Introduction
Meningiomas account for ∼30% of all central nervous
system neoplasms and are derived from the arachnoid cells covering
the brain. The majority of meningiomas are slow-growing and
correspond to grade I tumors, according to the World Health
Organization (WHO) classification. Grade II (atypical) and grade
III (anaplastic) meningiomas are considerably more aggressive
(1). Meningiomas may present as
sporadic solitary tumors. In the context of neurofibromatosis type
2 disease (NF2), the tumors may also be characterized by the
presence of bilateral acoustic schwannomas (1,2).
Molecular genetic alterations that are linked to meningioma
tumorigenesis indicate the inactivation of at least one member of
the 4.1 super-family of genes, primarily involving the
neurofibromin gene, NF2, that is located at 22q12.2 or the
4.1B (DAL-1) gene, located at 18p11.32 (1,2).
Chromosomal losses at other genomic regions, including 1p, 6q, 9p,
10, 14q and 18q, as well as gains in multiple regions, including
1q, are frequently identified in grade II and III tumors (1,3–5).
Accordingly, genetic alterations in those specific genomic regions
are likely to be associated with the progression of meningioma.
Controversial data have been obtained from a low sequence mutation
rate that has been identified by the analysis of potential target
genes in these locations (1,6–7).
The low mutation rate may be due to alternative gene silencing
mechanisms, since aberrant promoter hypermethylation of the CpG
islands may contribute to changes in the expression status of those
genes (8–11). The loss of tumor suppressor genes,
including CDKN2A and CDKN2B, and the involvement of
MEG3, NDRG2, TSLC1, LMO4, osteopontin and LEPR, have
been proposed as candidates for progression or recurrence markers
in meningiomas (1,12–13).
Transcriptomic expression profiling studies using array analyses
have been performed in meningiomas (14) and certain meningioma-specific genes
or those associated with the WHO grades have been identified.
Recently, common genes that are responsible for the
recurrence and progression of meningiomas, or for the existence of
differential expression patterns among spinal and intracranial
meningiomas, have been identified (15,16).
Analyses of low-density microarrays have previously identified two
expression subgroups in meningiomas. The tumors exhibited abnormal
patterns with deletions at 1p and 14q, in addition to a 22q
deletion/NF2 inactivation (17,18).
Attention has been focused on the aberrant signaling pathways in
meningiomas. The findings confirm that the most likely implication
involves the loss of NF2. A number of growth factors and
cytokines, and the deregulation of the calcium signaling system,
have been proposed to be involved in the development and
progression of meningiomas, but no firm conclusions are available
(19). This study presents
additional data to those that have been previously reported
(17,18) with regard to the expression
variations of 96 tumor-related genes included in six key signaling
pathways in meningiomas. In order to obtain new findings, the
present study focused on the signaling pathways that may be altered
in these brain tumors. The data show that the pathways that are
involved in cell cycle control, DNA damage repair, apoptosis and
angiogenesis were more commonly deregulated.
Materials and methods
The patterns of gene expression levels were analyzed
in 42 meningiomas, including 32 grade I, four recurrent grade I and
six grade II tumors, using the GE Array Q Series HS-006
(SuperArray, Bethesda, MD, USA) for the analysis of 96 genes
corresponding to various biological pathways that are frequently
deregulated in tumorigenesis. Group 1 was associated with cell
cycle control and DNA damage repair (15 genes), group 2 with
apoptosis and cell senescence (14 genes), group 3 with signal
transduction molecules and transcription factors (17 genes), group
4 with adhesion (15 genes), group 5 with angiogenesis (22 genes)
and group 6 with invasion and metastasis (13 genes).
Detailed information with regard to specific
tumor-related genes, housekeeping genes and negative controls is
available at http://www.superarray.com and http://www.sabiosciences.com. A control comprising
three commercial human adult normal (non-tumoral) RNA from cerebral
meninges (U.S. Biological Biochain, Hayward, CA, USA) was used. The
manufacturer's instructions were used with small variations, as
described (18). Previous array
experiment information and data are available at ArrayExpresss
(http://www.ebi.ac.uk/arrayexpress).
Quantitative polymerase chain reactions (qPCR) were used to
validate the microarray data and statistical analyses were
performed as previously described (18). A total of four genes (BCL2L1,
FOS, MDM2 and TIMP1) that were included in the
microarray were selected for the analysis and GAPDH was used
as an endogenous control gene. Expression data from the arrays and
q-PCR were adjusted to a normal distribution to compare the sets of
data and expression results were consistent between the two
methodologies. A particular gene was considered to be overexpressed
if the expression levels were ≥5 times the values obtained in the
control samples. A gene was considered to be underexpressed if the
expression levels were ≤0.2 of the mean values found in the
non-tumoral control samples.
Results and Discussion
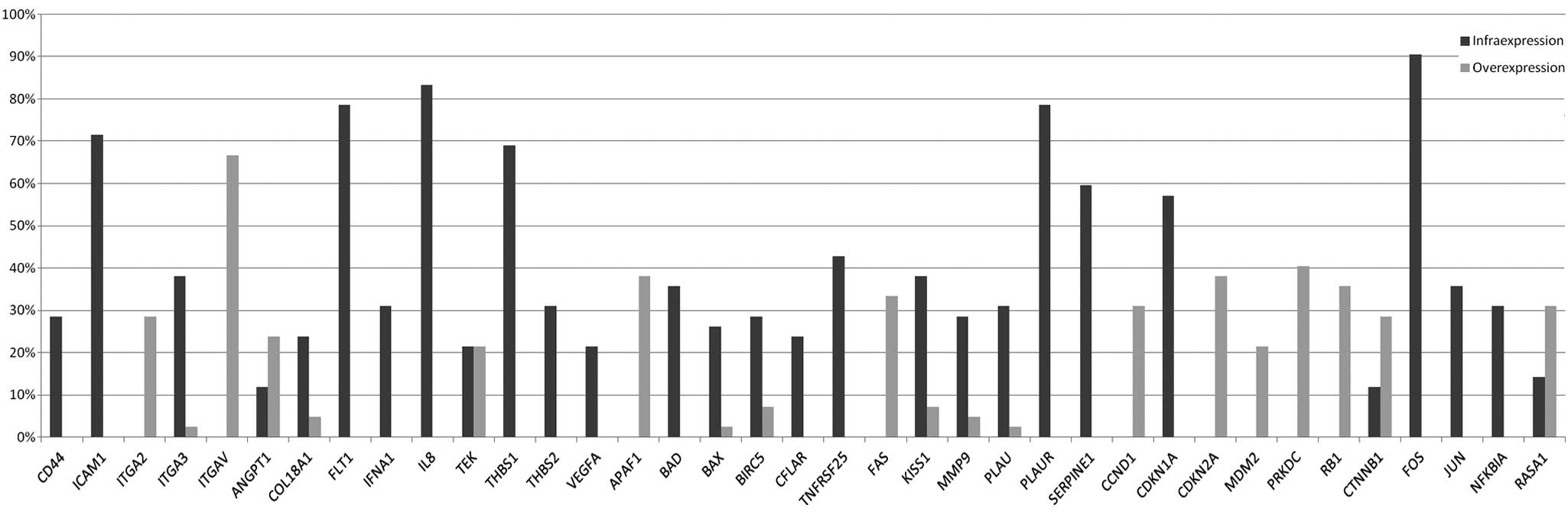
A total of 25 of the 96 genes were downregulated in
at least 20% of samples. These genes correspond to: CDKN1A (gene
group 1); BAD, BAX, BIRC5, CFLAR and TNFRSF25 (group
2); FOS, JUN and NFKBIA (group 3); CD44, ICAM1
and ITGA3 (group 4); COL18A1, FLT1, IFNA1, IL8, TEK,
THBS1, THBS2 and VEGFA (group 5); and KISS1, MMP9,
PLAU, PLAUR and SERPINE1 (group 6). An additional 56
genes were downregulated in <20% of samples and 15 genes
presented no underexpression. However, 14 of these 15 genes were
upregulated at variable levels and one remained constant. In 13
genes, the expression values were consistent with the criteria for
upregulation in at least 20% of samples, including those of grade I
and II. These genes were distributed as follows: CCND1, CDKN2A,
MDM2, PRKDC and RB1 (group 1); APAF1 and
FAS (group 2); CTNNB1 and RAS1 (group 3);
ITGA2 and ITGAV (group 4); and ANGPT1 and
TEK (group 5). No genes from group 6 (invasion and
metastasis) were observed to be overexpressed, which is consistent
with the characteristic low-grade malignancy (WHO grade I) of the
majority of meningiomas that generally do not exhibit metastasis. A
total of 29 genes presented no overexpression, but 28 were
underexpressed in at least one sample and involved grade I and II
tumors. Of all the meningiomas analyzed, 54 genes were upregulated
in <9 samples (<20%). A summary of the main findings is shown
in Table I and Fig 1.
 | Table I.Genes that were derugulated in at
least a 20% of samples. |
Table I.
Genes that were derugulated in at
least a 20% of samples.
| Gene Symbol | Unigene | Localization | Infra-expression
(%) | Overexpression
(%) | Functional group |
|---|
| CDKN1A | Hs.370771 | 6p21.2 | 57 | 0 | 1 |
| BAD | Hs.370254 | 11q13.1 | 36 | 0 | 2 |
| BAX | Hs.624291 | 19q13.3–q13.4 | 26 | 2 | 2 |
| BIRC5 | Hs.514527 | 17q25 | 29 | 7 | 2 |
| CFLAR | Hs.390736 | 2q33–q34 | 24 | 0 | 2 |
| TNFRSF25 | Hs.462529 | 1p36.2 | 43 | 0 | 2 |
| FOS | Hs.731317 | 14q24.3 | 90 | 0 | 3 |
| JUN | Hs.696684 | 1p32–p31 | 36 | 0 | 3 |
| NFKBIA | Hs.81328 | 14q13 | 31 | 0 | 3 |
| CD44 | Hs.502328 | 11p13 | 29 | 0 | 4 |
| ICAM1 | Hs.643447 | 19p13.2 | 71 | 0 | 4 |
| ITGA3 | Hs.265829 | 17q21.33 | 38 | 2 | 4 |
| COL18A1 | Hs.517356 | 21q22.3 | 24 | 5 | 5 |
| FLT1 | Hs.594454 | 13q12 | 79 | 0 | 5 |
| IFNA1 | Hs.37026 | 9p22 | 31 | 0 | 5 |
| IL8 | Hs.624 | 4q13–q21 | 83 | 0 | 5 |
| TEK | Hs.89640 | 9p21 | 21 | 21 | 5 |
| THBS1 | Hs.164226 | 15q15 | 69 | 0 | 5 |
| THBS2 | Hs.371147 | 6q27 | 31 | 0 | 5 |
| VEGFA | Hs.73793 | 6p12 | 21 | 0 | 5 |
| KISS1 | Hs.95008 | 1q32 | 38 | 7 | 6 |
| MMP9 | Hs.297413 | 20q11.2–q13.1 | 29 | 5 | 6 |
| PLAU | Hs.77274 | 10q24 | 31 | 2 | 6 |
| PLAUR | Hs.466871 | 19q13 | 79 | 0 | 6 |
|
SERPINE1 | Hs.414795 | 7q22.1 | 60 | 0 | 6 |
| CCND1 | Hs.523852 | 11q13 | 0 | 31 | 1 |
| CDKN2A | Hs.512599 | 9p21 | 0 | 38 | 1 |
| MDM2 | Hs.484551 | 12q14.3–q15 | 0 | 21 | 1 |
| PRKDC | Hs.491682 | 8q11 | 0 | 40 | 1 |
| RB1 | Hs.408528 | 13q14.2 | 0 | 36 | 1 |
| APAF1 | Hs.552567 | 12q23 | 0 | 38 | 2 |
| FAS | Hs.244139 | 10q24.1 | 0 | 33 | 2 |
| CTNNB1 | Hs.476018 | 3p21 | 12 | 29 | 3 |
| RASA1 | Hs.664080 | 5q13.3 | 14 | 31 | 3 |
| ITGA2 | Hs.482077 | 5q11.2 | 0 | 29 | 4 |
| ITGAV | Hs.436873 | 2q31–q32 | 0 | 67 | 4 |
| ANGPT1 | Hs.369675 | 8q23.1 | 12 | 24 | 5 |
| TEKa | Hs.89640 | 9p21 | 21 | 21 | 5 |
The genes that were included in group 2 (apoptosis
and cell senescence) were the most frequently altered in the
present meningioma study, as 7 out of 14 (50%) genes were
abnormally expressed, of which five were found to be downregulated
and two upregulated. Accordingly, the low expression levels of the
five genes in this functional group may be involved in meningioma
development. However, it is difficult to understand the role that
is played by the upregulation of the remaining two genes,
APAF1 and FAS. Alterations of at least 40% of genes
exhibiting significant expression variation, according to the
criteria for under- or overexpression present in at least 20% of
samples, were identified in two additional functional groups: 10 of
22 (45%) genes in group 5 (angiogenesis) and six of 15 (40%) genes
associated with cell cycle control and DNA damage repair (group 1).
Although the genes in group 5 were downregulated, those included in
group 1 appeared to be overexpressed. Due to the small number of
genes that were analyzed, internet databases such as DAVID
(http://david.abcc.ncifcrf.gov/) did not
show any enriched pathways or gene grouping. Thus, the findings
suggest that the alteration of regulatory pathways involved in
apoptosis, angiogenesis and DNA repair may be involved in
meningioma development as alterations that are secondary to
NF2 inactivation, or in general to the aberrant signaling
pathways involving membrane-associated 4.1 family proteins. Not all
the deregulated genes mapped to the chromosomal regions are
frequently altered in meningiomas.
The proportional gene loss, which may be a result of
underexpression due to hemizygosity, in the genomic regions that
were frequently deleted (22q, 14q, 1p, 6q, 9p, 10, 18q), may
contribute to the deregulation of other genes that are located in
surrounding genomic regions. An aberrant gene expression pattern
associated with allelic losses at 1p and 14q has been described in
meningiomas and the majority of the identified deregulated genes
are located outside those genomic regions (18). Moreover, although no statistical
analysis was performed due to the small number of cases considered
as recurrent (four tumors) or grade II (six cases) in the present
study, the more aggressive meningiomas and the grade I tumors were
associated with an abnormal cDNA expression pattern, indicating
that a subgroup of grade I meningiomas may exhibit a predisposition
to evolve into more aggressive forms. The genes that were found to
be deregulated concur with those that have been identified in
previous studies (14). The
aberrant signaling pathway that is predominantly involved in
meningioma tumorigenesis thus far identified involves the loss of
chromosome 22 and therefore NF2 gene inactivation (19). Less frequently, cellular signal
transduction pathways, including Hedgehog, MAPK, PI3K and Notch,
growth factors and cytokines, or calcium signaling pathways also
exhibit abnormal deregulation (19). The present data suggest that
apoptosis, angiogenesis, cell cycle control and DNA damage repair
pathways are commonly altered in meningiomas.
Due to the close molecular genetic association
(NF2 inactivation) between meningioma and schwannoma, the
results from the present study were compared with those that were
previously reported using the same methods in a schwannoma series
(20), in order to identify genes
that were equally or differentially expressed in the two entities
(Tables II and III). The most frequently altered
functional groups differ between the tumor types. The genes that
code for proteins involved in apoptosis and cell senescence (group
2) were found to be deregulated in up to 50% of meningioma samples
compared with the genes from group 4 (adhesion proteins) that were
abnormally regulated in 53% of schwannoma samples). In addition, a
concurrent expression alteration of genes included in groups 1 (up
to 40%) and 5 (45%) was identified in the meningioma and schwannoma
samples. These groups of genes correspond to cell cycle control and
DNA damage repair (group 1) and angiogenesis (group 5). With regard
to the involvement of specific genes, certain inter-tumoral group
variations were identified, including BRC5, JUN and
SERPINE1, which were underexpressed in >30% of
meningiomas compared with <10% of schwannomas. Low EGFR
levels were identified in up to 40% of schwannomas but <10% of
meningiomas. PIK3R1, ITGA4 and ITGA6 were
overexpressed in >35% of schwannomas versus <10% of
meningiomas. By contrast, CTNB1 was upregulated in the
majority of meningiomas but only in a few schwannoma tumors.
Significant differences were noted for three additional genes.
NCAM1 was overexpressed in 30% of schwannomas and
downregulated in a few meningioma tumors. CD44 and
ITGA3 were underexpressed in meningiomas and over-expressed
in schwannomas. FOS, ICAM1 and FLT1 were
underexpressed and ITGAV was upregulated in the two tumor
types.
 | Table II.Genes with differing trends between
meningiomas and schwannomas. |
Table II.
Genes with differing trends between
meningiomas and schwannomas.
| Expression | Gene | Meningioma (%) | Schwannoma (%) |
|---|
| Downregulation | BIRC5 | ∼30 | <10 |
| JUN | ∼30 | <10 |
|
SERPINE1 | ∼30 | <10 |
| EGFR | <10 | 40 |
| CD44 | 28 | - |
| ITGA3 | 36 | - |
| NCAM | <10 | - |
| Upregulation | PIK3R1 | <10 | >30 |
| ITGA4 | <10 | >30 |
| ITGA6 | <10 | >30 |
| CTNB1 | 28 | <10 |
| CD44 | - | 13 |
| ITGA3 | - | 30 |
| NCAM | - | 30 |
 | Table III.Genes showing a similar trend between
meningiomas and schwannomas. |
Table III.
Genes showing a similar trend between
meningiomas and schwannomas.
| Expression | Gene | Meningioma (%) | Schwannoma (%) |
|---|
| Downregulation | FOS | 90 | 65 |
| ICAM1 | 71 | 61 |
| FLY1 | 78 | 48 |
| Upregulation | ITCAV | 66 | 61 |
The present results thus complete the previous data
to classify gene expression in neurogenic neoplasms (17). In addition, the findings are
associated with the characterization of genes that are abnormally
expressed in meningiomas, as described previously, and are
concurrent with 1p and 14q deletions. However, the deregulated
genes are not always located in those specific genomic regions
(18). The findings from the
present study show that apoptosis, angiogenesis, cell cycle control
and DNA damage repair pathways are commonly deregulated in
meningiomas.
In conclusion, the identification of genes that are
abnormally expressed in meningiomas and schwannomas may be useful
for determining common therapeutic target strategies, primarily in
NF2 patients who frequently present with the two types of
neoplasms.
Acknowledgements
This study was partially supported by
Fondo de Investigaciones Sanitarias, Ministerio de Ciencia e
Innovación, Spain, Grants PI-08/1849 and PI-10/1972; and by grant
PI-10-045 from the Fundación Sociosanitaria de Castilla-La Mancha,
Spain.
References
|
1.
|
Perry A, Louis DN, Scheithauer BW, et al:
Meningiomas. Pathology and Genetics of Tumours of the Nervous
System. World Health Organization Classification of Tumors.
Kleihues P and Cavenee WK: 2nd edition. IARC Press; Lyon, France:
pp. 164–172. 2007
|
|
2.
|
Stemmer-Rachaminov AO, Wiestler OD and
Louis DN: Neurofibromatosis type 2. Pathology and Genetics of
Tumours of the Nervous System. World Health Organization
Classification of Tumors. Kleihues P and Cavenee WK: 2nd edition.
IARC Press; Lyon, France: pp. 210–214. 2007
|
|
3.
|
Bello MJ, de Campos JM, Kusak ME, et al:
Allelic loss at 1p is associated with tumor progression of
meningiomas. Genes Chromosomes Cancer. 9:296–298. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Weber R, Boström J, Wolter M, et al:
Analysis of genomic alterations in benign, atypical, and anaplastic
meningiomas: toward a genetic model of meningioma progression. Proc
Natl Acad Sci USA. 94:14719–14724. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Leone PE, Bello MJ, de Campos JM, et al:
NF2 gene mutations and allelic status of 1p, 14q, and 22q in
sporadic meningiomas. Oncogene. 18:2231–2239. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Mendiola M, Bello MJ, Alonso J, et al:
Search for mutations of the hRAD54 geen in sporadic meningiomas
with deletion at 1p32. Mol Carcinog. 24:300–304. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Lomas J, Bello MJ, Arjona D, et al:
Analysis of p73 gene in meningiomas with deletion at 1p. Cancer
Genet Cytogenet. 129:88–91. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Bello MJ, Amiñoso C, Lopez-Marin I, et al:
DNA methylation of multiple promoter-associated CpG islands in
meningiomas: relationship with the allelic status at 1p and 22q.
Acta Neuropathol. 108:413–421. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Liu Y, Pang JC, Dong S, et al: Aberrant
CpG island hyper-methylation profile is associated with atypical
and anaplastic meningiomas. Hum Pathol. 36:416–425. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Lomas J, Amiñoso C, Gonzalez-Gomez P, et
al: Methylation status of TP73 in meningiomas. Cancer Genet
Cytogenet. 148:148–151. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Lomas J, Bello MJ, Arjona D, et al:
Genetic and epigenetic alteration of the NF2 gene in sporadic
meninigomas. Genes Chromosomes Cancer. 42:314–319. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Pérez-Magán E, Rodríguez de Lope A,
Ribalta T, et al: Differential expression profiling analyses
identifies downregulation of 1p, 6q, and 14q genes and
overexpression of 6p histone cluster 1 gene as markers of
recurrence in meningiomas. Neuro Oncology. 12:1278–1290. 2010.
|
|
13.
|
Tseng KY, Chung MH, Sytwu HK, et al:
Osteopontin expression is a valuable marker for prediction of
short-term recurrence in WHO grade I benign meningiomas. J
Neurooncol. 100:217–223. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Aarhus M, Lund-Juhansen M and Knappskog
PM: Gene expression profiling of meningiomas: current status after
a decade of microarray-based transcriptomic studies. Acta Neurochir
(Wien). 153:447–456. 2011. View Article : Google Scholar
|
|
15.
|
Pérez-Magán E, Campos-Martín Y, Mur P, et
al: Genetic alterations associated with progression and recurrence
in meningiomas. J Neuropathol Exp Neurol. 71:882–893.
2012.PubMed/NCBI
|
|
16.
|
Sayagués JM, Tabernero MD, Maillo A, et
al: Microarray-based analysis of spinal versus intracranial
meningiomas: different clinical, biological, and genetic
characteristics associated with distinct patterns of gene
expression. J Neuropathol Exp Neurol. 65:445–454. 2006.
|
|
17.
|
Martinez-Glez V, Franco-Hernandez C,
Alvarez L, et al: Meningiomas and schwannomas: Molecular subgroup
classification found by expression arrays. Int J Oncol. 34:493–504.
2009.PubMed/NCBI
|
|
18.
|
Martínez-Glez V, Alvarez L,
Franco-Hernández C, et al: Genomic deletions at 1p and 14q are
associated with an abnormal cDNA microarray gene expression pattern
in meningiomas but not in schwannomas. Cancer Genet Cytogenet.
196:1–6. 2010.PubMed/NCBI
|
|
19.
|
Ragel BT and Jensen RL: Aberrant
signalling pathways in meningiomas. J Neurooncol. 99:315–324. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Torres-Martín M, Martinez-Glez V,
Peña-Granero C, et al: Expression analysis of tumor-related genes
involved in critical regulatory pathways in schwannomas. Clin
Transl Oncol. 15:409–411. 2013.
|