Introduction
Gastric cancer is the fourth most common worldwide
cancer and the second most common cause of cancer-related mortality
in Asia and worldwide (1). In
China, the incidence and mortality rates of gastric cancer have
been recorded as 33.12 and 22.64, respectively, per 100,000
individuals (2). The prognosis of
gastric cancer is extremely poor, with a 5-year survival rate of
<30% (3). Human mitochondrial
DNA (mtDNA) is a 16.6 kb closed circular duplex species of 16,569
bases, encoding 13 polypeptides that are involved in respiration
and oxidative phosphorylation, in addition to 2 rRNAs and 22 tRNAs
that are essential for protein synthesis in the mitochondria
(4). In addition, mtDNA contains a
non-coding region called the displacement loop (D-loop), which
controls the replication and the transcription of mtDNA (4,5). DNA
hypermethylation occurs at the CpG islands and is typically
associated with gene silencing. Human mtDNA is present in high
levels at 103–104 copies per cell (6). The mitochondria content and mtDNA copy
number of an individual cell may vary with the type of cell and
tissue, and the number may also change during cell differentiation
and the aging process (7).
Alterations have been reported in the mtDNA copy number in a
variety of human carcinomas (8).
However, alterations in the mtDNA content in gastric cancer and the
corresponding non-tumorous gastric tissue remain elusive.
The present study determined the copy number of
mtDNA in gastric cancer tissues and corresponding non-tumorous
gastric tissues from male and female gastric cancer patients, in
order to evaluate the alteration in the quantity of mtDNA and to
reveal the correlation between the mtDNA copy number and the
clinicopathological stages of gastric cancer. A demethylation
experiment was performed on rat gastric mucosa cells to determine
if DNA methylation and demethylation is involved in the alteration
of the mtDNA copy number.
Materials and methods
Patients and specimens
A total of 76 gastric cancer tissues and
corresponding non-cancerous tissues were surgically resected from
patients at the West China Hospital (Sichuan University, Chengdu,
Sichuan, China) between 2009 and 2011. This study was approved by
the ethics committee of West China Hospital, Sichuan University
(Chengdu, China). Informed consent was obtained from all the
patients. Samples were obtained from 47 males and 29 females, aged
34–75 years (median, 51.9±8.7; Table
I). All the specimens were fresh-frozen and maintained in
liquid nitrogen immediately. The specimens of the tumor tissues
were cut from the edge of the tumors and the corresponding
non-cancerous tissues were collected from >5 cm away from the
tumors. The investigation conforms to the principles outlined in
the Declaration of Helsinki.
 | Table IClinical data of 76 gastric cancer
patients. |
Table I
Clinical data of 76 gastric cancer
patients.
| Characteristic | Gastric cancer
patients, n (%) | Age, years
(range) |
|---|
| Gender |
| Male | 47 (61.8) | 50 (27–71) |
| Female | 29 (38.2) | 53 (34–73) |
| Clinicopathological
stage |
| Male |
| I | 5 (6.6) | 49 (27–69) |
| II | 9 (11.8) | 47 (37–70) |
| III | 14 (18.4) | 52 (28–71) |
| IV | 19 (25.0) | 51 (37–70) |
| Female |
| I | 4 (5.3) | 50 (41–63) |
| II | 6 (7.9) | 48 (34–61) |
| III | 10 (13.2) | 56 (41–70) |
| IV | 9 (11.8) | 54 (39–73) |
DNA extraction and quantitative PCR
analysis
A quantitative PCR method was used to determine the
mtDNA copy number. The DNA specimens were prepared using the
TIANamp Genomic DNA kit (DP304; Tiangen Biotech Co., Ltd., Beijing,
China). The primers of β-actin (product length, 138 bp) were
forward, 5′-CGGGAAATCGTGCGTGACAT-3′ and reverse, 5′-GAA
GGAAGGCTGGAAGAGTG-3′. The PCR cycling conditions consisted of an
activation step at 95°C for 10 min, followed by 40 cycles for 25
sec at 95°C, 1 min at 45°C and 20 sec at 72°C. The primers of mtDNA
(product length, 487 bp) were forward, 5′-TACTCACCAGACGCCTCAACCG-3′
and reverse, 5′-TTA TCGGAATGGGAGGTGATTC-3′. The PCR cycling
conditions consisted of an activation step at 95°C for 10 min,
followed by 40 cycles for 25 sec at 95°C, 1 min at 35°C and 35 sec
at 72°C. All PCRs were performed on a 7900HT Fast Real-Time PCR
system (Applied Biosystems, Carlsbad, CA, USA) and each specimen
was run in triplicate. The average of all three measurements was
calculated. A negative control was included in each reaction.
Primary rat gastric mucosa cell culture
and demethylation experiment
Primary rat gastric mucosa cells were cultured as
previously described (9).
5-Aza-2′-deoxycytidine (5-Aza; Sigma, St Louis, MA, USA) was used
to induce mtDNA demethylation. Cells (5×105) were seeded
into 6-well plates in 2 ml medium. Following a 24-h incubation
period, the medium was removed and the cells were incubated in 2 ml
fresh medium containing a final concentration of 5 μM 5-Aza for 96
h. Following the treatment, the medium was removed and the cells
were subjected to an additional 24-h incubation.
Statistical analysis
The qualitative and quantitative changes in the
mtDNA were analyzed using SPSS version 15.0 (SPSS, Inc., Chicago,
IL, USA). The relative mtDNA copy numbers (mtDNA1/β-actin) of every
specimen were calculated and Student’s t-test was used to analyze
the difference in the mtDNA copy numbers between the tumor and the
corresponding non-cancerous tissues and also the difference between
genders. The quantitative data are expressed as the mean ± standard
deviation.
Results
Decreased relative mtDNA copy number in
gastric cancer
To identify the alteration in the mtDNA copy number
in the gastric cancer tumor tissues, the mtDNA copy number was
quantified in the 76 gastric cancer and corresponding non-cancerous
stomach tissues. Following the statistical analysis, the results
revealed that compared with the non-cancerous tissues, the relative
mtDNA copy numbers of the tumor tissues were markedly decreased
(Fig. 1B; P<0.05). The average
relative mtDNA copy numbers were 94.71±28.11 in the tumor tissues
and 111.67±21.84 in the corresponding non-cancerous tissues. No
significant difference was noted between the male and female
patients in the tumor and corresponding non-cancerous tissues
(Fig. 1C; P>0.05).
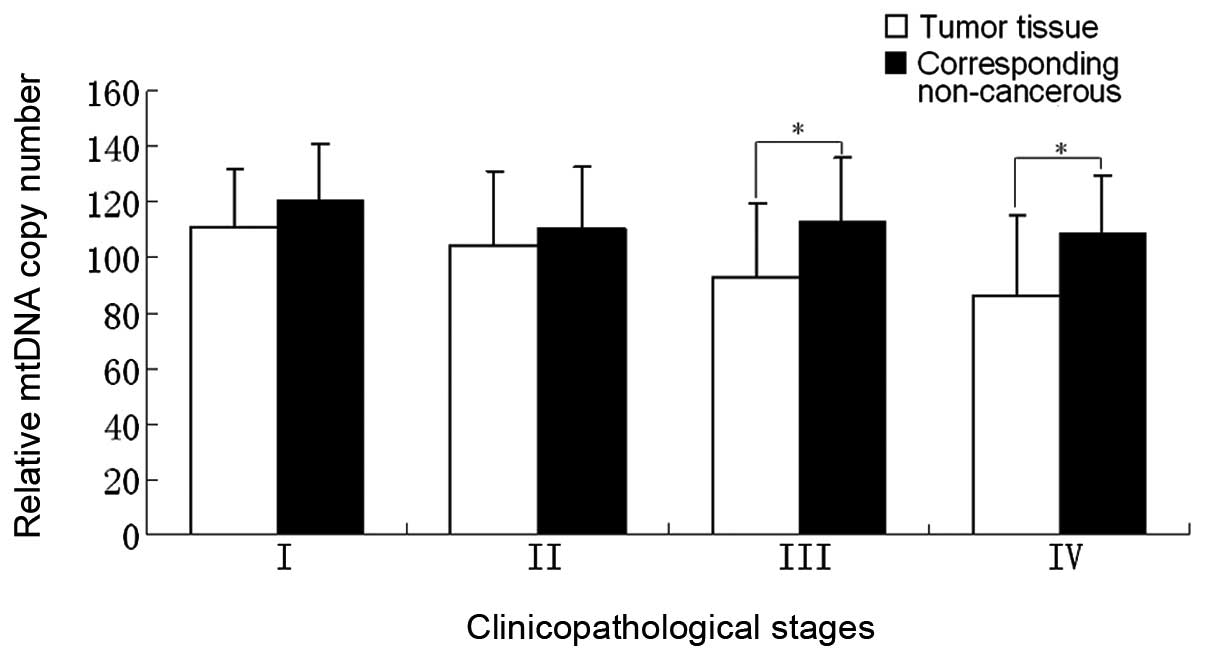
Correlation between relative mtDNA copy
number and clinicopathological stages
The correlation between the clinicopathological
stage and relative mtDNA copy number of 76 cancer cases was
analyzed and the average mtDNA copy number of each stage of the
tumor tissues and their corresponding non-cancerous tissues are
shown in Table II. From stage I to
stage IV, the relative mtDNA copy number of tumor tissues was lower
than that of the corresponding non-cancerous tissues in each stage
and these differences exhibited a marked difference in stages III
and IV (P<0.05). While in stages I and II there were no marked
differences (P>0.05; Fig. 2).
Stages I and II were defined as group 1 and stages III and IV as
group 2 (Table III). A marked
difference was noted between groups 1 and 2 with regard to the
relative mtDNA copy number between the tumor tissues (P<0.05).
However, no significant difference was observed between the
corresponding non-cancerous tissues (P=0.427).
 | Table IIRelative mtDNA copy number of the
various clinicopathological stages. |
Table II
Relative mtDNA copy number of the
various clinicopathological stages.
| Tissue type | Stage |
|---|
|
|---|
| I | II | III | IV |
|---|
| TT | 111±20.89 | 104.38±26.79 | 92.85±26.71 | 85.9±29.34 |
| CNT | 120.2±20.62 | 109.9±22.43 | 113.10±23.17 | 108.66±21.05 |
 | Table IIIAverage relative mtDNA copy number of
groups 1 (stages I and II) and 2 (stages III and IV). |
Table III
Average relative mtDNA copy number of
groups 1 (stages I and II) and 2 (stages III and IV).
| Tissue type | Group 1 | Group 2 |
|---|
| TT | 106.86±24.49a | 89.11±28.10 |
| CNT | 113.76±21.91b | 110.71±21.95 |
Demethylation treatment and increased
mtDNA copy number
Prior to and following the treatment with 5-Aza, the
primary rat gastric mucosal cells were collected for PCR analysis
of the mtDNA copy number. As shown in Fig. 3, the mtDNA copy number of rat
gastric mucosal cells was increased significantly subsequent to
being treated with 5-Aza (P<0.05).
Discussion
A growing number of experiments have revealed that
mtDNA content variations may affect the behaviors of malignant
cells, including cell growth, apoptosis, anticancer drug
sensitivity and the invasive and metastatic potentials (10). To date, the characterization of
mtDNA by utilizing the power of quantitative PCR assays has
revealed the quantitative abnormalities of the mtDNA content in a
multitude of human cancers. The alterations in the mtDNA copy
number in tumor specimens are usually retained within a relatively
stable range in comparison with those in the adjacent non-cancerous
tissues (11). The mtDNA content
increases in a large number of malignant tumors, including acute
lymphoblastic leukemia (12),
colorectal carcinoma (13,14) and esophageal squamous cell carcinoma
(15,16–18),
but decreases in numerous other tumors, including breast cancer
(19), hepatocellular carcinoma
(20) and Ewing’s sarcoma (21–23).
Several studies in colorectal carcinoma (24,25),
breast cancer (25) and lung cancer
(26) reported the positive
association between an increased mtDNA content in peripheral blood
specimens and an elevated cancer risk. Further studies on the
epigenetic alterations of mtDNA and its downstream products are
beneficial to understanding the function of mtDNA in malignancy
(27,28).
In the present study, quantitative PCR was performed
to measure the relative mtDNA copy number of the tumor and
corresponding non-cancerous tissues. Fig. 1A and B show the tendencies of the
relative mtDNA copy number between the tumor and corresponding
non-cancerous tissues in gastric cancer. The average mtDNA copy
numbers of the tumor and corresponding non-cancerous tissues
exhibited statistically significant differences.
Furthermore, the mtDNA copy numbers of the tumor and
corresponding non-cancerous tissues were compared individually,
between males and females. No significant difference was observed
between gender. Thus, it appeared that there was no gender effect
on the mtDNA copy number in gastric cancer (Fig. 1C).
Advanced gastric cancer is classified into Borrmann
types I–IV, with an increasing degree of malignancy (29). Gastric cancers of Borrmann types I
and type II are well-defined tumors, while type III and IV are
ill-defined tumors with little or no gland-forming capability. The
majority of the patients that have tumors of types III and IV have
a poor prognosis and low 5-year survival rates following gastric
resection (30). In nasal polyp
tissue, Park et al reported that the change in the mtDNA
copy number is correlated with tumor progression (31). The correlation between the relative
mtDNA copy number and clinicopathological stages was also analyzed
in the present study. Fig. 2 shows
that the differences in the relative mtDNA copy numbers between the
tumor tissues and the corresponding non-cancerous tissues of stages
III and IV are more evident than those of stages I and II. This
result is consistent with the phenomenon of the ‘Warburg effect’,
which indicates that cancer cells exhibit an enhanced generation of
ATP mainly by glycolysis, but not by oxidative phosphorylation
(32).
However, mtDNA also contains a non-coding region
called the D-loop, which controls the replication and transcription
of mtDNA (15,33). Our previous study on colorectal
cancer showed that the demethylation of the D-loop is an early
molecular event in colorectal cancer. The demethylation of the
D-loop may also have an effect on the expression of the ND1 gene,
which is encoded by mtDNA (28). In
the present study, a demethylation experiment was performed to
determine whether the change of mtDNA copy number was associated
with D-loop demethylation in gastric cancer. The results (Fig. 3) revealed that following the
demethylation treatment, the mtDNA content increased in the gastric
cells, suggesting that demethylation of the D-loop may be one of
the mechanisms that leads to a decrease in the mtDNA content of
gastric cancer cells. However, further studies are required to
confirm this hypothesis.
In conclusion, the present study shows that the
mtDNA copy number decreases in gastric cancer. This decrease is
particularly notable in ill-defined tumors of clinicopathological
stages III and IV. The decreased mtDNA copy number is a late
molecular event during the progression of gastric cancer. This
finding may be used to determine the clinicopathological stage of
gastric cancer.
Acknowledgements
This study was supported by the Science Research
Foundation of Sichuan University for Fresh Teachers (no.
2012SCU11018).
References
|
1
|
Thun MJ, DeLancey JO, Center MM, Jemal A
and Ward EM: The global burden of cancer: priorities for
prevention. Carcinogenesis. 31:100–110. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen WQ, Zheng RS, Zhang SW, Li N, Zhao P,
Li GL, Wu LY and He J: Report of incidence and mortality in china
cancer registries, 2008. Chin J Cancer Res. 24:171–180. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu J and Chen L: Current status and
progress in gastric cancer with liver metastasis. Chin Med J
(Engl). 124:445–456. 2011.PubMed/NCBI
|
|
4
|
Anderson S, Bankier AT, Barrell BG, et al:
Sequence and organization of the human mitochondrial genome.
Nature. 290:457–465. 1981. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Taanman JW: The mitochondrial genome:
structure, transcription, translation and replication. Biochim
Biophys Acta. 1410:103–123. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lightowlers RN, Chinnery PF, Turnbull DM
and Howell N: Mammalian mitochondrial genetics: heredity,
heteroplasmy and disease. Trends Genet. 13:450–455. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Barrientos A, Casademont J, Cardellach F,
Estivill X, Urbano-Marquez A and Nunes V: Reduced steady-state
levels of mitochondrial RNA and increased mitochondrial DNA amount
in human brain with aging. Brain Res Mol Brain Res. 52:284–289.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu M: Generation, function and diagnostic
value of mitochondrial DNA copy number alterations in human
cancers. Life Sci. 89:65–71. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Honda K, Kato K, Dairaku N, et al: High
levels of intracellular ATP prevent nitric oxide-induced apoptosis
in rat gastric mucosal cells. Int J Exp Pathol. 84:281–288. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Parr RL, Dakubo GD, Thayer RE, McKenney K
and Birch-Machin MA: Mitochondrial DNA as a potential tool for
early cancer detection. Hum Genomics. 2:252–257. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee HC, Yin PH, Lin JC, et al:
Mitochondrial genome instability and mtDNA depletion in human
cancers. Ann NY Acad Sci. 1042:109–122. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Egan K, Kusao I, Troelstrup D, Agsalda M
and Shiramizu B: Mitochondrial DNA in residual leukemia cells in
cerebrospinal fluid in children with acute lymphoblastic leukemia.
J Clin Med Res. 2:225–229. 2010.PubMed/NCBI
|
|
13
|
Chen T, He J, Shen L, et al: The
mitochondrial DNA 4,977-bp deletion and its implication in copy
number alteration in colorectal cancer. BMC Med Genet. 12:82011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Feng S, Xiong L, Ji Z, Cheng W and Yang H:
Correlation between increased copy number of mitochondrial DNA and
clinicopathological stage in colorectal cancer. Oncol Lett.
2:899–903. 2011.PubMed/NCBI
|
|
15
|
Lin CS, Chang SC, Wang LS, et al: The role
of mitochondrial DNA alterations in esophageal squamous cell
carcinomas. J Thorac Cardiovasc Surg. 139:189–197. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kusao I, Agsalda M, Troelstrup D,
Villanueva N and Shiramizu B: Chemotoxicity recovery of
mitochondria in non-Hodgkin lymphoma resulting in minimal residual
disease. Pediatr Blood Cancer. 51:193–197. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mambo E, Chatterjee A, Xing M, et al:
Tumor-specific changes in mtDNA content in human cancer. Int J
Cancer. 116:920–924. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mizumachi T, Muskhelishvili L, Naito A, et
al: Increased distributional variance of mitochondrial DNA content
associated with prostate cancer cells as compared with normal
prostate cells. Prostate. 68:408–417. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fan AX, Radpour R, Haghighi MM, et al:
Mitochondrial DNA content in paired normal and cancerous breast
tissue samples from patients with breast cancer. J Cancer Res Clin
Oncol. 135:983–989. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vivekanandan P, Daniel H, Yeh MM and
Torbenson M: Mitochondrial mutations in hepatocellular carcinomas
and fibrolamellar carcinomas. Mod Pathol. 23:790–798. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu M, Wan Y and Zou Q: Decreased copy
number of mitochondrial DNA in Ewing’s sarcoma. Clin Chim Acta.
411:679–683. 2010.PubMed/NCBI
|
|
22
|
Lin CS, Wang LS, Tsai CM and Wei YH: Low
copy number and low oxidative damage of mitochondrial DNA are
associated with tumor progression in lung cancer tissues after
neoadjuvant chemotherapy. Interact Cardiovasc Thorac Surg.
7:954–958. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Meierhofer D, Mayr JA, Foetschl U, et al:
Decrease of mitochondrial DNA content and energy metabolism in
renal cell carcinoma. Carcinogenesis. 25:1005–1010. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qu F, Liu X, Zhou F, et al: Association
between mitochondrial DNA content in leukocytes and colorectal
cancer risk: a case-control analysis. Cancer. 117:3148–3155. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen J, Platek M, Mahasneh A, Ambrosone CB
and Zhao H: Mitochondrial copy number and risk of breast cancer: a
pilot study. Mitochondrion. 10:62–68. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bonner MR, Shen M, Liu CS, Divita M, He X
and Lan Q: Mitochondrial DNA content and lung cancer risk in Xuan
Wei, China. Lung Cancer. 63:331–334. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chinnery PF, Elliott HR, Hudson G, Samuels
DC and Relton CL: Epigenetics, epidemiology and mitochondrial DNA
diseases. Int J Epidemiol. 41:177–187. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Feng S, Xiong L, Ji Z, Cheng W and Yang H:
Correlation between increased ND2 expression and demethylated
displacement loop of mtDNA in colorectal cancer. Mol Med Rep.
6:125–130. 2012.PubMed/NCBI
|
|
29
|
Hamilton SR and Aaltonen LA: World Health
Organization Classification of Tumors. Pathology and Genetics of
Tumors of the Digestive System. IARC Press; Lyon, France: 2000
|
|
30
|
Tanigawa N, Amaya H, Matsumura M, Lu C and
Iki M: Association between tumor angiogenesis and Borrmann type 4
carcinomas of the stomach. Oncology. 55:461–467. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Park SY, Shin MG, Kim HR, et al:
Alteration of mitochondrial DNA sequence and copy number in nasal
polyp tissue. Mitochondrion. 9:318–325. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim MM, Clinger JD, Masayesva BG, et al:
Mitochondrial DNA quantity increases with histopathologic grade in
premalignant and malignant head and neck lesions. Clin Cancer Res.
10:8512–8515. 2004. View Article : Google Scholar : PubMed/NCBI
|