Introduction
Our knowledge of the pathogenesis of leukemia,
including an understanding of its molecular mechanisms, has
progressed as numerous studies have been undertaken with regard to
the various aspects of gene therapy. The prognosis of patients with
leukemia is closely associated with the invasion and metastasis of
the malignant cells. The nm23-H1 gene is a tumor metastasis
suppressor gene. The effect of the gene on the prognosis and tumor
metastasis have been described in studies of solid tumors,
including those of gall bladder (1,2) liver
(3,4) and gastric (5) cancer.
For malignant tumors of the blood system, the
expression of the nm23-H1 gene is a poor prognostic factor
(6,7). Magyarosy et al(8) studied nm23-H1 expression in acute
lymphoblastic leukemia and revealed that the expression of nm23-H1
in low-differentiated cells was higher than that in relatively
well-differentiated cells. Therefore, the nm23-H1 gene was
considered to be a prognostic marker for a variety of cancers of
the blood system. The K562 cell line originates from chronic
myeloid leukemia (CML). Currently, studies on the nm23-H1 gene in
CML are rare. Therefore, in the present study, the RNAi technique
was used to inhibit nm23-H1 gene expression in the K562 cell line
to investigate the affects of nm23-H1 gene expression on the
proliferation and migration of the K562 cells and to further
clarify its correlation with prognosis for the molecular targeted
treatment of CML.
Materials and methods
Cell lines
K562 cells were obtained from the Shanghai Institute
of Cell Biology, Chinese Academy of Sciences (Shanghai, China). The
cells were cultured at 37°C in humidified 5% CO2 in
RPMI-1640 medium (Sigma, St. Louis, MO, USA), supplemented with 10%
fetal bovine serum and 100 units/ml penicillin and
streptomycin.
Short hairpin RNA (shRNA) preparation and
plasmid construction
Two pairs of shRNA sequences were designed, one
according to the nm23-H1 sequence in GenBank (D1734) and the other
sequence with no homology to the human sequence, which was used as
a control. Each pair contained a unique 19-nt double-stranded
sequence that was separated by a loop of 9-nt sequences
(ttcaagaga). The oligonucleotide sequences of siRNA contained a
BamHI and HindIII site. Subsequent to the
purification and restriction digestion, the oligonucleotides were
ligated into the pGCsi plasmid (GeneChem Inc., Shanghai, China)
with the polymerase III U6 promoter. The nm23-H1 recombinant
plasmid was confirmed by sequencing and named pGCsi-nm23-H1.
RNA extraction and semi-quantitative
RT-PCR
Total RNA extraction was performed using TRIzol
reagent (Takara, Shiga, Japan). The reverse transcription reaction
was performed using 2 μg total RNA with a first strand cDNA kit
(Takara), according to the manufacturer's instructions. PCR was
performed in a 25-μl reaction volume containing 2 μl cDNA template,
10X buffer, 0.15 mM dNTP, 0.1 mM of each primer and 0.5 U Ex Taq
Hot Start Version (Takara). The primers and the amplification
conditions that were used in the PCR are listed in Table I. The final products were identified
in 1.7% agarose gel and stained with ethidium bromide.
 | Table IList of primer sequences and
amplification conditions used in the PCR. |
Table I
List of primer sequences and
amplification conditions used in the PCR.
| Gene | Primer sequences
(5′-3′) | PCR conditions | Product size
(bp) |
|---|
| nm23-H1 |
5′-TTAATCAGATGGTCGGGGAT-3′
5′-GATCTATGAATGACAGGAGG-3′ | 94°C, 30 sec; 56°C,
30 sec, 72°C, 30 sec; 32 cycles | 186 |
| β-actin |
5′-CGTGGCCTTAGCTGTGCT-3′
5′-TGTGCATAAAGTGTAAGTGTATAAGCA-3′ | 94°C, 30 sec; 54°C,
30 sec; 72°C, 30 sec; 32 cycles | 457 |
Transfection assay
To generate the nm23-H1 siRNA-transfected K562
cells, 3 μg plasmid DNA was transfected into 1×105 cells
in a 60-mm dish using lipofectamine 2000 (Invitrogen, Carlsbad, CA,
USA), according to the manufacturer's instructions. The transfected
cells were selected in a medium containing 400 μg/ml G418
(Geneticin; Invitrogen) and the stable nm23-H1 siRNA-transfected
cells were named pGCsi-nm23-H1 K562 cells. The control K562 cells
were transfected with liposome and named the liposome K562
cells.
Western blotting
Each group of cells was washed twice with
phosphate-buffered saline (PBS), lysed for 10 min in hot water and
centrifuged at 20,000 × g for 10 min. Total proteins (10 μl) were
separated by 5% sodium dodecyl sulfate polyacrylamide gel
electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene
fluoride (PVDF) membrane. Subsequent to being immersed in 10 ml 5%
skimmed milk in TBST solution for 1 h, the membrane was incubated
with primary and secondary antibodies. Human monoclonal
anti-nm23-H1 (1:300; BD Biosciences Pharmingen, San Diego, CA, USA)
and β-actin (1:500; Invitrogen) antibodies were used as the primary
antibodies. Bovine anti-mouse IgG (1:2500; Santa Cruz
Biotechnologies, Santa Cruz, CA) was used as the secondary
antibody. Finally, images of the results were captured with an
enhanced chemiluminescence (ECL) substrate.
MTT assay
For the cell proliferation assays, each group of
cells was plated in triplicate in 96-well plates at a density of
1×104 cells/well and grown for 1, 2, 3, 4, 5, 6 and 7
days, respectively. A total of 20 μl 5 mg/ml MTT was added.
Following a 4-h incubation period, the number of metabolically
active cells was quantified.
Colony formation assay
The cells (1×103) were seeded into 6-well
plates with 2 ml culture medium. Subsequent to a two-week
incubation period in RPMI-1640 medium supplemented with 10% fetal
bovine serum at 37°C and 5% CO2, the colonies were
washed twice with PBS, stained with Giemsa, counted, visualized
microscopically and had their images captured.
Transwell assay
For the migration assays, the pGCsil-nm23-H1 K562
and control liposome K562 cells were cultured in RPMI-1640 medium
supplemented with 10% fetal bovine serum at 37°C and 5%
CO2. When the cells had grown to 80% confluence, they
were incubated for 24 h in medium without fetal bovine serum. The
cell culture supernatants were collected and preserved at −20°C for
further use as epidermal growth factor (EGF). The undersides of the
Transwell chamber membranes (BD Biosciences Pharmingen) were coated
with 250 μl Matrigel gels mixed with 250 μl RPMI-1640 medium. Each
group contained 1×105 cells and was seeded on the
Transwell chamber. Following this, 800 μl NIH3T3 EGF that was
prepared previously was added to the 6-well plates. Following 24 h,
the Matrigel gel on the upper sides of the membranes was removed
using cotton swabs. The Transwell chamber membranes were fixed in
95% ethanol for 15 min. The cells that had migrated to the
undersides of the membranes were stained with hematoxylin and eosin
(HE) and counted by microscopy (x200). The results were determined
by averaging the cell counts in five fields.
Statistics
The data were analyzed using the SPSS software
program (v 11.0; SPSS, Inc., Chicago, IL, USA). Non-parametric
tests were performed using independent samples. The mean values
were compared by a one-way ANOVA. P<0.05 was considered to
indicate a statistically significant difference.
Results
Transfection assay
Following the transfection with the pGCsi-nm23-H1
plasmids, a change was observed in the morphology of the cells, and
green fluorescence in the nucleus was visualized by fluorescence
microscope. Following 48 h, the efficiency of the plasmid
transfection was calculated. Plasmid transfection efficiency =
number of fluorescent cells per high power field (HPF) / number of
cells in the same field × 100 (3).
The transfection efficiency of the pGCsi-nm23-H1-transfected K562
cells was ~40%. (Fig. 1)
Inhibition of nm23-H1 gene expression by
shRNA expression vectors
The knockdown efficiencies of nm23-H1-specific shRNA
in the K562 cells were analyzed using semiquantitative PCR and
western blotting. The relative nm23-H1 mRNA levels were normalized
by internal control β-actin and the western blot assay for nm23-H1
protein expression was normalized by β-tubulin. Following
transfection, the mRNA and protein expression levels of nm23-H1
were reduced in the pGCsi-nm23-H1 K562 cells. (Figs. 2 and 3)
MTT assay
For the cell proliferation assays on the inhibition
of nm23-H1 gene expression by the shRNA expression vectors, the
number of metabolically active cells was quantified. The
quantification of the metabolic activity of the cells that were
transfected with pGCsil-nm23-H1 was significantly higher than that
in the cells of the control groups, particularly following 4 days
of transfection. (Fig. 4)
nm23-H1-specific shRNAs induce cell
forming colonies
To evaluate the tumor suppression function of
nm23-H1 in CML, the anchorage-independent growth abilities of the
pGCsi-nm23-H1 K562 and liposome control K562 cells were compared in
soft agar culture. The stably-transfected pGCsi-nm23-H1-siRNA K562
cells exhibited a dramatically increased ability to form colonies
on soft agar. The number and sizes of the colonies that were formed
by the pGCsi-nm23-H1 K562 cells were significantly increased
compared with those that were formed by the liposome control group
(Fig. 5).
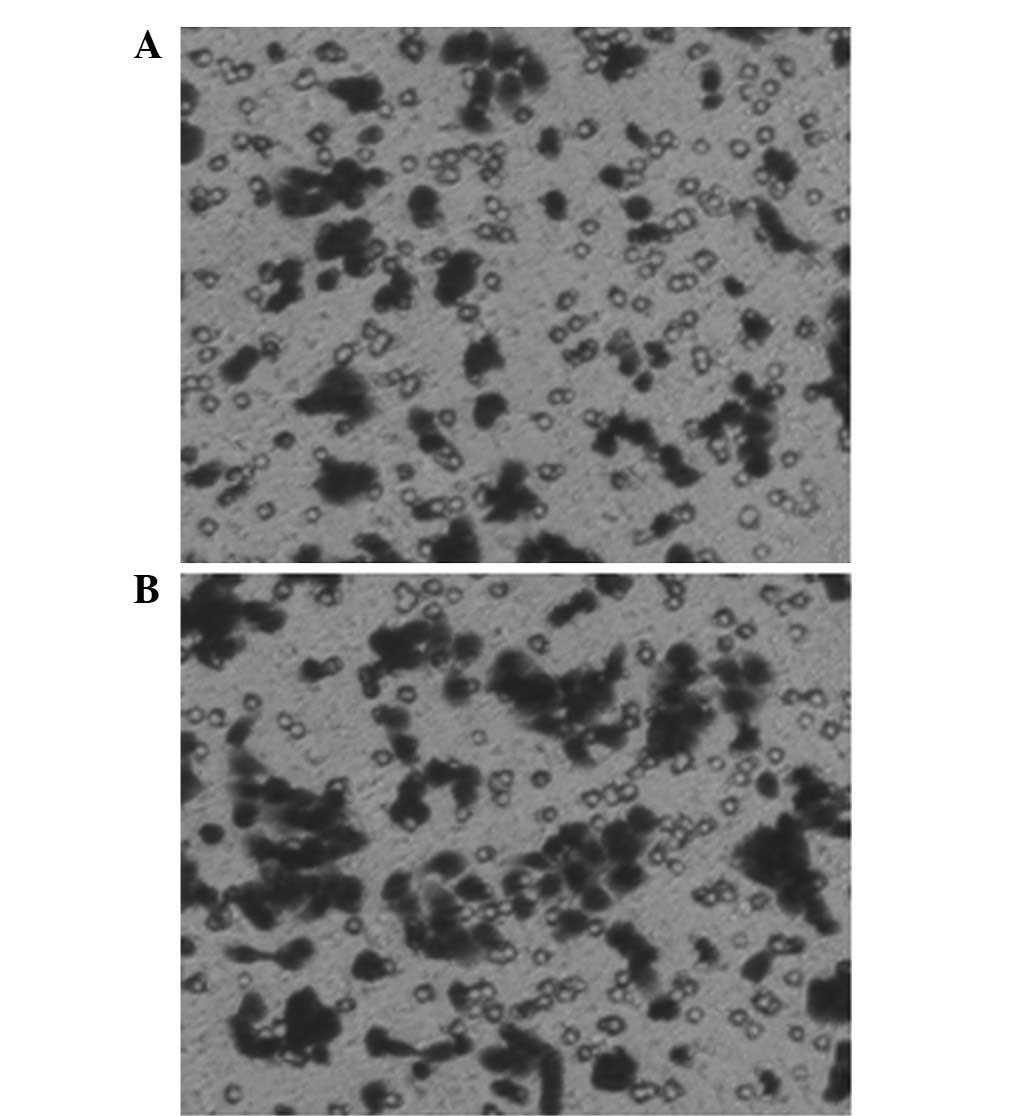
nm23-H1-specific shRNAs induce cell
migration in vitro
To analyze whether the nm23-H1 gene was involved in
the migration of the K562 cells, the effect of the invasiveness of
the pGCsi-nm23-H1 K562 cells was examined in vitro. The
cells migrated through a Matrigel-coated membrane during a 20-h
incubation period. The results revealed that the number of the
pGCsi-nm23-H1 K562 cells that migrated into the lower compartment
of the invasion chamber was markedly increased compared with the
number of the liposome control K562 cells. Fig. 6 shows the mean ± standard deviation
(SD) of three independent experiments (pGCsi-nm23-H1 group,
112.4±4.56; and control group, 68.4±2.40).
Discussion
CML is a clonal myeloproliferative disorder that is
characterized by the presence of the fusion oncogene, BCR-ABL. The
constitutive expression of BCR-ABL leads to the unregulated
production of mature myeloid cells in the bone marrow and their
subsequent release into the blood (9). If untreated, CML will progress from a
chronic to an accelerated phase over a number of years, prior to
quickly proceeding to a terminal blast crisis phase, reminiscent of
acute leukemia (10). The advent of
tyrosine kinase inhibitors has led to an improved management of the
disease. However, these drugs do not provide a cure as they are
unable to eradicate the most primitive, quiescent fraction of CML
stem cells (11).
The nm23-H1 gene is a metastatic suppressor that was
identified in a melanoma cell line and is expressed in various
tumors where their levels of expression are associated with a
reduced or increased metastatic potential. nm23-H1 is one of >20
metastasis suppressor genes (MSGs) that have been confirmed in
vivo. The gene is highly conserved from yeast to humans,
implying a critical developmental function. Cell surface nm23-H1
has been previously observed in non-Hodgkin lymphoma (NHL) cells
(12,13) and certain myeloid cell lines
(14,15). Specific studies (13,14)
have demonstrated that tumors with a reduced expression of the nm23
gene are more prone to metastasis. It has been also previously
documented that the expression of nm23-H1 transcripts and, more so,
the levels of nm23-H1 protein in serum, provide strong indicators
of prognosis, with higher values being associated with poorer
overall survival (13–15).
The present study revealed a strong association
between nm23-H1 gene expression and K562 cell survival in
vitro. The MTT assay demonstrated that the stably-transfected
pGCsi-nm23-H1 K562 cells exhibited a markedly increased ability to
form colonies on soft agar. The number and sizes of the colonies
that were formed by the pGCsi-nm23-H1 K562 cells were significantly
increased compared with those of the liposome control group.
Furthermore, to test whether the nm23-H1 gene was involved in the
migration of the K562 cells, the effect of the invasiveness of the
pGCsi-nm23-H1 K562 cells was examined in vitro. The results
revealed that the number of the pGCsi-nm23-H1 K562 cells that
migrated into the lower compartment of the invasion chamber was
markedly increased compared with the liposome control K562 cells.
This suggests that the behavior of the nm23-H1 gene affects the
biology of the CML cell lines, including growth, proliferation and
invasiveness (16). These
observations are consistent with other studies of solid tumors
(17–19). However, the data from a study by
Okabe-Kado et al(20)
strongly indicated that the nm23-H1 gene may act as a tumor-derived
survival factor in acute myeloid leukemia (AML). However, the study
was unable to delineate between nm23-H1-binding AMLs and normal
AMLs, in which the mechanism is likely to be active (20).
The experimental results from the present study
suggest that the nm23-H1 gene is closely associated with the
inhibition of metastasis. To assess the ultimate therapeutic
potential of peptide vaccines derived from nm23, it will be
necessary to determine, firstly, whether or not aberrant nm23-H1
expression is a widespread feature of CMLs and, secondly, whether
the protein generates peptides that are able to act as functional
antigens in HLA backgrounds other than HLA-A32. Given the
widespread involvement of nm23 proteins in tumorigenesis, it will
also be noteworthy to investigate the potential relevance of
nm23-H2 as a therapeutic target in other cancers. The regulatory
interdependence of nm23-H2 and c-myc provides a basis from which to
design specific studies to elucidate the function of nm23 proteins
in normal and leukemic cells, which may contribute to our
understanding of the molecular mechanisms underlying the
development and progression of CML (21). Future studies should investigate the
association between nm23-H1 binding and responses to CML therapies,
and aim to determine the nature of the nm23-H1 receptor in CML,
which may provide a novel target for adjunctive therapies.
Acknowledgements
This study was supported by a grant from the
Shanghai Bureau of Health, China (no. 2010052).
References
|
1
|
Chang HJ, Yoo BC, Kim SW, et al:
Significance of PML and p53 protein as molecular prognostic markers
of gallbladder carcinomas. Pathol Oncol Res. 13:326–335. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jiang WX, Song BG and Wang PJ: Expression
of nm23, KAI1 and spiral computed tomography findings in primary
gallbladder carcinoma. Chin Med J (Engl). 122:2666–2668.
2009.PubMed/NCBI
|
|
3
|
Marshall JC, Collins JW, Nakayama J, et
al: Effect of inhibition of the lysophosphatidic acid receptor 1 on
metastasis and metastatic dormancy in breast cancer. J Natl Cancer
Inst. 104:1306–1319. 2012.PubMed/NCBI
|
|
4
|
Boissan M, De Wever O, Lizarraga F, et al:
Implication of metastasis suppressor NM23-H1 in maintaining
adherens junctions and limiting the invasive potential of human
cancer cells. Cancer Res. 70:7710–7722. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu HK, Wang Q, Li Y, et al: Inhibitory
effects of gamma-tocotrienol on invasion and metastasis of human
gastric adenocarcinoma SGC-7901 cells. J Nutr Biochem. 21:206–213.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Niitsu N, Hayama M, Yoshino T, et al:
Multicentre phase II study of the CyclOBEAP regimen for patients
with peripheral T-cell lymphoma with analysis of biomarkers. Br J
Haematol. 153:582–588. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Niitsu N, Nakamine H, Okamoto M, et al;
Adult Lymphoma Treatment Study Group, ALTSG. Expression of nm23-H1
is associated with poor prognosis in peripheral T-cell lymphoma. Br
J Haematol. 123:621–630. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Magyarosy E, Sebestyén A and Timár J:
Expression of metastasis associated proteins, CD44v6 and nm23-H1,
in pediatric acute lymphoblastic leukemia. Anticancer Res.
21:819–823. 2001.
|
|
9
|
Bozkurt S, Uz B, Buyukasik Y, et al:
Prognostic importance of additional cytogenetic anomalies in
chronic myeloid leukemia. Med Oncol. 30:4432013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Piccaluga PP, Paolini S, Bertuzzi C, et
al: First-line treatment of chronic myeloid leukemia with
nilotinib: critical evaluation. J Blood Med. 3:151–156. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sawyers CL: The 2011 Gordon Wilson
lecture: overcoming resistance to targeted cancer drugs. Trans Am
Clin Climatol Assoc. 123:114–125. 2012.PubMed/NCBI
|
|
12
|
Bircan S, Inamdar KV, Rassidakis GZ and
Medeiros LJ: nm23-H1 expression in non-Hodgkin and Hodgkin
lymphomas. Appl Immunohistochem Mol Morphol. 16:207–214. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Niitsu N, Honma Y, Iijima K, et al:
Clinical significance of nm23-H1 proteins expressed on cell surface
in non-Hodgkin's lymphoma. Leukemia. 17:196–202. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lilly AJ, Khanim FL, Hayden RE, et al:
Nm23-H1 indirectly promotes the survival of acute myeloid leukemia
blast cells by binding to more mature components of the leukemic
clone. Cancer Res. 71:1177–1186. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bach E, Krahl R, Lange T, et al: Delayed
processing of bone marrow samples reveals a prognostic pattern of
NME mRNA expression in cytogenetically normal acute myeloid
leukemia. Leuk Lymphoma. 53:1561–1568. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jin L, Liu G, Zhang CH, et al: Nm23-H1
regulates the proliferation and differentiation of the human
chronic myeloid leukemia K562 cell line: a functional proteomics
study. Life Sci. 84:458–467. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Boissan M and Lacombe ML: NM23, an example
of a metastasis suppressor gene. Bull Cancer. 99:431–440. 2012.(In
French).
|
|
18
|
Conery AR, Sever S and Harlow E:
Nucleoside diphosphate kinase Nm23-H1 regulates chromosomal
stability by activating the GTPase dynamin during cytokinesis. Proc
Natl Acad Sci USA. 107:15461–15466. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao XS, Song PL, Sun B, et al: Arsenic
trioxide inhibits metastatic potential of mouse hepatoma H22 cells
in vitro and in vivo. Hepatobiliary Pancreat Dis Int. 8:510–517.
2009.PubMed/NCBI
|
|
20
|
Okabe-Kado J, Kasukabe T, Honma Y, et al:
Extracellular NM23-H1 protein inhibits the survival of primary
cultured normal human peripheral blood mononuclear cells and
activates the cytokine production. Int J Hematol. 90:143–152. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tschiedel S, Gentilini C, Lange T, et al:
Identification of NM23-H2 as a tumour-associated antigen in chronic
myeloid leukaemia. Leukemia. 22:1542–1550. 2008. View Article : Google Scholar : PubMed/NCBI
|