Introduction
Breast cancer is the most significant worldwide
health problem in women >35–40 years of age. Additionally,
breast cancer accounts for ~1.35 million new cases and >450,000
cancer-related mortalities annually worldwide (1). Despite improved early detection,
treatment options and survival, the morbidity and mortality
continue to increase and numerous patients with invasive breast
cancer develop metastatic diseases that eventually lead to the
patient succumbing to their condition. Thus, there is an urgent
requirement to search for and identify novel biomarkers to predict
tumor recurrence and metastasis and to develop more novel treatment
strategies to effectively control aggressive breast cancers.
To this end, the present study focused on the
identification of biomarkers for breast cancer. One possible
biomarker of breast cancer is filamin A (FLNa), which is a
cytoskeletal protein with a molecular weight of 280 kDa (2,3) that
crosslinks actin filaments into orthogonal networks. FLNa also
interacts with >45 proteins and serves as the scaffold in
various signaling networks (4,5). The
FLNa protein, containing an integrin-β binding domain and an RAC1
binding domain, is localized in the edge of the cytoplasm, is able
to cross the membrane and may even appear in the nuclei (6,7). The
actin cytoskeleton is central to numerous cell functions, including
the maintenance of cell shape, cell division, adhesion, motility,
signal transduction and protein sorting. The alteration of FLNa
expression may contribute to cancer development and progression
(4,5,8,9), and
previous studies have identified FLNa overexpression in a number of
malignancies (10–12). For example, in lung cancer, FLNa
overexpression was shown to be associated with tumor metastasis
(11), and in melanoma,
FLNa-positive cells had higher migration and invasion abilities
compared with FLNa-negative tumor cells (12). In breast cancer, it was reported
that cyclin D1 interacted with the FLNa protein to affect the
migration and invasion potential of breast cancer cells (13). However, another study showed that
FLNa was able to regulate focal adhesion disassembly and suppress
breast cancer cell migration and invasion (14). The reason for this discrepancy or
conflict in the data is unclear. To date, there have been no
studies reporting FLNa expression in ex vivo breast cancer
tissue specimens; thus, the present study was proposed in order to
detect FLNa expression in breast cancer tissue samples and to
identify any associations between FLNa expression and
clinicopathological data. The present results may provide useful
information with regard to the function of FLNa in breast cancer
and the potential of FLNa as a biomarker to predict breast cancer
progression.
Materials and methods
Tumor tissue samples
Tissue samples were recruited from 96 consecutive
primary breast cancer patients who underwent surgical resection
between January 2008 and January 2010 at the Department of Breast
Surgery, First Hospital of Jilin University (Changchun, Jilin,
China). None of these patients received chemotherapy, radiotherapy
or immunotherapy prior to surgery. All the hematoxylin and
eosin-stained tissue sections were re-evaluated and confirmed by
two pathologists according to the World Health Organization
classifications (NCCN Breast Cancer Guidelines, version 1, 2013).
Of the 96 samples, 82 cases were classified as invasive breast
cancer and 14 as non-invasive cancer. Paraffinized and snap-frozen
tissues of distant normal breast tissue and 20 benign tumors were
also included in the study as controls. Normal skin tissues were
obtained from the healthy skin of the chest area of female patients
who underwent benign tumor resections and were used as a positive
control for FLNa expression. In addition, 30 cases of breast cancer
and distant non-tumor specimens were snap-frozen in liquid nitrogen
and stored at −80°C after resection for reverse transcription PCR
(RT-PCR) analysis. The institutional review board of Jilin
University approved the study and each patient signed a consent
form agreeing to their participation in the study.
Immunohistochemical staining
The formalin-fixed and paraffin-embedded tissue
sections were prepared for immunohistochemical analysis of FLNa
protein expression. Briefly, archival paraffin blocks were
retrieved and 3-μm thick tissue sections were prepared. For
immunohistochemistry, the sections were deparaffinized in xylene
and rehydrated in a series of graded ethanol solutions. The
sections were immersed in citrate buffer (0.01 mol/l citric acid,
pH 6.0) and heated for two 5 min intervals in a microwave oven for
antigen retrieval. Next, the sections were incubated with 0.3%
H2O2 for 15 min to block potential endogenous
peroxidase activity. Subsequent to the sections being rinsed in tap
water and washed with phosphate-buffered saline (PBS), the sections
were incubated with 20% normal goat serum for 30 min at room
temperature, then incubated with a monoclonal mouse anti-FLNa
antibody (Millipore, Bedford, MA, USA; 1:100) in PBS for 18 h at
4°C. The next day, the sections were washed with PBS three times,
then incubated with a goat anti-mouse polymer secondary antibody
(Zhongshan Goldenbridge Biotechnology Co., Ltd., Beijing, China)
for 30 min at room temperature. Next, DAB plus (Zhongshan
Goldenbridge Biotechnology Co.) was added to the sections once they
had briefly been washed with PBS and incubated for 5 min to
visualize the positive signal. Finally, the sections were
counterstained with hematoxylin. Normal skin tissue sections served
as the positive control, while normal mammary gland and breast
benign tumor tissues served as the negative controls. PBS was used
as a blank control instead of the first antibody. The stained
tissue sections were reviewed and scored under a light microscope:
the intensity of staining was scored as zero (no staining); 1+
(weak cytoplasmic staining in <10% of cells); 2+ (moderate
cytoplasmic staining in >10% of cells); and 3+ (marked
cytoplasmic staining in >10% of the cells). A score of 0 or 1
was considered to indicate a negative result for FLNa expression
(low), whereas scores of 2+ or 3+ were considered to show positive
(high) FLNa expression.
RT-PCR
An RT-PCR analysis was performed to detect FLNa mRNA
expression in the snap-frozen breast cancer and distant normal
breast tissues. Total RNA from these tissues was isolated using
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer’s instructions. After measuring the quantity and
quality using a spectrophotometer, the total RNA was subjected to
reverse transcription into cDNA with 1 μg RNA as a template, using
an RT kit (Fermentas, Glen Burnie, MD, USA) in a total volume of 20
μl. PCR amplification was then performed using primers derived from
the human FLNa sequence (5′-AGCCTCCACGAGACATCATC-3′ and
5′-CCAGTGTGT ACTCCCCCTTG-3′). The results were normalized to GAPDH,
which was used as an internal control. The primer sequences of
GAPDH were 5′-GGGTGATGCTGGTGCTGA GTATGT-3′ and
5′-AAGAATGGGAGTTGCTGTTGA AGTC-3′. A semi-quantitative determination
of the FLNa mRNA levels was achieved following 30 cycles of PCR
amplification. The PCR products were then analyzed using 1%
ethidium bromide agarose gel electrophoresis in comparison with the
DNA molecular weight marker (Takara, Dalian, China). Finally, a
quantitative analysis of the PCR target bands was performed using
Image J software (NIH, Bethesda, MD, USA).
Statistical analysis
SPSS software version 13.0 for Windows (SPSS Inc.,
Chicago, IL, USA) was used for the statistical analyses. Two-sample
t-tests or Mann-Whitney U tests, with respect to data distribution
in the measured data, were performed for comparisons between two
groups. χ2 tests were used for enumeration data.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Overexpression of FLNa protein in breast
cancer tissues
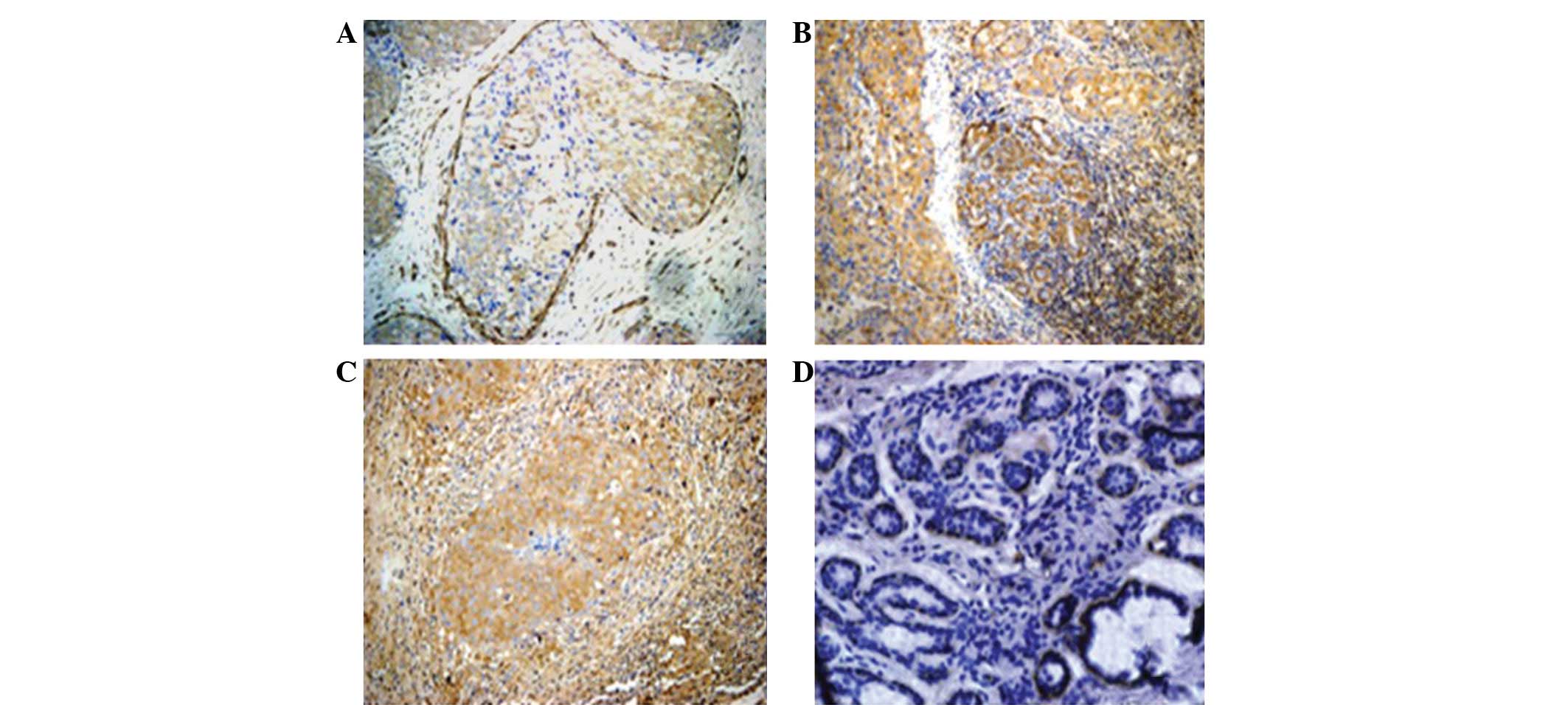
First, FLNa protein expression was assessed in the
breast cancer and distant normal and benign breast tumor tissue
specimens (Fig. 1). The data showed
that the FLNa protein was mainly detected in the cytoplasm of the
breast cancer cells, mostly at the edge of the cells and in the
basal cells or intercellular substance (Fig. 1 and 2).
The expression of the FLNa protein was detected in
63.54% of the breast cancer tissues, whereas the immunoreactivity
of the FLNa antibody was extremely low in certain distant normal
tissues. FLNa was undetectable in the most distant normal breast
tissues and benign tumor sections (Fig.
2). Furthermore, in the breast cancer samples, the positive
rate of FLNa staining was increased according to the tumor stage,
with 36, 66.7 and 84.6% in stage I, II and III breast cancer
tissues, respectively (P<0.05).
Association of FLNa protein expression
with clinicopathological features of patients with breast
cancer
Next, the associations between FLNa protein
expression and the clinicopathological features of the patients
with breast cancer were investigated. It was observed that the
expression of the FLNa protein was associated with TNM stage, lymph
node metastasis, vascular or neural invasion of the tumors,
menstruation state and other risk stratifications (P<0.05).
However, the expression of the FLNa protein was not associated with
any other clinicopathological features, including age, tumor size,
localization, histological type and the status of the estrogen and
progesterone receptors or the Her2/neu protein (P>0.05; Table I).
 | Table IAssociation of FLNa protein expression
with the clinicopathological characteristics of patients with
breast cancer. |
Table I
Association of FLNa protein expression
with the clinicopathological characteristics of patients with
breast cancer.
| Characteristic | n | +, n | −, n | χ2 | P-value |
|---|
| Age (years) |
| ≤35 | 5 | 4 | 1 | 3.154 | 0.207 |
| 35–55 | 68 | 45 | 23 | | |
| ≥55 | 23 | 11 | 22 | | |
| Tumor size (cm) |
| ≤2 | 44 | 23 | 21 | 4.76 | 0.120 |
| 2–5 | 49 | 34 | 15 | | |
| ≥5 | 3 | 3 | 0 | | |
| Tumor location |
| Left | 46 | 33 | 13 | 3.217 | 0.073 |
| Right | 50 | 27 | 23 | | |
| TNM stage |
| I | 25 | 9 | 16 | 13.360 | 0.010 |
| II | 45 | 30 | 15 | | |
| III | 26 | 22 | 4 | | |
|
Lymph-node-metastasis |
| Yes | 53 | 40 | 13 | 8.495 | 0.040 |
| No | 43 | 20 | 23 | | |
| Vascular/nerve
infiltration |
| Yes | 42 | 31 | 11 | 4.710 | 0.030 |
| No | 52 | 27 | 25 | | |
| Pathological
type |
| Invasive | 82 | 52 | 30 | 0.201 | 0.645 |
| Non-invasive | 14 | 8 | 6 | | |
| Histological
type |
| I | 3 | 2 | 1 | 0.998 | 1.000 |
| II | 47 | 29 | 18 | | |
| III | 19 | 12 | 7 | | |
| Others | 27 | 17 | 10 | | |
| Menstrual state |
|
Non-pausimenia | 60 | 42 | 18 | 4.313 | 0.038 |
| Pausimenia | 35 | 17 | 18 | | |
| ER |
| − | 32 | 21 | 11 | 0.316 | 0.574 |
| + | 62 | 37 | 25 | | |
| PR |
| − | 43 | 29 | 14 | 1.105 | 0.293 |
| + | 51 | 29 | 22 | | |
| CerB-b2 |
| 0 | 60 | 36 | 24 | 0.216 | 0.975 |
| 1+ | 3 | 2 | 1 | | |
| 2+ | 11 | 7 | 4 | | |
| 3+ | 20 | 13 | 7 | | |
| Risk |
| Low | 6 | 2 | 4 | 13.426 | 0.010 |
| Middle | 56 | 29 | 27 | | |
| High | 34 | 29 | 5 | | |
| MBNG |
| I | 10 | 6 | 4 | 0.725 | 0.867 |
| II | 49 | 32 | 17 | | |
| III | 25 | 14 | 11 | | |
| Others | 12 | 8 | 4 | | |
Differential expression of FLNa mRNA in
breast cancer and distant non-tumor breast tissues
To investigate whether FLNa expression was regulated
at the transcriptional level, a semi-quantitative RT-PCR analysis
of FLNa mRNA expression was performed on 30 cases of breast cancer
and distant normal tissues. After normalizing to the GAPDH mRNA
levels, the expression level of the FLNa mRNA was 0.634±0.53 in the
breast cancer tissues and 0.06±0.01 in the distant non-tumor breast
tissues (P<0.05; Fig. 3).
Discussion
In the present study, the differential expression of
FLNa mRNA and protein was analyzed in breast cancer tissue
specimens in order to provide ex vivo data on the potential
role of the FLNa protein in breast cancer. It was observed that the
FLNa protein was overexpressed in the breast cancer tissues
compared with the distant normal mammary gland and benign breast
tissues, and that this overexpression was associated with advanced
stages, lymph node metastasis and vascular or neural invasion of
breast cancer. Semi-quantitative RT-PCR data showed the
transcriptional regulation of FLNa overexpression in breast cancer.
The present data indicate that FLNa contributes to breast cancer
development and progression.
In the present study, FLNa expression was
demonstrated in the cytoplasm of the breast cancer cells, mainly at
the edge of the cells and in the basal cells or intercellular
substance. Until now, it was difficult to determine the association
between the function and the localization of FLNa. However, we
provide clinical evidence demonstrating that FLNa is frequently
overexpressed in breast cancer specimens.
Various other studies have demonstrated an
association between FLNa overexpression and tumor metastasis in
several types of cancer. For example, the expression of FLNa
protein was higher in hepatocellular carcinoma HCCLM9 cells with
high metastatic potential compared with hepatocellular carcinoma
MHCC97L cells with low metastatic potential (10). The overexpression of FLNa has been
associated with the invasion and metastasis of breast cancer
(15). Furthermore, FLNa has been
shown to affect the invasion and metastatic capacity of lung tumor
cells (11). FLNa was reported to
be essential for the locomotion of human melanoma cells, and the
suppression of FLNa expression inhibited melanoma cell migration
and induced apoptosis (12,16). The present study supported these
published data. However, two further studies showed contradicting
data. The first study by Zhong et al reported that the
knockdown of cyclinD1 expression suppressed breast cancer cell
invasion, which was associated with the downregulation of FLNa
protein phosphorylation (13). The
second study by Xu et al showed that FLNa suppressed the
migration and invasion capacity of breast cancer (14). The present ex vivo data
supported the hypothesis that FLNa expression is associated with
breast cancer development and progression. Structurally and
molecularly, the FLNa protein functions to crosslink actin
filaments into orthogonal networks and serves as the scaffold in
various signaling networks (4,5), which
in turn has a role in regulating cell shape, adhesion and motility.
During tumorigenesis, FLNa may regulate tumor cell invasion and
metastasis. Cancer metastasis is, however, a major obstacle for
cancer therapy and targeting it may effectively control advanced or
aggressive tumors. In addition, the development of novel biomarkers
to predict tumor progression, such as metastasis, may also improve
cancer survival and reduce fatalities. These biomarkers should
distinguish cancers with high metastatic potential from cancers
with less metastatic potential, thus optimizing individualized
therapeutic planning. However, whether the detection of FLNa
protein expression is likely to service as such a biomarker
requires further study and verification. Indeed, an additional
cohort of tissue samples is likely to aid in the confirmation of
the current data.
Previous studies have shown that FLNa expression is
positively associated with VEGF, an angiogenesis regulator, in lung
cancer (11). FLNa is an important
regulatory molecule in the TGF-β signal transduction pathways
(17) and is able to bind to SMAD2
to regulate actin polymerization reconciliation and interaction
with myosin via β1-integrin and RhoA GTPase. Kim et
al(18) reported that FLNa and
β1-integrin interacted together to mediate lung cancer
(A549) cell proliferation and prevent apoptosis. In addition, FLNa
was reported to interact with CEACAM1 (19), P311 (20) and FilGAP (21) to promote tumor cell migration. Ravid
et al(22) showed that
caveolin-1 expression in breast cancer MCF-7 cells upregulated FLNa
phosphorylation to induce MCF-7 cell migration through the PI3/AKT
pathway. The FLNa protein also interacted with the GTP-binding
protein R-Ras to promote metastasis of melanoma cells (23). Together, these findings and the
results of the present study clearly support the hypothesis that
FLNa is involved in tumor invasion and metastasis. However, it has
also been reported that FLNa is able to inhibit MMP-9 expression
through Ras/MAPK/ERK signaling to affect tumor cell invasion
(24). Thus, future studies should
further investigate the role of FLNa in human carcinogenesis and
cancer progression.
Acknowledgements
The authors would like to thank Medjaden Bioscience
Limited (Hong Kong, SAR, China) for assisting in the preparation of
the original manuscript.
References
|
1
|
Qiang S: The early diagnosis of breast
cancer. J Practical Med. 23:165–168. 2007.(In Chinese).
|
|
2
|
Wang K, Ash JF and Singer SJ: Filamin, a
new high-molecular-weight protein found in smooth muscle and
non-muscle cells. Proc Natl Acad Sci USA. 72:4483–4486. 1975.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang K: Filamin, a new
high-molecular-weight protein found in smooth muscle and nonmuscle
cells. Purification and properties of chicken gizzard filamin.
Biochemistry. 16:1857–1865. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Feng Y and Walsh CA: The many faces of
filamin: a versatile molecular scaffold for cell motility and
signalling. Nat Cell Biol. 6:1034–1038. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Popowicz GM, Schleicher M, Noegel AA and
Holak TA: Filamins: promiscuous organizers of the cytoskeleton.
Trends Biochem Sci. 31:411–419. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bachmann AS, Howard JP and Vogel CW:
Actin-binding protein filamin A is displayed on the surface of
human neuroblastoma cells. Cancer Sci. 97:1359–1365. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou X, Borén J and Akyürek LM: Filamins
in cardiovascular development. Trends Cardiovasc Med. 17:222–229.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Robertson SP, Twigg SR, Sutherland-Smith
AJ, et al: Localized mutations in the gene encoding the
cytoskeletal protein filamin A cause diverse malformations in
humans. Nat Genet. 33:487–491. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Keshamouni VG, Michailidis G, Grasso CS,
et al: Differential protein expression profiling by
iTRAQ-2DLC-MS/MS of lung cancer cells undergoing
epithelial-mesenchymal transition reveals a migratory/invasive
phenotype. J Proteome Res. 5:1143–1154. 2006. View Article : Google Scholar
|
|
10
|
Ai J, Huang H, Lv X, et al: FLNA and PGK1
are two potential markers for progression in hepatocellular
carcinoma. Cell Physiol Biochem. 27:207–216. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Uramoto H, Akyurek LM and Hanagiri T: A
positive relationship between filamin and VEGF in patients with
lung cancer. Anticancer Res. 30:3939–3944. 2010.PubMed/NCBI
|
|
12
|
Flanagan LA, Chou J, Falet H, Neujahr R,
Hartwig JH and Stossel TP: Filamin A, the Arp2/3 complex, and the
morphology and function of cortical actin filaments in human
melanoma cells. J Cell Biol. 155:511–517. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhong Z, Yeow WS, Zou C, et al: Cyclin
D1/cyclin-dependent kinase 4 interacts with filamin A and affects
the migration and invasion potential of breast cancer cells. Cancer
Res. 70:2105–2114. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu Y, Bismar TA, Su J, et al: Filamin A
regulates focal adhesion disassembly and suppresses breast cancer
cell migration and invasion. J Exp Med. 207:2421–2437. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu Y, Li J, Zhao R, Wang X, Shan B and Zhu
T: Expression of filamin A in invasive breast carcinoma and its
significance. Tumor. 29:659–662. 2009.(In Chinese).
|
|
16
|
Cunningham CC, Gorlin JB, Kwiatkowski DJ,
et al: Actin-binding protein requirement for cortical stability and
efficient locomotion. Science. 255:325–327. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang J, Fan Y, Guo L and Lu S: Search for
interacting proteins of esophageal cancer related gene-1 encoded
protein through the yeast two-hybrid system. Zhonghua Zhong Liu Za
Zhi. 24:219–221. 2002.(In Chinese).
|
|
18
|
Kim H, Sengupta A, Glogauer M and
McCulloch CA: Filamin A regulates cell spreading and survival via
beta1 integrins. Exp Cell Res. 314:834–846. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Klaile E, Muller MM, Kannicht C, Singer BB
and Lucka L: CEACAM1 functionally interacts with filamin A and
exerts a dual role in the regulation of cell migration. J Cell Sci.
118:5513–5524. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
McDonough WS, Tran NL and Berens ME:
Regulation of glioma cell migration by serine-phosphorylated P311.
Neoplasia. 7:862–872. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ohta Y, Hartwig JH and Stossel TP: FilGAP,
a Rho- and ROCK-regulated GAP for Rac binds filamin A to control
actin remodelling. Nat Cell Biol. 8:803–814. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ravid D, Chuderland D, Landsman L, Lavie
Y, Reich R and Liscovitch M: Filamin A is a novel
caveolin-1-dependent target in IGF-I-stimulated cancer cell
migration. Exp Cell Res. 314:2762–2773. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gawecka JE, Griffiths GS, Ek-Rylander B,
Ramos JW and Matter ML: R-Ras regulates migration through an
interaction with filamin A in melanoma cells. PLoS One.
5:e112692010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu TN, He HJ, Kole S, et al: Filamin
A-mediated down-regulation of the exchange factor Ras-GRF1
correlates with decreased matrix metalloproteinase-9 expression in
human melanoma cells. J Biol Chem. 282:14816–14826. 2007.
View Article : Google Scholar : PubMed/NCBI
|