Introduction
Mitochondrial transcription factor A (TFAM), a
member of the high mobility group (HMG) box protein family, is
required for mitochondrial DNA (mtDNA) replication and
transcription. HMG proteins are often overexpressed in cancer cells
and are involved in apoptosis (1).
Furthermore, mitochondria are critical for cancer cell metabolism.
Mitochondrial uncoupling regulates the metabolic shift to aerobic
glycolysis, which is termed the Warburg Effect and is essential for
the survival and proliferation of cancer cells (2,3). As a
result, mitochondria are involved in the regulation of cancer cell
survival and growth.
Bladder cancer is the most prevalent form of cancer
among males. Urinary bladder cancers are detected in 3.71% of
elderly males in the USA, indicating that the cancer is associated
with aging and oxidative stress (4). The high expression of TFAM has been
reported to be related with a poor prognosis in multiple malignant
tumors, including ovarian cancer, endometrial carcinoma and colon
cancer (5–7). TFAM is expressed not only in
mitochondria, but also in nuclei (7). Furthermore, as well as being a
transcription factor in mitochondria, TFAM also regulates the
expression of nuclear genes. It has been reported that the number
of mitochondria correlates with the growth rate of cancer cells and
that the TFAM protein multimerizes and binds to mtDNA, suggesting
that the TFAM levels may be increased in cancer cells and be
associated with the malignant progression and proliferative
activity (8). However, the roles of
TFAM have not been fully identified in cancer cells.
microRNAs (miRNAs) are small, endogenous and
non-coding RNAs that inhibit gene expression via the interaction
with target sites in the 3′-untranslated regions (UTRs) of mRNA
(9). miRNAs play significant roles
in regulating multiple biological processes, including the
pathogenesis of a variety of human cancers (10–12).
miR-590-3p has been reported to be involved in mediating the
expression of autoimmune genes and neuronal death (13,14).
However, whether or not miR-590-3p is associated with malignant
tumors remains unclear.
The present study aimed to elucidate the roles of
TFAM and miR-590-3p and their association in bladder cancer
cells.
Materials and methods
Cell culture
The human bladder carcinoma 5637 cell line
(Institute of Cell Biology, Chinese Academy of Sciences, Shanghai,
China) was cultured in RPMI-1640 medium with 10% FBS and 1%
penicillin/streptomycin at 37°C, with 5% CO2.
RNA extraction and quantitative (q)PCR
analysis
RNA was extracted from the tissues or cell lines
using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to
the manufacturer’s instructions. The RNA integrity was then
assessed. A TaqMan qRT-PCR miRNA assay (Applied Biosystem,
Carlsbad, CA, USA) was performed to detect the mature miR-590-3p
expression levels. The relative expression of mature miR-590-3p
levels normalized to U6 endogenous control was determined using the
2−ΔΔCt method. Each measurement was performed in
triplicate. To detect the target genes (mRNA expression), 1
μg total RNA was reverse transcribed to cDNA using
SuperScript™ III First-Strand Synthesis SuperMix (Invitrogen). For
TFAM expression, SYBR Green qPCR Mix (Toyobo, Osaka, Japan) was
used. SYBR green q-PCR was performed using the Bio-Rad iQ5 PCR
detection system (Bio-Rad, Hercules, CA, USA) with the following
gene-specific primers: forward, 5′-AAAGATTCCAAGAAGCTAAGGGTG-3′ and
reverse, 5′-CCTAACTGGTTTCCTGTGCCTA-3′.
Western blotting
The cells were solubilized in cold RIPA lysis buffer
and then separated with 10% SDS-PAGE. Following this, the proteins
were transferred to PVDF membranes. The membranes were blocked in
5% skimmed dried milk in PBS for 3 h and then incubated overnight
with primary antibodies for TFAM (rabbit polyclonal), Akt (rabbit
polyclonal), p-Akt (rabbit polyclonal), matrix metalloproteinase
(MMP)-2 (mouse monoclonal), MMP-9 (rabbit polyclonal) (Abcam, San
Francisco, CA, USA), phosphatidylinositol-3-kinase (PI3K; rabbit
polyclonal), p-PI3K (rabbit polyclonal) and β-actin (mouse
monoclonal) (Abzoom Biolabs, Dallas, TX, USA). Subsequent to being
incubated with goat polyclonal anti-rabbit and goat anti-mouse
secondary antibodies (Abcam), the immune complexes were detected
using the enhanced chemiluminescence (ECL) method. The results were
visualized by autoradiography using preflashed Kodak XAR film
(Kodak, Tokyo, Japan).
MTT assays
The cells were plated at a density of 5,000 cells
per well. MTT (Promega, Madison, WI, USA) was added to the medium
at a final concentration of 0.5 μg/ml. The cells were then
incubated at 37°C with 5% CO2 for 3 h. The medium was
removed and 100 μl DMSO was added into each well. The plate
was gently rotated on an orbital shaker for 10 min to completely
dissolve the precipitation. The absorbance was detected at 570 nm
with a microplate reader (Bio-Rad).
Dual Luciferase reporter assays
To generate the reporter vectors bearing
miRNA-binding sites, a normal and mutated 3′-UTR of TFAM was
subcloned using PCR-based methods. The constructs were inserted
into the multiple cloning sites downstream of the luciferase gene
in the psiCHECK-2 lucif-erase miRNA expression reporter vector.
For the luciferase assay, 105 cells were
cultured to ∼70–80% conf luence in 24-well plates and cotransfected
with psiCHECK-2-TFAM-3′-UTR or psiCHECK2-mut-TFAM-3′-UTR vector
plus 50 nM miR-590-3p or 100 mM miR-590-3p inhibitor using
Lipofectamine 2000 (Invitrogen), according to the manufacturer’s
instructions. The cells were incubated with transfection
reagent/DNA complex for 5 h and refreshed with fresh medium
containing 10% FBS. At 48 h post-transfection, firefly and renilla
luciferase activities were evaluated using the dual-luciferase
reporter assay system (Promega) and the renilla luciferase activity
was normalized to firefly luciferase activity.
Plasmid construction and
transfection
TFAM, miR-590-3p and scramble miRNA [miR-SCR;
negative control (NC)] plasmids were obtained from Auragene Bio
(Changsha, China). The retroviral supernatants were prepared using
Eco-Phoenix packaging cells and the 5673 cells were transduced
using 20 mg/ml polybrene over 48 h.
Colony formation assay
The colony formation rate was measured by a plate
colony formation assay. In total, ∼200 cells were added to each
well of a 6-well plate. The plates were incubated at 37°C for 14
days and gently washed and stained with crystal violet. Viable
colonies that contained at least 50 cells were counted.
Flow cytometric analyses
For the apoptosis analysis, 2×105 cells
were collected, washed twice with PBS and resuspended in 400
μl 1X binding buffer. According to the manufacturer’s
instructions, 5 μl Annexin V-FITC and propidium iodide (PI)
solution was then added. The samples were incubated for 15 min at
room temperature and analyzed using flow cytometry (FACSCalibur;
Beckman Coulter, High Wycombe, UK).
For the cell cycle analysis, the cells were
collected in 1X PBS and resuspended in 70% ethanol to fix overnight
at −20°C. The cells were pelleted, washed twice in 3% BSA in 1X PBS
and pelleted again. The cells were resuspended and incubated for 30
min at room temperature in a PI staining buffer containing 3% BSA,
40 μg/ml PI and 0.2 mg/ml RNase in 1X PBS. DNA content
analyses were carried out using flow cytometry (FACSCalibur;
Beckman Coulter).
Transwell
For all the groups, migration was measured in
24-well Transwell chambers (Chemicon, Rosemont, IL, USA). Following
a 24-h incubation period at 37°C, the migrated cells were stained
with 0.04% trypan blue and counted with ×200 magnification.
Statistical analysis
The data are shown as the mean ± SD of at least
three determinations. The two-sided Student’s t-test was used to
analyze the differences in the experiments. P<0.05 was
considered to indicate a statistically significant difference. All
statistical analyses were conducted using SPSS 17.0 (SPSS, Inc.,
Chicago, IL, USA).
Results
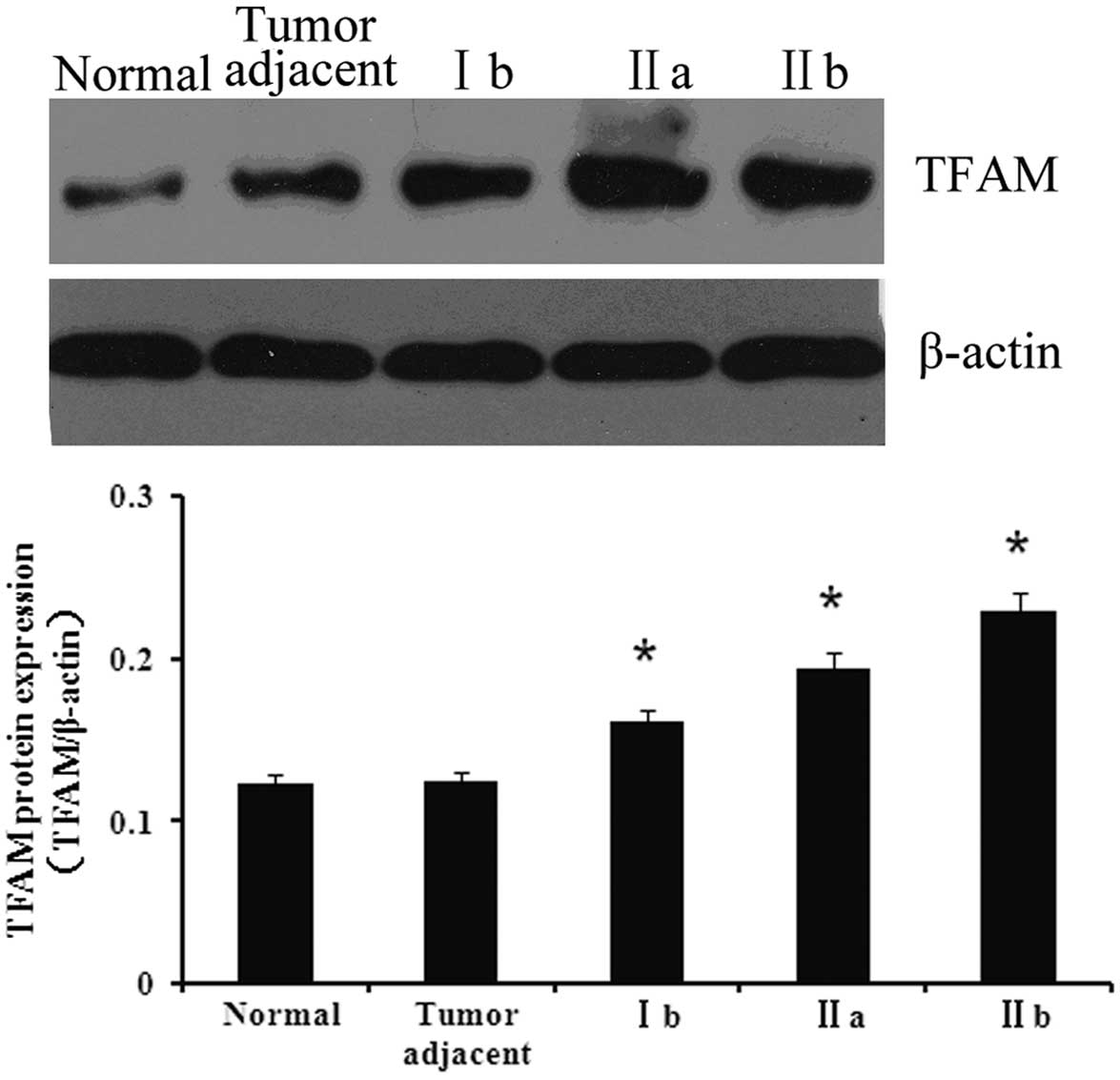
Protein expression of TFAM in the various
tissues
To investigate the association between TFAM and
bladder cancer, the expression of TFAM was first detected in
normal, adjacent and bladder cancer tissues. As shown in Fig. 1, the expression of TFAM in the
bladder cancer tissues was significantly higher compared with the
normal and adjacent tissues (P<0.05). Furthermore, the tissues
of the various cancer stages showed differing expression levels of
TFAM. The expression of TFAM was at its highest in stage IIb
tissues, whereas its expression in stage Ib tissues was the
lowest.
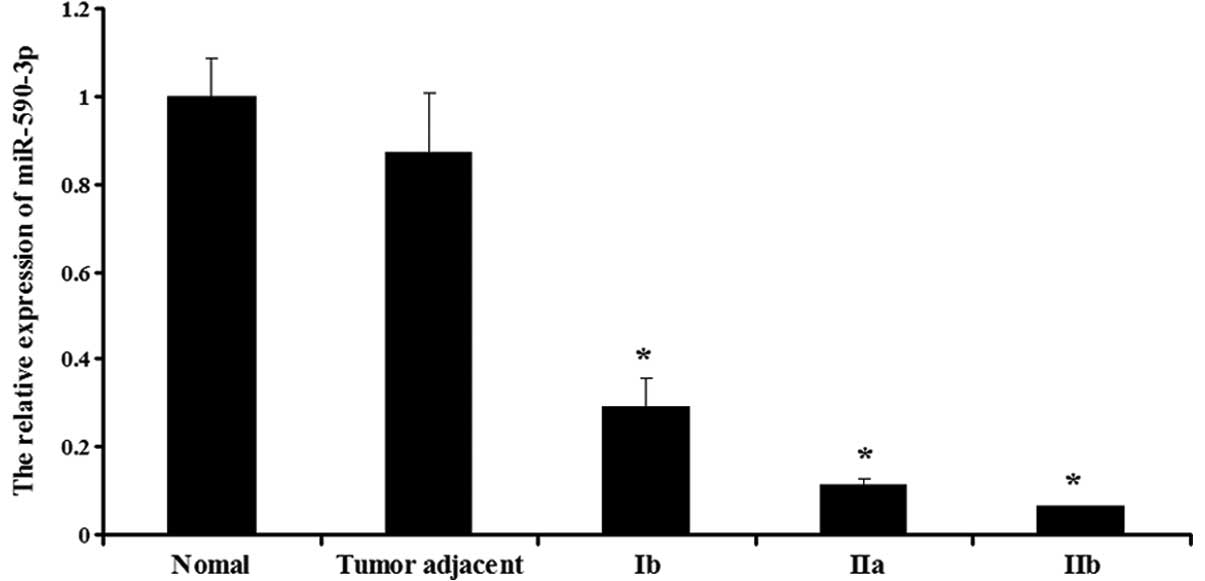
Expression of miR-590-3p in the various
tissues
The expression of miR-590-3p was then determined in
the normal, adjacent and bladder cancer tissues. As shown in
Fig. 2, the bladder cancer tissues
showed a lower expression of miR-590-3p compared with the normal
and adjacent tissues (P<0.05). Furthermore, miR-590-3p displayed
different expression levels in the various stages of bladder
cancer. The expression in the stage Ib tissues was the highest,
whereas the expression in the stage IIb tissues was the lowest
(P<0.05). However, in the normal and adjacent tissues, the
expression of miR-590-3p was not significantly different
(P>0.05).
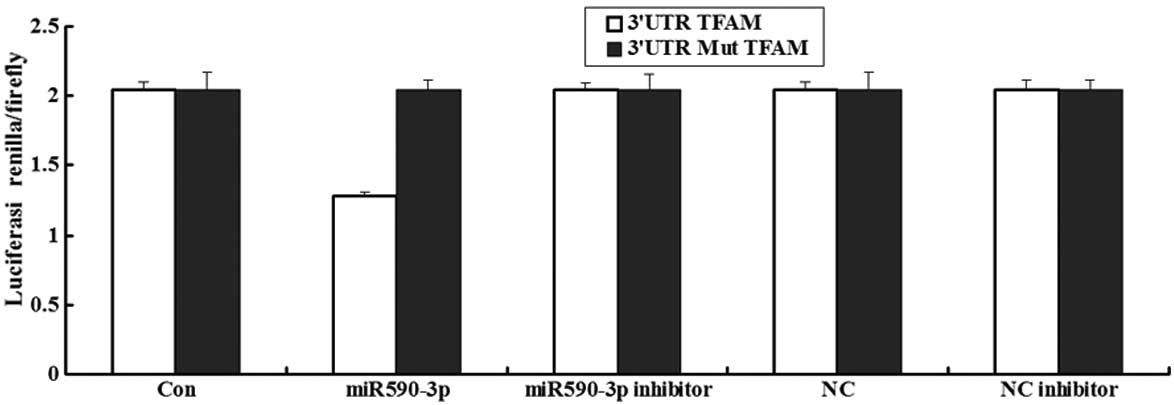
Luciferase assay of miR-590-3p
A luceriferase assay was performed to detect if TFAM
was the direct target of miR-590-3p. The data showed that the
renilla/firefly value of luciferase was significantly lower in the
miR-590-3p treatment cells following transfection with the 3′UTR of
the TFAM gene, while the renilla/firefly value of luciferase showed
no difference following transfection with the mutated 3′UTR of TFAM
compared with the control (Fig. 3).
These data suggested that miR-590-3p may downregulate TFAM gene
expression and that the 3′UTR of TFAM is the target of
miR-590-3p.
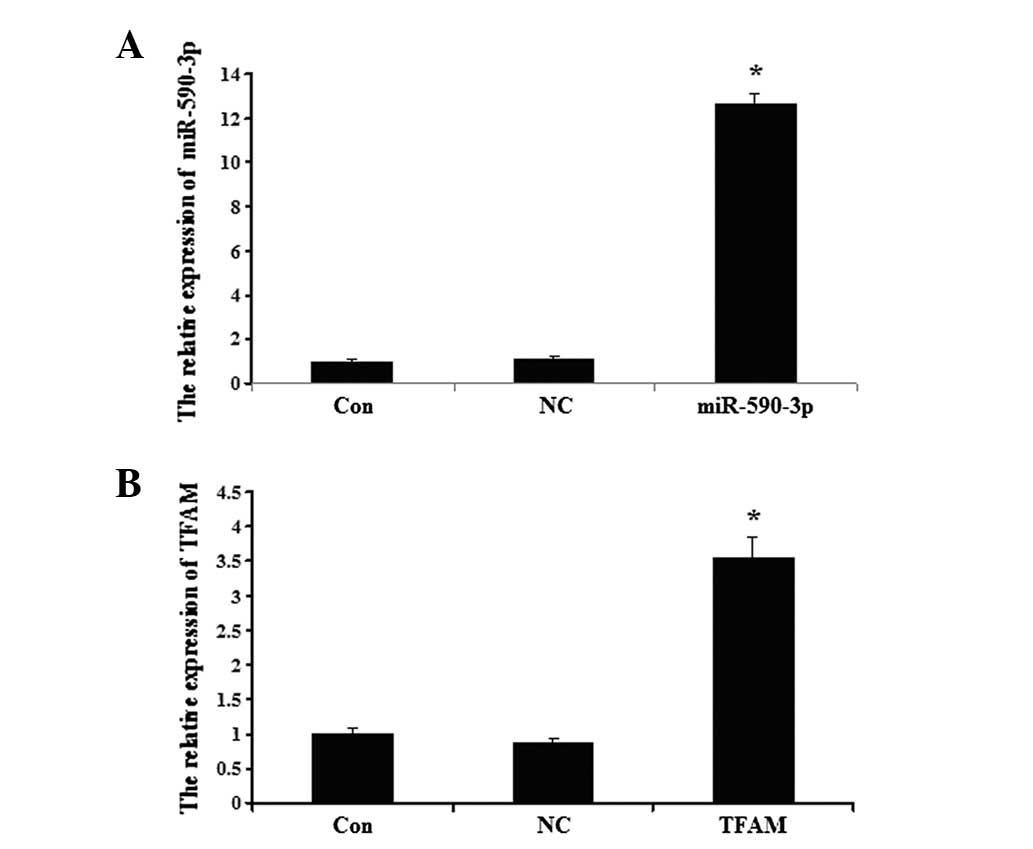
Detection of miR-590-3p and TFAM
following transfection
The data showed that the miR-590-3p expression level
was significantly higher in the 5637 cells that were transfected
with miR-590-3p lentiviral vectors compared with the controls
(P<0.05; Fig. 4A). As shown in
Fig. 4B, the expression of TFAM in
the 5637 cells was upregulated following transection with the TFAM
overexpression plasmid compared with the controls (P<0.05;
Fig. 4B). No significant
differences were observed between the expression of TFAM in the
control cells and the cells that were transfected with the NC virus
(P>0.05).
Effect of miR-590-3p and TFAM
overexpression on the proliferation of 5637 cells
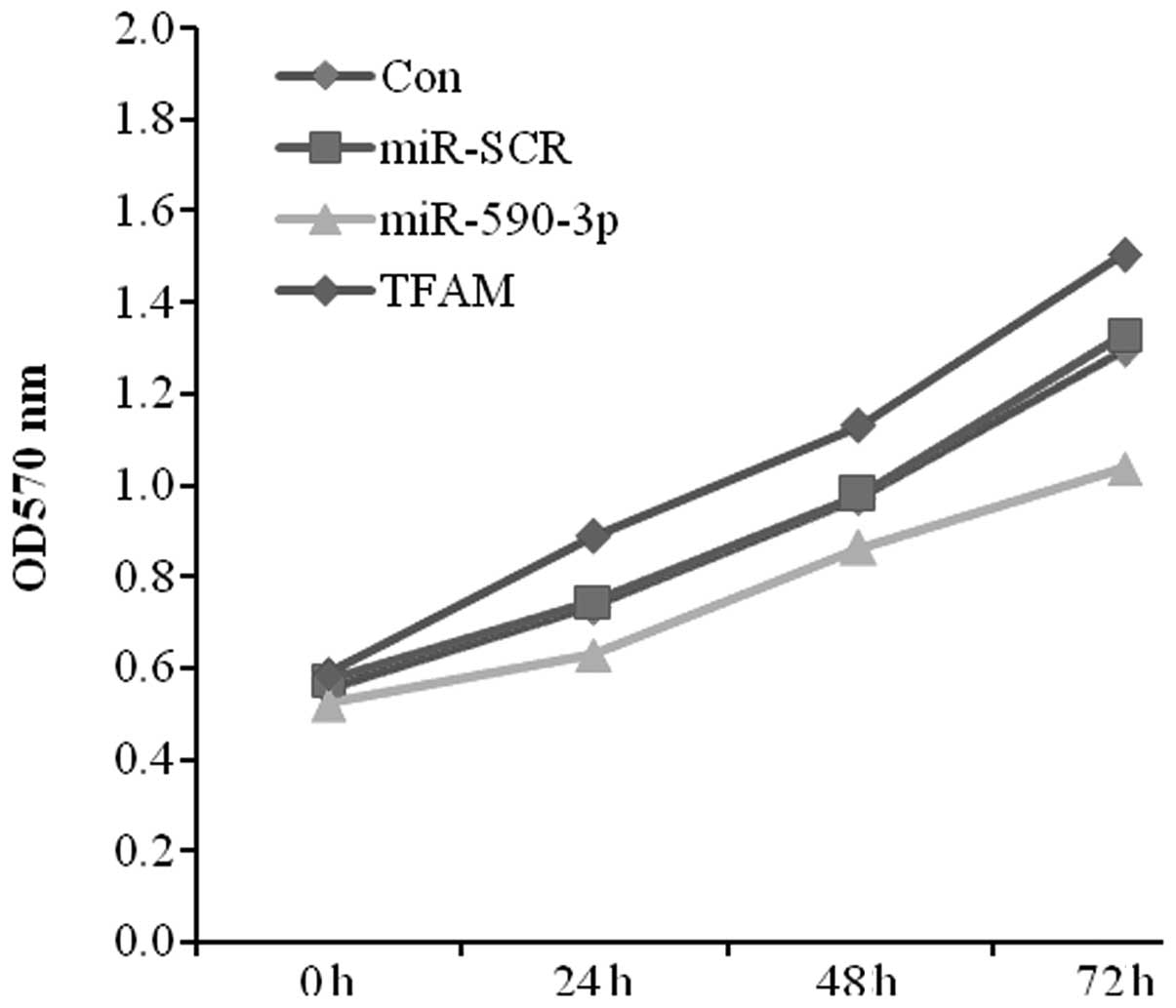
As the expression level of miR-590-3p was lower in
the bladder cancer tissues and the expression of TFAM was higher,
the effect of the transfection of miR-590-3p and TFAM on 5637 cell
proliferation was studied using MTT assays. The data showed
significant cell growth inhibition in the miR-590-3p transfectant
compared with the control from the 5367 cell lines. In contrast,
the TFAM transfectant displayed a positive effect on cell growth
(Fig. 5).
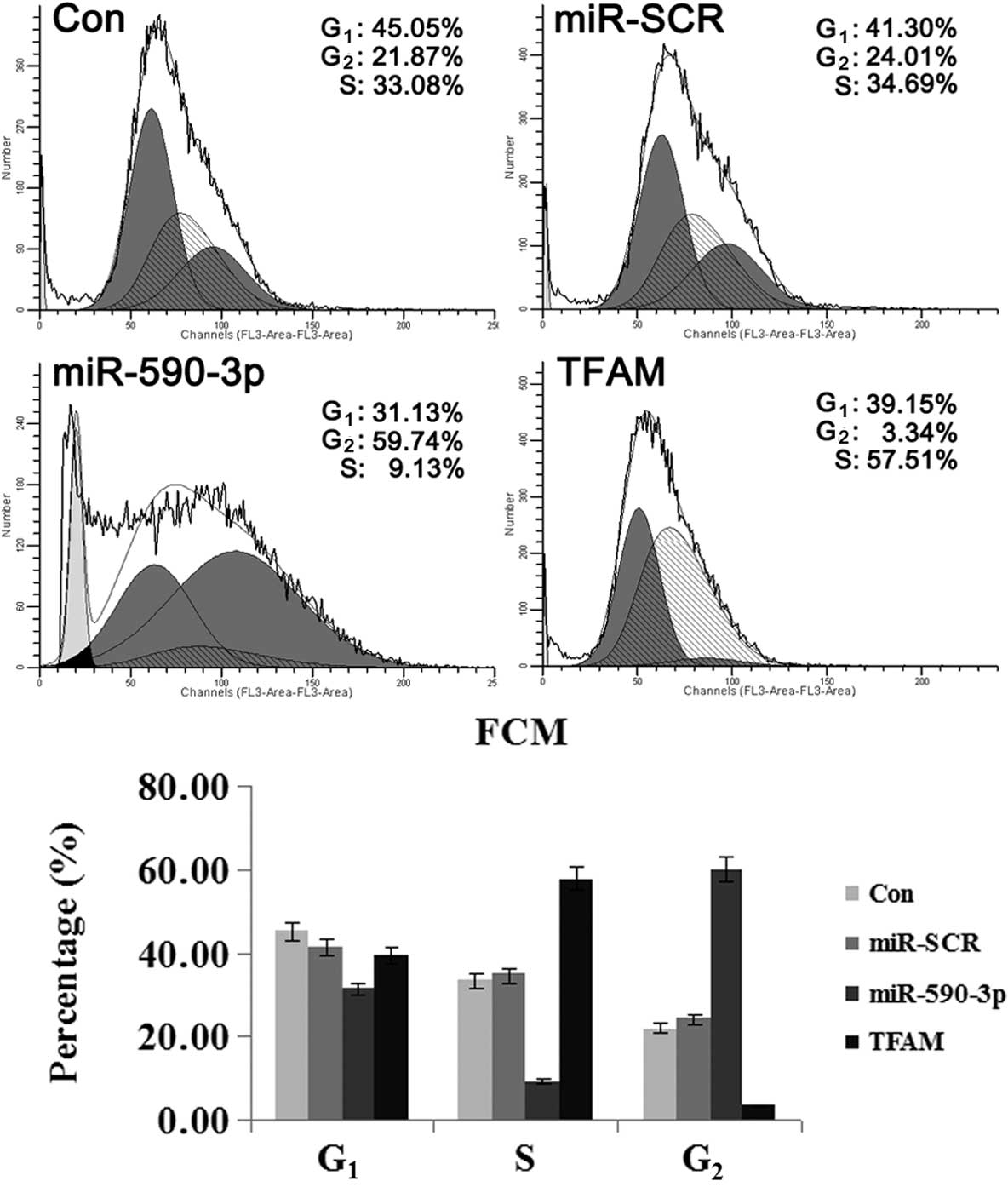
Effect of miR-590-3p and TFAM
overexpression on the cell cycle of 5637 cells
As shown in Fig. 6,
the cells that were transfected with the various vectors showed
differences in the percentage of cells in the varying phases of the
cell cycle. The cells that were transfected with miR-590-3p
lentiviral vectors showed the highest percentage of cells in the
G2 phase, suggesting that mitosis was blocked. For the
cells transfected with TFAM over-expression vectors, the majority
were in the G1 and S phases and only a few cells were in
the G2 phase, indicating that cell division was active.
These results indicated that TFAM promoted the mitosis of the 5637
cells, while miR-590-3p suppressed it.
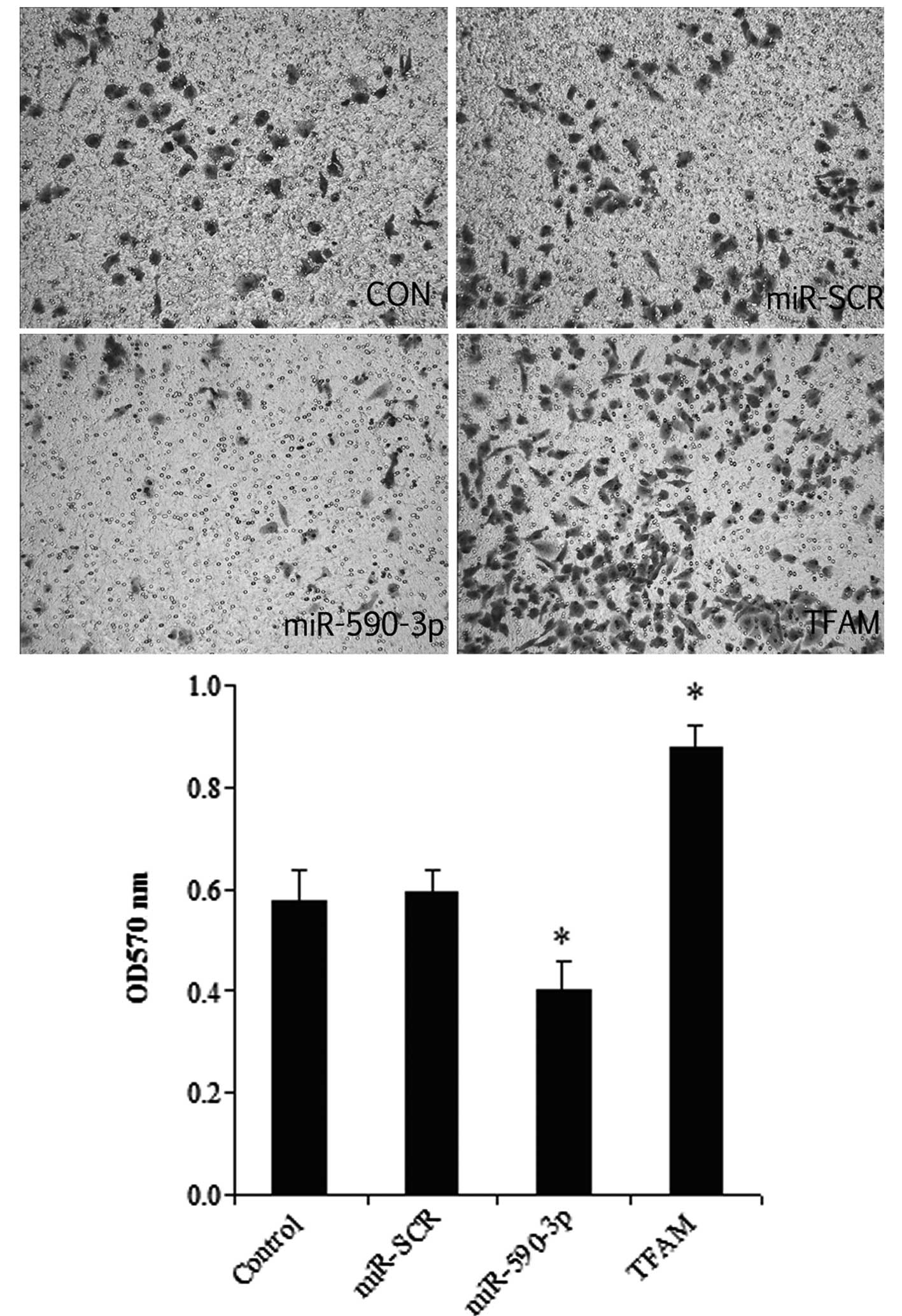
Effect of miR-590-3p and TFAM
overexpression on the migration of 5637 cells
As shown in Fig. 7,
the overexpression of TFAM significantly promoted cell migration,
while the miR-590-3p groups had the lowest migration level among
all the groups. In addition, a histological analysis revealed no
significant difference between the control and miR-SCR groups.
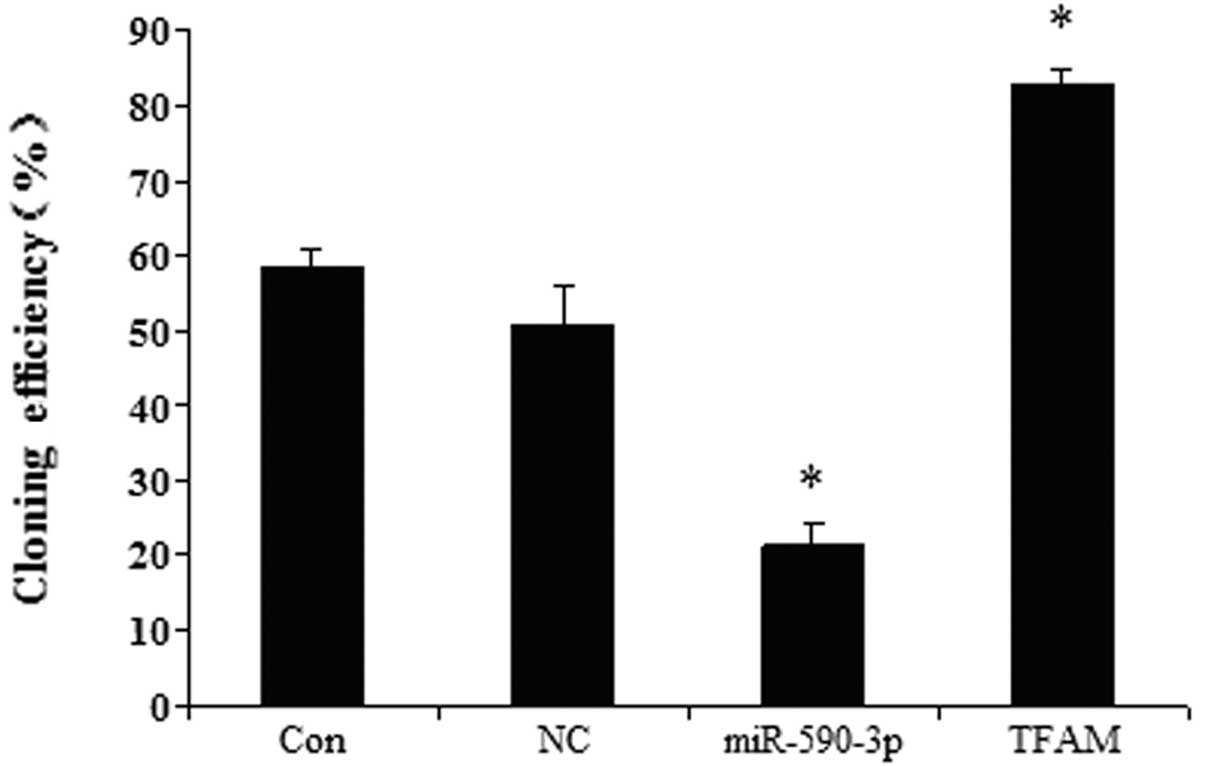
Effect of miR-590-3p and TFAM
overexpression on the colony-formation efficiency of the 5637
cells
The effects of TFAM and miR-590-3p on the
colony-formation efficiency were examined in the human bladder 5637
cell line. Fig. 8 shows that the
cells transfected with the different vectors had different
colony-formation efficiencies. The cells that were transfected with
the miR-590-3p lentiviral vectors showed the lowest
colony-formation efficiency. The cells that were transfected with
the TFAM overexpression vectors demonstrated the highest
colony-formation efficiency. The control and miR-SCR groups showed
no significant difference.
Effect of miR-590-3p and TFAM on the
expression of proliferation- and migration-related genes
The protein expression of certain proliferation- and
migration-related genes was detected in the 5637 cells, which were
transfected with the miR-590-3p or TFAM vectors, respectively. As
demonstrated in Fig. 9, the western
blot analysis indicated that the expression of PI3K, p-PI3K, Akt,
p-Akt, MMP2 and MMP9 was decreased in the cells that were
transfected with miR-590-3p lentiviral vectors, while the
expression was increased in the cells that were transfected with
TFAM overexpression vectors when compared with the controls
(P<0.05). No significant difference in the gene expression was
identified between the cells that were transfected with NC and the
control cells. (P>0.05).
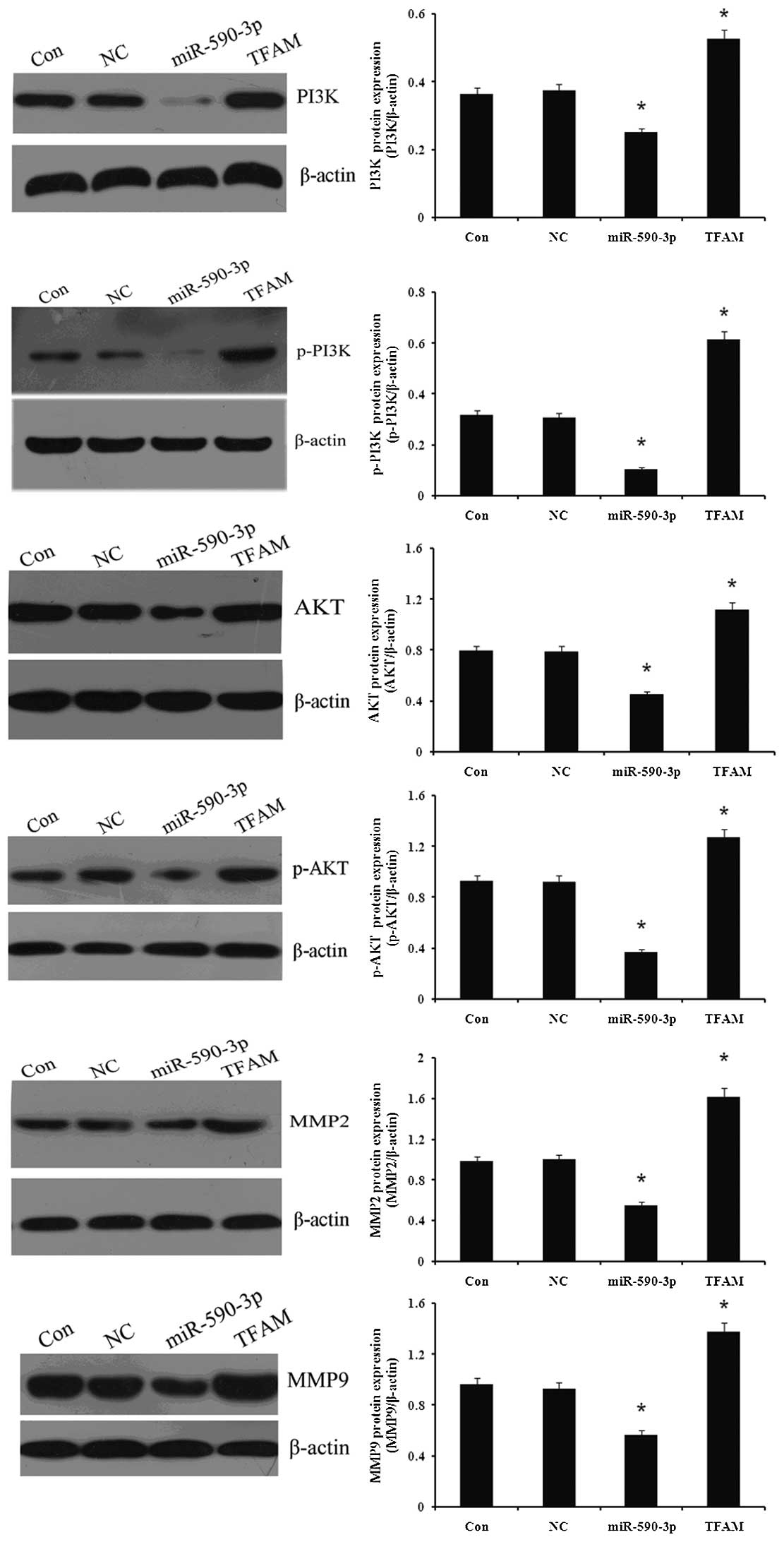 | Figure 9.Effect of miR-590-3p and TFAM
overexpression on proliferation- and migration-related genes
expression in 5637 cells. Western blotting was used to determine
the effect of miR-590-3p and TFAM overexpression on proliferation-
and migration-related genes expression in 5637 cells. Protein
expression levels of TFAM, PI3K, p-PI3K, Akt, p-Akt, MMP2 and MMP9
were examined and β-actin was used as an internal reference. Con,
normal 5637 cells; NC, cells transfected with NC virus; miR-590-3p,
5637 cells transfected with miR-590-3p lentiviral vectors; TFAM,
5637 cells transfected with mitchondrial transcription factor A
(TFAM) lentiviral vectors. *P<0.05 vs. control. miR,
microRNA; PI3K, phosphotidite-3-kinase; MMP, matrix
metalloproteinase; NC, negative control. |
Discussion
miRNAs represent a class of snRNAs that have central
roles in gene silencing and function as part of large gene
regulatory networks (15). In
animals, miRNAs hybridize to partially complementary binding sites
that are typically located in the 3-UTR of target mRNAs and repress
their expression (16). Recently,
miRNAs were identified to have significant roles in tumorigenesis
(15). Previous studies have
demonstrated that certain miRNAs are significantly deregulated in
bladder cancer and may function as tumor suppressors (17,18).
However, the role of miRNA-159-3p in the development of bladder
cancer remains unknown. The present study reported that the
expression of miRNA-159-3p was decreased in bladder cancer tissues
compared with normal and adjacent tissues. Furthermore, the data
showed that the higher the cancer grade, the lower the miRNA-159-3p
expression in the bladder cancer tissues.
TFAM was first purified and cloned as a
transcription factor for mtDNA. TFAM is able to enhance mtDNA
transcription using mitochondrial RNA polymerase in a
promoter-specific fashion (19).
Since the replication of mammalian mtDNA is proposed to be coupled
with transcription, TFAM is thought to be essential for the
replication of mtDNA (20). It has
been reported that TFAM may function to promote cell survival and
proliferation. TFAM-null mice have been demonstrated to possess an
embryonic lethal phenotype and exhibit apoptosis in heart cells
(21). The knockdown of TFAM
expression has been shown to induce p21-dependent G1
cell cycle arrest (22). Indeed,
the present data showed that in bladder cancer, the expression of
TFAM was significantly higher compared with the normal and adjacent
tissues, indicating that TFAM may be associated with the growth of
bladder cancer.
Notably, the expression of miRNA-159-3p and TFAM in
cancer tissues was shown to be negatively correlated, which
indicated that miRNA-159-3p may inhibit the expression of TFAM. The
results of the luciferase assay demonstrated that miRNA-159-3p was
able to directly downregulate TFAM expression, whereas the mutated
miRNA-159-3p did not. The present data demonstrated that TFAM is
the direct target of miRNA-159-3p. To the best of our knowledge,
there has been no earlier study reporting the interaction between
TFAM expression and these particular miRNAs. The cause of TFAM
overexpression is unknown, but one possible mechanism is through
the regulation by miRNAs. The loss of miRNA-159-3p, the endogenous
TFAM inhibitor, may promote the aberrant expression of TFAM,
contributing to the pathogenesis and progression of bladder
cancer.
Furthermore, the present study investigated the
molecular function of miR-590-3p and TFAM in the bladder cancer
5637 cell line. The MTT data showed that TFAM transfection
significantly promoted cell proliferation in the 5637 cells, while
miR-590-3p markedly decreased the level of cell proliferation. A
previous study demonstrated that TFAM in the colorectal carcinoma
cell line RKO carrying a TFAM truncating mutation suppressed cell
proliferation and inhibited RKO cell-induced xenograft tumor growth
(23). miR-590-3p suppressed the
growth rate of the 5637 cells in the present study. The cell cycle
results showed that the 5637 cells that were transfected with
miR-590-3p had the highest percentage of cells in the G1
phase and the lowest percentage of cells in the S and G2
phases. For the 5637 cells that were transfected with TFAM
overexpression vectors, the majority were in the S and
G2 phases and only a few were in the G1
phase. To detect the molecular regulatory pathway of miR-590-3p and
TFAM in the 5637 cells, western blotting was performed to determine
the protein expression of PI3K, p-PI3K, Akt, p-Akt, MMP2 and MMP9
after the forced overexpression of miR-590-3p or TFAM. TFAM
transfection significantly promoted PI3K, p-PI3K, Akt, p-Akt, MMP2
and MMP9 protein expression. In contrast, miR-590-3p showed
significant deregulation of protein expression of PI3K, p-PI3K,
Akt, p-Akt, MMP2 and MMP9. Metastasis is a complex and multi-step
dispersion process of malignant tumor cells from the primary tumor
site to a secondary site within the body. Therefore, the activation
of the zinc-binding endopeptidases, the MMPs, is considered to play
a crucial role in the process of cancer invasion and metastasis
(24). MMPs are primarily regulated
at the transcriptional level through AP-1 or NF-κB via
mitogen-activated protein kinase (MAPK) or PI3K-Akt pathways, at
post-transcriptional levels, at the protein level via their
activators or inhibitors or at their cell surface localization
(25–27). The overexpression of MMP-2 and MMP-9
in malignant tumors has been demonstrated to develop a vasculature
by angiogenesis (28). In the
present study, TFAM was shown to be able to promote this pathway.
miR-590-3p was able to downregulate TFAM expression. Thus, the
deregulatory effect of miR-590-3p on the pathway should be
associated with TFAM.
In summary, the present study identified miR-590-3p
and TFAM transfection expression patterns in bladder cancer. Using
the luciferase assay, TFAM was identified as a target of
miR-590-3p. The study was also extended into the molecular
functions of miR-590-3p and TFAM. miR-590-3p was able to deregulate
the metastasis pathway and may be used a new target for the therapy
of bladder cancer.
References
|
1.
|
Krynetskaia NF, Phadke MS, Jadhav SH and
Krynetskiy EY: Chromatin-associated proteins HMGB1/2 and PDIA3
trigger cellular response to chemotherapy-induced DNA damage. Mol
Cancer Ther. 8:864–872. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the Warburg effect: the metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009.PubMed/NCBI
|
|
3.
|
Ponisovskiy MR: Warburg effect mechanism
as the target for theoretical substantiation of a new potential
cancer treatment. Crit Rev Eukaryot Gene Expr. 21:13–28. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar
|
|
5.
|
Toki N, Kagami S, Kurita T, et al:
Expression of mitochondrial transcription factor A in endometrial
carcinomas: clinicopathologic correlations and prognostic
significance. Virchows Arch. 456:387–393. 2010. View Article : Google Scholar
|
|
6.
|
Yoshida Y, Hasegawa J, Nezu R, et al:
Clinical usefulness of mitochondrial transcription factor A
expression as a predictive marker in colorectal cancer patients
treated with FOLFOX. Cancer Sci. 102:578–582. 2011. View Article : Google Scholar
|
|
7.
|
Kurita T, Izumi H, Kagami S, et al:
Mitochondrial transcription factor A regulates BCL2L1 gene
expression and is a prognostic factor in serous ovarian cancer.
Cancer Sci. 103:239–444. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Tatarkova Z, Kuka S, Petráš M, et al: Why
mitochondria are excellent targets for cancer therapy. Klin Onkol.
25:421–426. 2012.PubMed/NCBI
|
|
9.
|
Li X, Zhang G, Luo F, et al:
Identification of aberrantly expressed miRNAs in rectal cancer.
Oncol Rep. 28:77–84. 2012.PubMed/NCBI
|
|
10.
|
He L, He X, Lim LP, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
12.
|
Volinia S, Calin GA, Liu CG, et al: A
microRNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Villa C, Fenoglio C, De Riz M, et al: Role
of hnRNP-A1 and miR-590-3p in neuronal death: genetics and
expression analysis in patients with Alzheimer disease and
frontotemporal lobar degeneration. Rejuvenation Res. 14:275–281.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Vinuesa CG, Rigby RJ and Yu D: Logic and
extent of miRNA-mediated control of autoimmune gene expression. Int
Rev Immunol. 28:112–138. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Hermeking H: MicroRNAs in the p53 network:
micromanagement of tumour suppression. Nat Rev Cancer. 12:613–626.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Papaconstantinou IG, Lykoudis PM, Gazouli
M, Manta A, Polymeneas G and Voros D: A review on the role of
microRNA in biology, diagnosis, and treatment of pancreatic
adenocarcinoma. Pancreas. 41:671–677. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Majid S, Dar AA, Saini S, et al:
MicroRNA-1280 inhibits invasion and metastasis by targeting ROCK1
in bladder cancer. PLoS One. 7:e467432012. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Nordentoft I, Birkenkamp-Demtroder K,
Agerbæk M, et al: miRNAs associated with chemo-sensitivity in cell
lines and in advanced bladder cancer. BMC Med Genomics. 5:402012.
View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Vernochet C, Mourier A, Bezy O, et al:
Adipose-specific deletion of TFAM increases mitochondrial oxidation
and protects mice against obesity and insulin resistance. Cell
Metab. 16:765–776. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Uchiumi T and Kang D: The role of
TFAM-associated proteins in mitochondrial RNA metabolism. Biochim
Biophys Acta. 1820:565–570. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Wallace DC and Fan W: The pathophysiology
of mitochondrial disease as modeled in the mouse. Genes Dev.
23:1714–1736. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Han B, Izumi H, Yasuniwa Y, et al: Human
mitochondrial transcription factor A functions in both nuclei and
mitochondria and regulates cancer cell growth. Biochem Biophys Res
Commun. 408:45–51. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Guo J, Zheng L, Liu W, et al: Frequent
truncating mutation of TFAM induces mitochondrial DNA depletion and
apoptotic resistance in microsatellite-unstable colorectal cancer.
Cancer Res. 71:2978–2987. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Groblewska M, Siewko M, Mroczko B and
Szmitkowski M: The role of matrix metalloproteinases (MMPs) and
their inhibitors (TIMPs) in the development of esophageal cancer.
Folia Histochem Cytobiol. 50:12–19. 2012. View Article : Google Scholar
|
|
25.
|
Rietz A and Spiers J: The relationship
between the MMP system, adrenoceptors and phosphoprotein
phosphatases. Br J Pharmacol. 166:1225–1243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Mannello F and Medda V: Nuclear
localization of matrix metal-loproteinases. Prog Histochem
Cytochem. 47:27–58. 2012. View Article : Google Scholar
|
|
27.
|
Bauvois B: New facets of matrix
metalloproteinases MMP-2 and MMP-9 as cell surface transducers:
outside-in signaling and relationship to tumor progression. Biochim
Biophys Acta. 1825:29–36. 2012.PubMed/NCBI
|
|
28.
|
Siefert SA and Sarkar R: Matrix
metalloproteinases in vascular physiology and disease. Vascular.
20:210–216. 2012. View Article : Google Scholar : PubMed/NCBI
|