2 and 37°C.
Bacteria, plasmids and reagents
Escherichia coli (E. coli) DH5α was
obtained from the Key Laboratory of Virology, College of Life
Sciences, Wuhan University, China.
The pGL3 basis of the hTERT promoter was constructed
by Dr Liao at the Key Laboratory of Tumor Biological Behaviours,
Zhongnan Hospital of Wuhan University, China. PGL4.17(1uc2/Neo) and
pGL4.51 (luc2/CMV/Neo) were purchased from Promega Corporation
(Madison, WI, USA).
T4 DNA ligase, restriction enzymes HindIII
and SacI and DNA markers were purchased from Takara Bio Inc.
(Dalian, China, Japan). Plasmid DNA extraction and gel extraction
kits were purchased from TianGen Biotech Co., Ltd. (Beijing,
China). A Lipofectamine 2000 kit was purchased from Invitrogen Life
Technologies (Carlsbad, CA, USA). The antibiotic, G418, was
purchased from Amresco (Solon, OH, USA) and the luciferase
substrate was purchased from Kaisheng Medical Technology Co., Ltd.
(China).
Experimental animals
Female BALB/c nude mice (4–6 weeks old) were
purchased from the Disease Control Center of Hubei Province
(Certificate of Conformity: 0042029), their weights were ∼14–18 g.
All the animals were raised in a specific pathogen-free (SPF)
environment.
Construction of recombinant plasmid
phTERTp-luc-neo
Restriction enzymes SacI and HindIII
were used to digest plasmid pGL3 basic hTERT promoter to pGL4.17
(luc/neo) and run in gel electrophoresis. The recovered fragments
of phTERTp-luc-neo and pGL4.17 from gel extraction were then
connected with T4 ligase (4°C overnight), transformed into DH5α,
and then the plasmid was extracted from selected clones using a
plasmid DNA extraction kit. Preliminarily the plasmid was
identified by electrophoresis after double digestion (SacI
and the HindIII), and then the identified plasmid was
sequenced (sequenced by invtrogen company).
hTERT promoter expression detection in
vivo
Telomerase-positive (HeLa, MCF-7 and 293T),
telomerase-negative (U2OS and SaOS) and normal human embryonic lung
(MRC-5) cell lines were separately seeded in 24-well plates, with
each cell line being inoculated with 12 holes. Each group was
inoculated with three holes following transfection with the
recombinant vector phTERTp-luc-neo, positive control vector pGL4.51
(luc2/CMV/Neo) and negative control vector pGL4.17 (luc2/ neo). A
blank control was set up for non-transfected plasmids. After 48 h
of transfection, the cells were digested to the state of suspension
(100 μl per well), and then transferred to 96-well cell
plates. One microliter of 15 mg/ml luciferase substrate was added
to each hole, mixed and incubated at 37°C for 5 min. Images were
then captured using the in vivo bio-optical imaging system
(Kai Sheng Branch in vivo bioluminescence imaging optical
system), white light imaging for 0.1 sec and fluorescence imaging
for 1–3 min. Bioluminescence intensity was recorded for each cell
line.
Stable transfection with
hTERTp-luc-neo
HeLa cells were adjusted to 10,000/ml for detection
of the minimum lethal concentration of G418 in the HeLa cell line,
and 0.5 ml/well was added to the 24-well plates. Eight
concentrations (300, 400, 500, 600, 700, 800, 900 and 1000/ml) of
G418 were used for the selection of HeLa cell lines, with each
concentration added to three wells. The minimum concentration in
which all the cells had died after 10–14 days was selected for
screening of HeLa cells.
The logarithmic phase of HeLa cells for recombinant
vector transfection and monoclonal screening was selected, and
seeded into 6-well culture plates (2×105 cells/well) 24
h prior to transfection. Transfection was conducted according to
the manufacturer’s instructions for the Lipofectamine 2000 Kit.
G418 was used to screen for the optimal concentration 24 h after
transfection, and monoclonal cell lines were screened with a
limited cloning dilution method when there was no futher cell
death.
Suspension (107 cells/ml) with the
initial screening of monoclonal cell lines was generated to
identify the positive clone by the in vivo bioluminescent
imaging system, by adding 100 μl/well to the 96-well plates,
with three wells per group. Luciferase substrate (1 μl; 15
mg/ml) was added to each well and mixed for 5 min at 37°C. Images
were then captured using the in vivo bioluminescence imaging
system.
The selected screened positive monoclonal (HeLa-luc)
cells and the HeLa cells that were used as a control for the single
cell suspension were then added to 24-well plates at
2×104 cells/well. The following day, cells (per three
wells) were digested and counted. The cell doubling time
(tD) was calculated over the subsequent six consecutive
days, using the formula: tD = t × lg2 / lg
(N/N0) (t, time interval in hours; N0, cell
number at start; N, cell number in the end. The experiment was
repeated three times and a cell growth curve was generated.
The screened monoclonal cells were diluted in a
number of gradient concentrations to determine the fluorescence
value of each gradient concentration, with cell counts of
106, 2×105, 105 and
104, per 100 μl medium. The image was then
captured by the in vivo bioluminescence imaging system. The
bioluminescence intensity of each clone was compared, and the
correlation between bioluminescence intensity and the cell number
was analyzed. The cell line which demonstrated the highest
correlation and the highest high luciferase activity was selected
for determination of the fluorescence value of the cell line in
various gradient concentrations.
In vivo observation stably expressing
luciferase tumor cell growth
The logarithmic growth phase of the screened
monoclonal cells was selected, and the concentrations were adjusted
with phosphate-buffered saline (PBS) to 104,
105, 106 and 107 cells/ml.
Gradient concentrations of cell suspensions (100 μl) were
implanted subcutaneously into each side of the dorsal axillary and
groin regions of a nude mouse, at a total of 4 points. In another
nude mouse, 107 cells were subcutaneously inoculated
into one side of the dorsal axillary region. The mice were
administered an intraperitoneal luciferase substrate injection (150
mg/kg) 5–6 min prior to imaging. Subsequently, the mice were
administered an intraperitoneal 1% pentobarbital sodium injection
(100 mg/kg) during imaging. The duration of the imaging process was
1–3 min.
Statistical analysis
Experimental data were recorded as mean ± standard
deviation. Data were analyzed by a Dixon’s Q test using the
Statistical Package for the Social Sciences (SPSS) software,
version 13.0. P<0.05 and P <0.01 were used to indicate
statistically significant differences.
Results
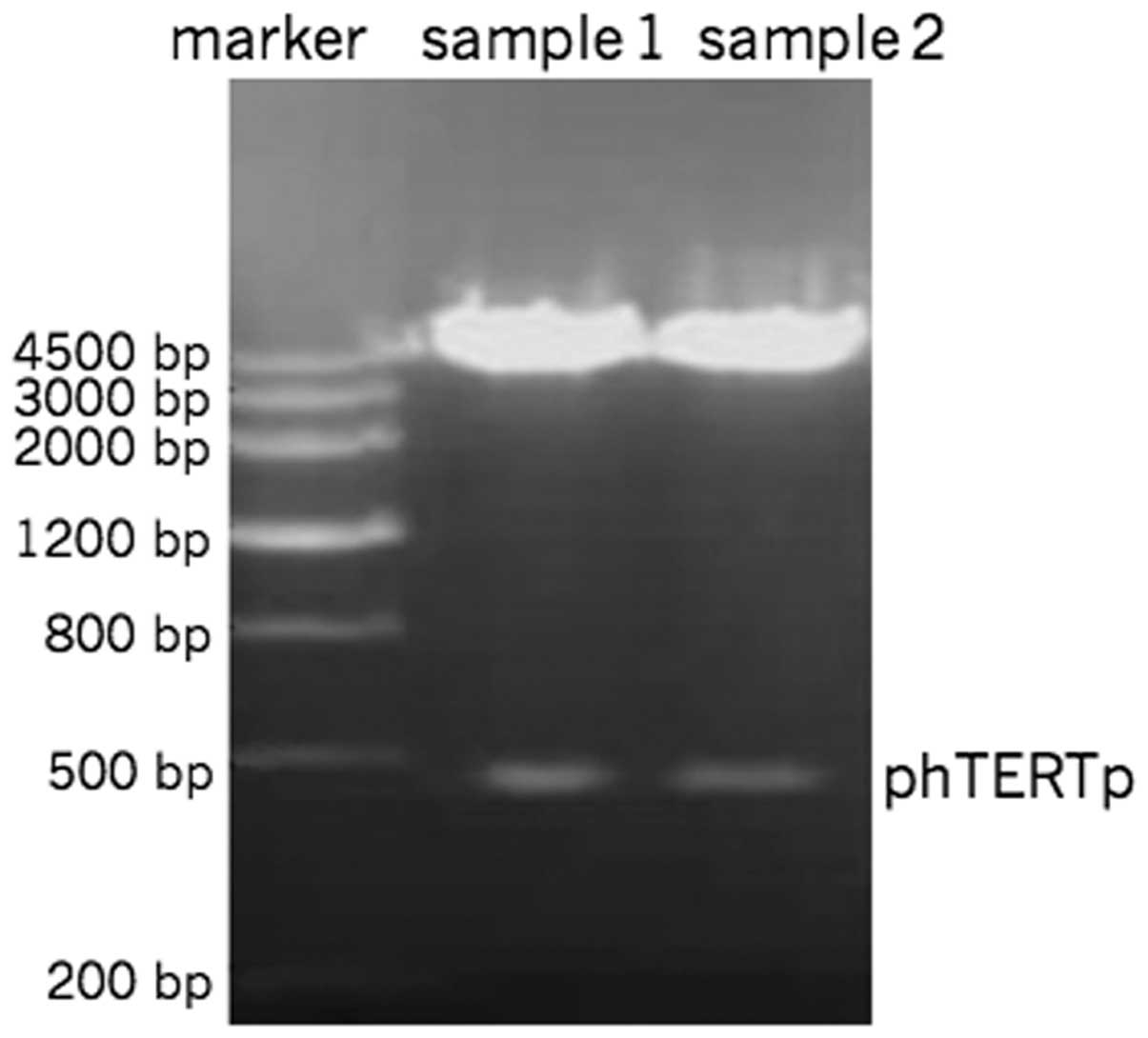
Restriction endonuclease
PhTERTp-luc-neo was double-digested with SacI
and HindIII. The digestion products revealed clear bands at
500 bp by gel electrophoresis (Fig.
1). The hTERT promoter sequence was identical to that of
Genbank.
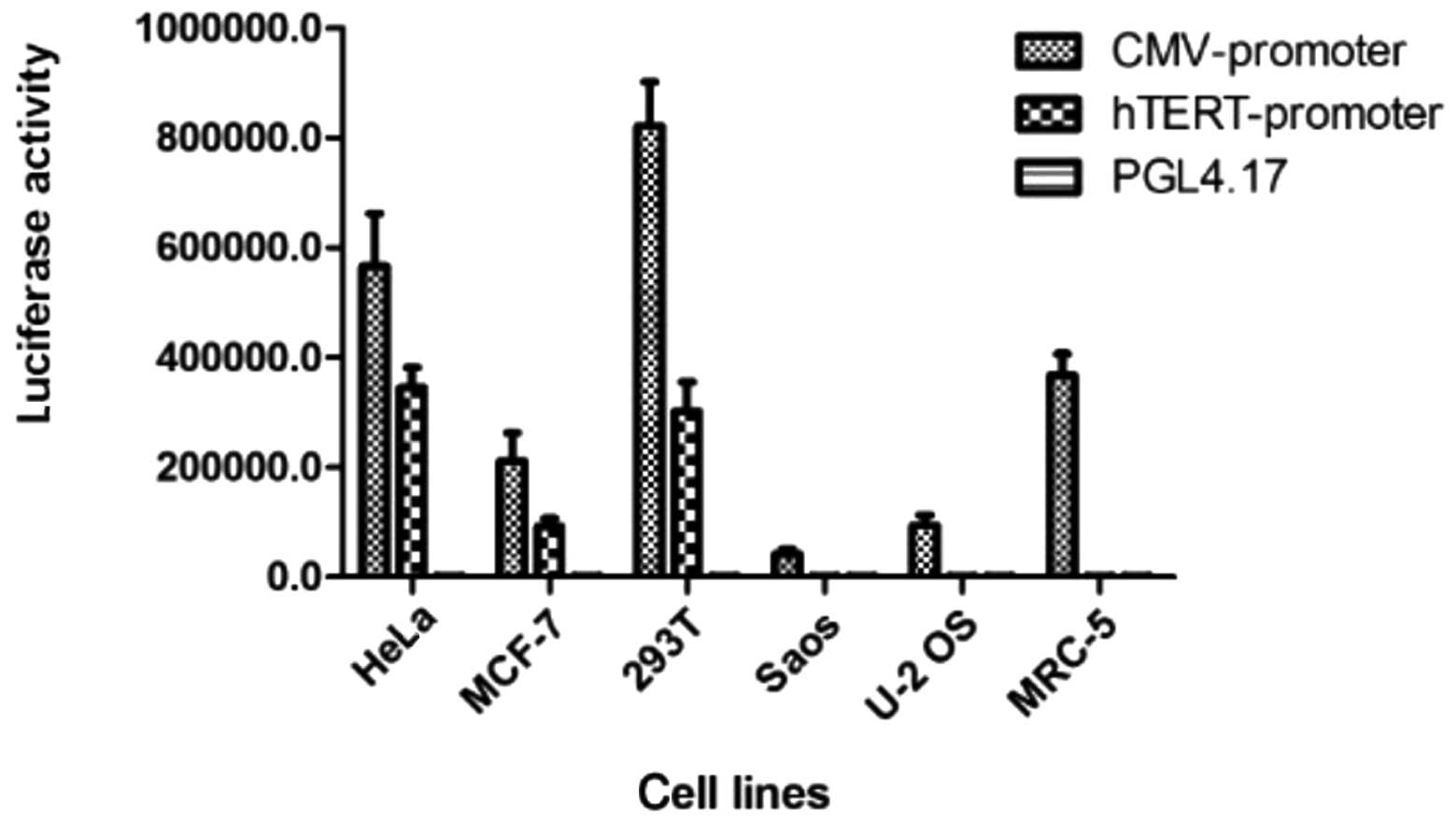
Expression of the hTERT promoter
following transient transfection
Significant luciferase expression was demonstrated
by pGL4.51 (luc2/CMV/Neo) in each cell line, although this
expression was not tumor-specific. The expression activity of
luciferase regulated by hTERT in the telomerase-positive cell lines
(HeLa, MCF-7, U251 and 293T) was significantly higher than that of
the telomerase-negative (U2OS and Saos) and normal (MRC-5) cell
lines. This result confirmed that the constructed vector,
phTERTp-luc-neo, was tumor-specific (Fig. 2).
Identification of positive clones
The optimal concentration of G418 selection in HeLa
cells was determined to be 800 μg/l by the G418 gradient
concentration filter.
Initial screening of 30 clones
The transfected plasmid vector was randomly
integrated into the chromosome, according to the luciferase
activity of the different clones. The luciferase-expressing clones
2, 6 and 15 were determined to be the positive clones and were
designated as HeLa-luc-2, -6 and -15, respectively.
Doubling time of clone cells cultured in
vitro
According to the growth curves of the positive
clonal cells, HeLa-luc-2, -6 and -15, the doubling times were
28.41, 22.37 and 30.20 h, respectively, whereas the doubling time
of HeLa control cells was 22.11 h. We found that the growth of
clonal HeLa-luc-6 cells was similar to that of HeLa control cells;
no significant difference was observed (P>0.05).
Determination of the cell fluorescence
value of the gradient concentration in vitro
Cells were diluted to four gradient concentrations
(104, 5×104, 1×105 and
1×106 cells/100 μl) and cell fluorescence values
of the four gradient concentrations were determined. The data
showed that there was a higher correlation between the cell number
and fluorescence in the HeLa-luc-6 monoclone; the correlation
coefficient was 0.9937 and an extremely high luciferase activity
was observed (Fig. 3).
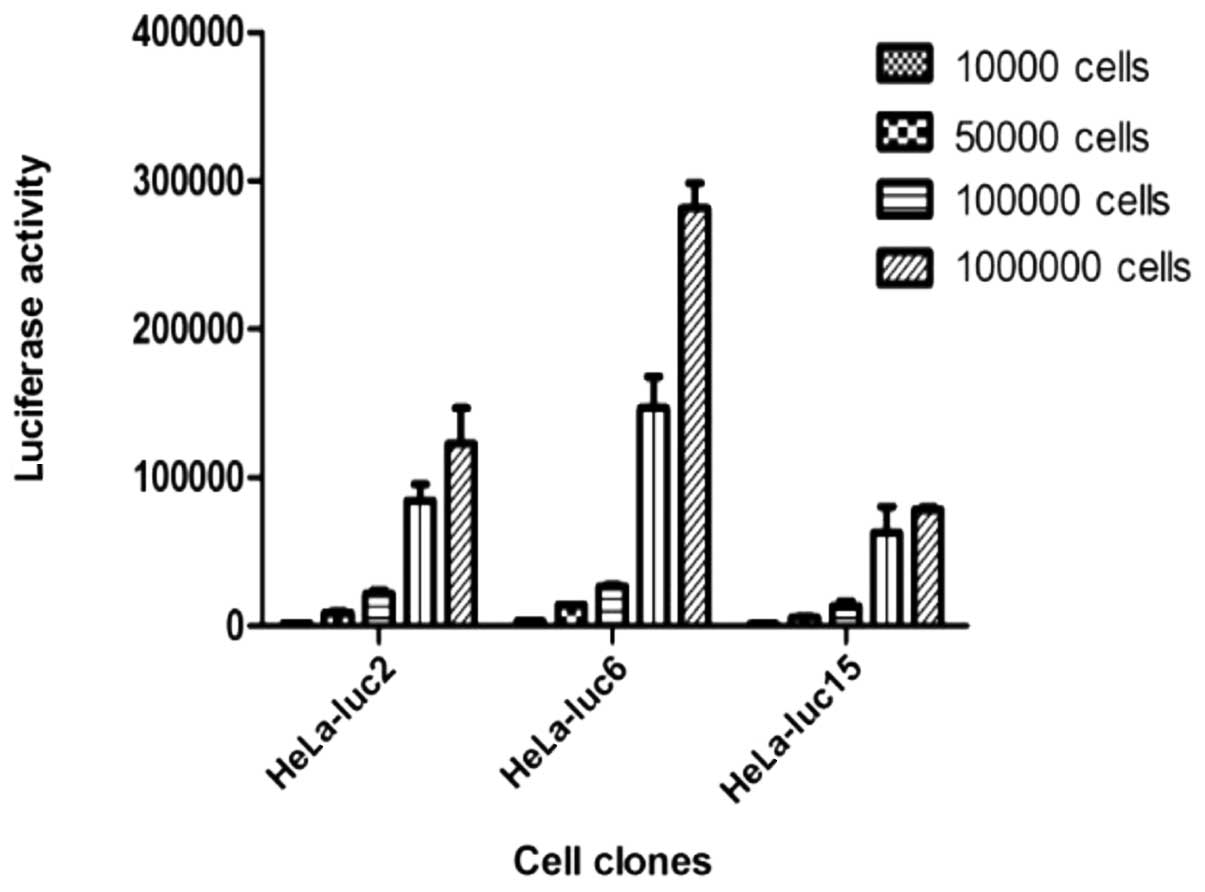 | Figure 3.The recombinant vector,
phTERTp-luc-neo, was transfected into a HeLa cell line. Antibiotic
G418 was used to select neomycin-positive cell clones.
Bioluminescent imaging was applied to screen and verify whether
selected neomycin-positive cell clones exhibited specific, high
luciferase expression. A HeLa-luc cell clone, which constantly
expressed both neomycin and luciferase, was obtained by selection
with G418 and bioluminescent imaging. According to the luciferase
activity of the different clones, the lucif-erase-expressing clones
2, 6 and 15 were determined to be the positive clones, and were
termed HeLa-luc-2, -6, and -15, respectively. Cell fluorescence
values of the four gradient concentrations (104,
5×104, 1×105 and 1×106 cells/100
μl) were determined. Data showed that the highest
correlation between the cell number and fluorescence was in the
HeLa-luc-6 monoclone; the correlation coefficient was 0.9937 and an
extremely high luciferase activity was observed. |
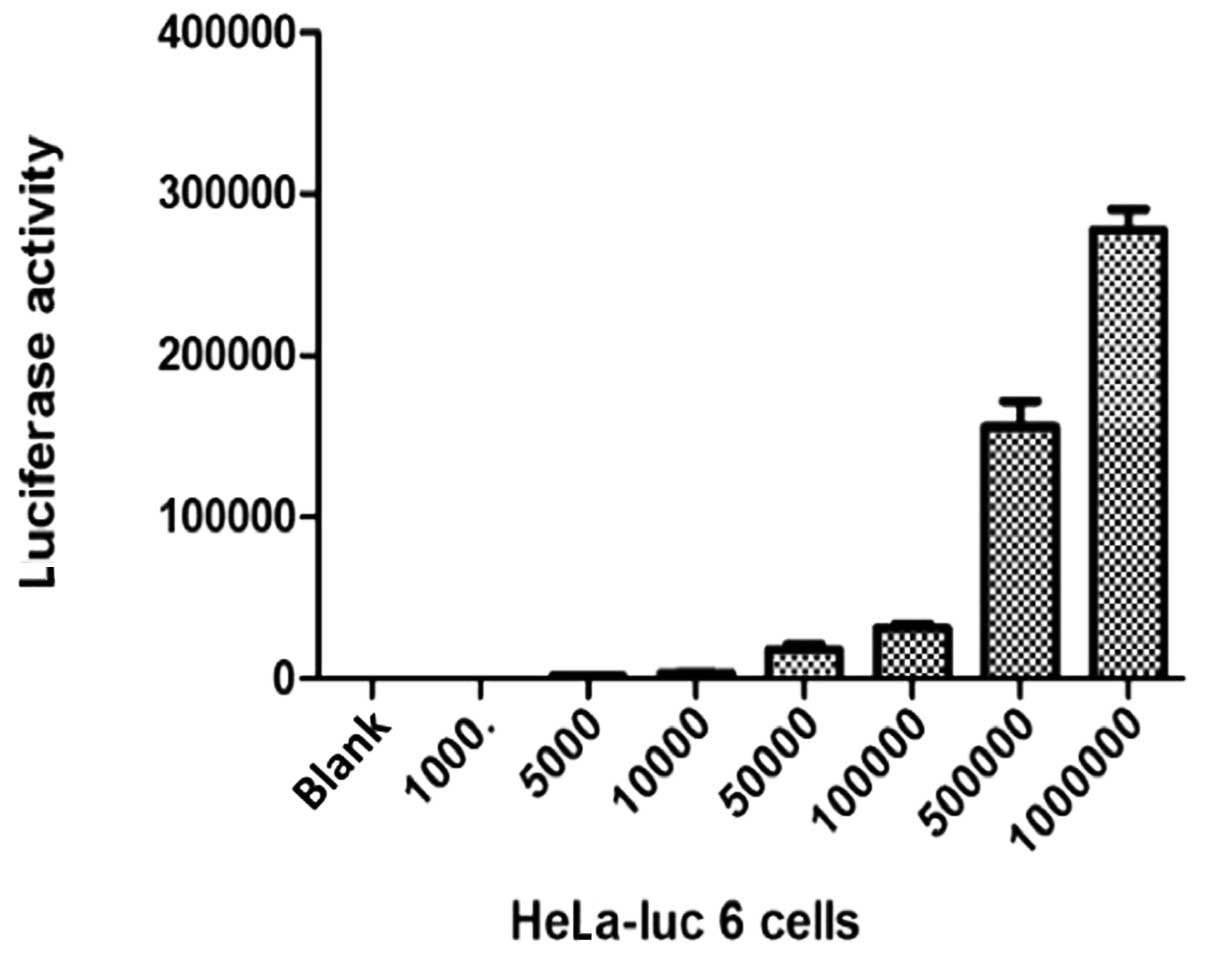
Determination of cell fluorescence of
HeLa-luc 6 in gradient concentration
The cells were diluted into seven gradient
concentrations (1000, 5000, 104, 5×104,
1×105, 5×105 and 1×106 cells/ml).
Cell fluorescence values of the seven gradient concentrations were
subsequently determined (Fig.
4).
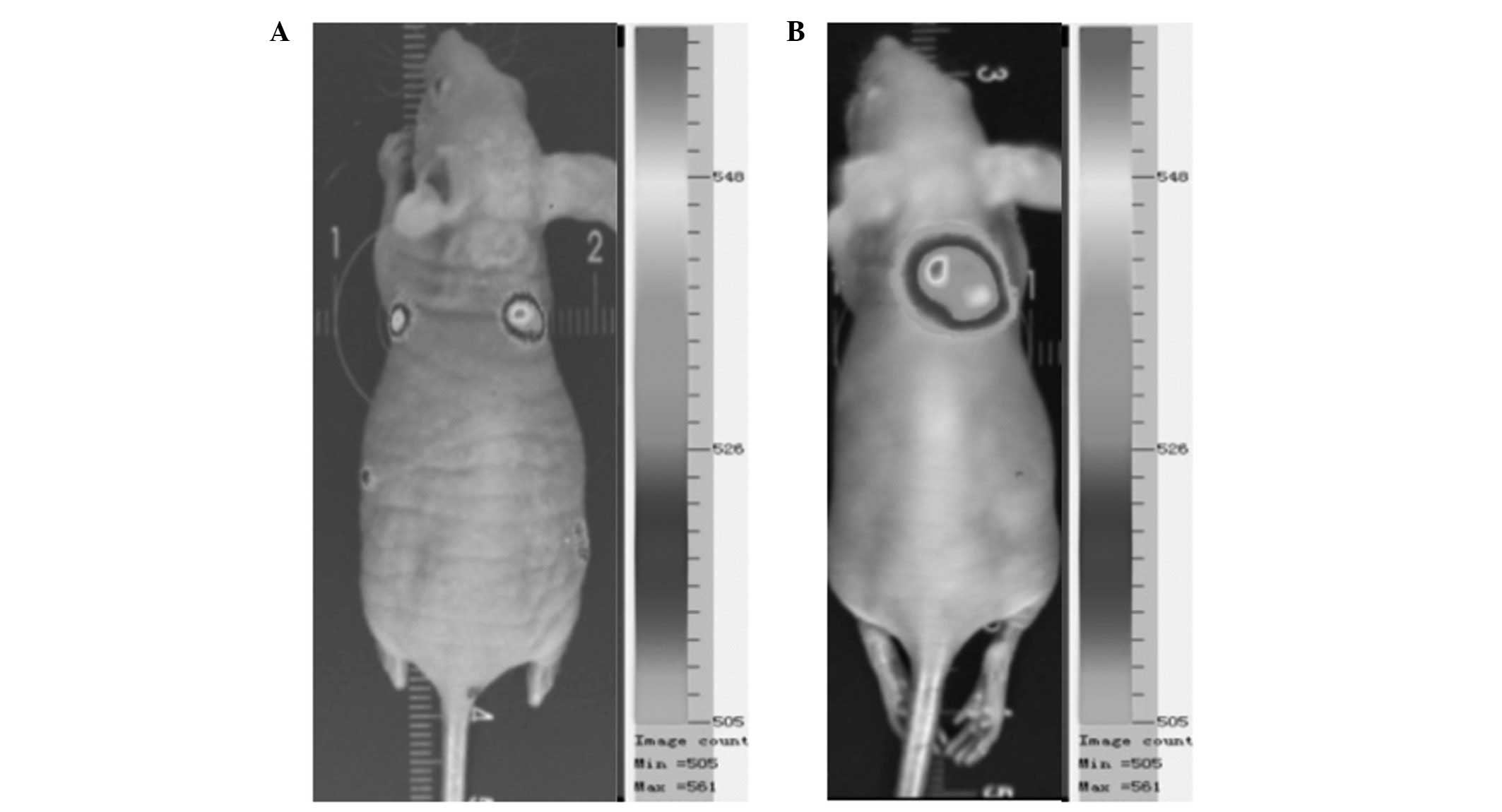
Animal experiments
Tumor growth was clearly visible subcutaneously in
the mouse implanted with HeLa-luc-6 cells, and the tumor was
detected on the fifth day following implantation by bioluminescent
imaging, demonstrating that the HeLa-luc-6 cells were tumorigenic.
During imaging of the mouse subcutaneously implanted with
HeLa-luc-6 cells, the expression of luciferase was clearly detected
(Fig. 5).
Discussion
The aim of the present study was to develop methods
for testing the usage of in vivo bioluminescence imaging
technology in tumor detection, for this purpose. Therefore, we
constructed the hTERT tumor-specific bioluminescence eukaryotic
expression vector and established a stable HeLa-luc cell line that
expressed the luciferase gene. Nude mice were then inoculated with
the constructed vector in order to observe the tumor growth in
vivo.
The specific promoter was selected to be 480 bp
located at the proximal region of the hTERT gene. The eukaryotic
expression vector phTERTp-luc-neo was constructed and was regulated
by the hTERT promoter. The recombinant vector was characterized by
this in that the hTERT promoter regulated the expression of the
downstream luc gene, and showed highly specific expression in
telomerase-positive tumor cells. In addition, its expression was
observed both in vitro and in vivo using in
vivo bioluminescence imaging technology.
The bioluminescence generated in cells labeled with
the vector, on addition of the luciferase substrate, was detected
by a charge-coupled device (CCD) camera without the existence of an
exogenous excitation light source.
Both the experimental and control plasmids were
transiently transfected into telomerase-positive, -negative and
normal cell lines by lipofection. The tumor-specific characteristic
of the plasmid was confirmed by detecting the fluorescence
intensity with the in vivo bioluminescence imaging system.
The in vivo bioluminescence imaging system was found to be
more intuitive and convenient than dual luciferase reporter gene
detection for the detection of luciferase gene expression. The
in vivo bioluminescence imaging technology is predicted to
become increasingly applied to the field of cancer research.
A study by Jenkins et al applied the in
vivo bioluminescence imaging technique to the observation of
tumor cell growth and metastasis (7). Additionally, Gupta investigated breast
cancer metastasis, gene expression and the tumor microenvironment
by an in vivo bioluminescence imaging technique (8). The high sensitivity of in vivo
bioluminescence imaging technology has been widely used to
construct tumor models (9–11); however, its usage in combination
with the hTERT promoter in tumor diagnosis and gene therapy is not
common. In the present study, the tumor-specific eukaryotic
expression vector, phTERTp-luc-neo, an integration of the
luciferase reporter and neo genes, was transfected into HeLa cells
by lipofection. The transfected cells were monoclonal due to the
implementation of a limited cloning dilution method a period of
time after G418 selection, to ensure that the cells were obtained
from the same ancestors, and that the genetic traits were
consistent. Luciferase expression of monoclonal cells was then
detected by an in vivo bioluminescence imaging system. Three
cell lines demonstrated luciferase expression. Dilution of cells to
gradient concentration and determination of luciferase expression
of each gradient concentration, were conducted, and HeLa-luc-6
cells were selected for further animal experiments. BALB/c mice
were subcutaneously inoculated with HeLa-luc-6 cells. The results
showed that the cells were tumorigenic and the bioluminescence
signal was detected.
In summary, we constructed a tumor-specific
bioluminescence eukaryotic expression vector regulated by an hTERT
promoter. The vector was visual, intuitive and highly sensitive,
and demonstrated potential for the study of gene therapy with
telomerase or hTERT as the target. In addition, HeLa-luc cell lines
that stably expressed luciferase were established. This study has
provided an intuitive, convenient, sensitive and reliable basis for
investigation of the expression and regulation of hTERT, and with
the early diagnosis of tumors. It also promotes the use of the
in vivo bioluminescence imaging technique in subsequent
experiments.
Acknowledgements
This study was supported by the
Science and Technology Department (2008CDA065) and the Key
Laboratory of Tumor Biological Behaviors, Hubei Province,
China.
References
|
1.
|
Rubin H: The disparity between human cell
senescence in vitro and lifelong replication in vivo.
Nat Biotechnol. 20:675–681. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Vega LR, Mateyak MK and Zakian VA: Getting
to the end: telomerase access in yeast and humans. Nat Rev Mol Cell
Biol. 4:948–959. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Bryce LA, Morrison N, Hoare SF, Muir S and
Keith WN: Mapping of the gene for human telomerase reverse
transcriptase, hTERT, to chromosome 5p15.33 by fluorescence in
situ hybridization. Neoplasia. 2:197–201. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Blackburn EH, Chan S, Chang J, et al:
Molecular manifestations and molecular determinants of telomere
capping. Cold Spring Harb Symp Quant Biol. 65:253–263. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Kim NW, Piatyszek MA, Prowse KR, et al:
Specific association of human telomerase activity with immortal
cells and cancer. Science. 266:2011–2015. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Edinger M, Cao Y A, Verneris M R, et al:
Revealing lymphoma growth and the efficacy of immune cell therapies
using in vivo bioluminescence imaging. Blood. 101:640–648.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Jenkins DE, Oei Y, Hornig YS, et al:
Bioluminescent imaging (BLI) to improve and refine traditional
murine models of tumor growth and metastasis. Clin Exp Metastasis.
20:733–744. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Gupta GP, Nguyen DX, Chiang AC, et al:
Mediators of vascular remodelling co-opted for sequential steps in
lung metastasis. Nature. 446:765–770. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Nyati MK, Symon Z, Kievit E, et al: The
potential of 5-fluorocytosine/cytosine deaminase enzyme prodrug
gene therapy in an intrahepatic colon cancer model. Gene Ther.
9:844–849. 2002.PubMed/NCBI
|
|
10.
|
Kemper EM, Leenders W, Küsters B, et al:
Development of luciferase tagged brain tumour models in mice for
chemotherapy intervention studies. Eur J Cancer. 42:3294–3303.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Choy G, O’Connor S, Diehn FE, et al:
Comparison of noninvasive fluorescent and bioluminescent small
animal optical imaging. Biotechniques. 35:1022–1026. 1028–1030.
2003.PubMed/NCBI
|