Introduction
Of all breast cancer patients, those with
triple-negative tumors that lack the expression of the estrogen
receptor (ER), progesterone receptor and Her2 receptor, bear the
poorest prognoses (1). This is due
to the aggressive behavior of triple-negative breast cancer (TNBC)
cells and the lack of targeted therapies for this particular
subgroup. TNBC is, however, a highly heterogeneous disease and more
specific information concerning the biology of the various subtypes
is required for future targeted therapies (1,2).
Toll-like receptor-9 (TLR9) is an innate immunity
DNA receptor that was first identified in cells of the immune
system (3). Agonistic TLR9 ligands,
such as microbial and vertebrate DNA or synthetic
CpG-sequence-containing oligonucleotides, induce an inflammatory
reaction in cells that express TLR9. In addition to inducing the
release of cytokines (4,5) in cancer cells, TLR9 agonists also
induce invasion in vitro, which is mediated via the
activation of matrix metalloproteinases (MMPs), such as MMP-13
(6–8). We previously demonstrated that TLR9
has an important role in TNBC (9)
and showed that, while low tumor TLR9 expression was associated
with significantly shortened breast cancer-specific survival in
patients with TNBC, TLR9 had no prognostic value in breast cancer
patients with ER+ tumors (9). This is likely to be, at least partly,
explained by the hypoxia-associated behavior of TNBC cells that
express low TLR9 levels. We revealed that, although decreased TLR9
expression in TNBC cells results in decreased invasion when the
tumor cells are in normoxia, the cells become highly invasive in
hypoxia (9). These results
suggested that TLR9 also has ligand-independent effects on invasion
and that in the absence of TLR9 expression in hypoxia, another
pathway is the actual mediator of invasion. The pathway that
mediates invasion in hypoxia in the absence of TLR9 is not
currently known. It is also not known whether possible impaired
TLR9-mediated inflammation at the site of the tumor contributes to
the poor prognosis in this subgroup of TNBC.
Since the hypoxia-induced in vitro invasion
and viability of TNBC cells expressing low levels of TLR9 was
inhibited in vitro by chloroquine (9), a well-established malaria and
rheumatoid arthritis drug that is known to interfere with endosomal
signaling, the present study aimed to further characterize the
anti-tumor efficacy of chloroquine against TNBC cells with
differences in TLR9 expression.
Materials and methods
Cell culture
Parental MDA-MB-231 breast cancer cells and D54MG,
U373MG, Caco-2 and AGS cells were cultured in Dulbecco’s modified
Eagle’s medium (Gibco BRL, Life Technologies, Carlsbad, CA, USA)
supplemented with 10% heat-inactivated fetal bovine serum,
L-glutamine, penicillin/streptomycin and non-essential amino acids
(all from Gibco BRL, Life Technologies) (10). The cells were cultured in incubators
at 37°C with an atmosphere of 5% CO2/95% air with ~21%
pO2 or in a hypoxia incubator with 5% pO2
(I-Glove; BioSpherix, Ltd., Lacona, NY, USA). The stable control
siRNA and TLR9 siRNA MDA-MB-231 cells have been described
previously and were cultured in the presence of G418 (800 μg/ml)
(9). Chloroquine was purchased from
Sigma (St. Louis, MO, USA).
RNA isolation and quantitative
(q)PCR
Total RNA was isolated from the cells using the
TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA)
and purified with RNeasy mini kits (Qiagen, Hilden, Germany). All
reagents for the qPCR experiments were purchased from Applied
Biosystems (Foster City, CA, USA). cDNA was synthesized from 0.2 μg
total RNA, using Multiscribe Reverse Transcriptase and random
hexamers. Quantification of TLR9 mRNA expression was performed as
previously described (11). The
other primer and probe sets that were used (MMP-2, MMP-9, MMP-13
and TIMP-3) were purchased from Applied Biosystems as ready-made
primer/probe sets. A standard amplification program was used for
all amplifications (1 cycle of 50°C for 2 min, 1 cycle of 95°C for
10 min, 40 cycles of 95°C for 15 sec and 60°C for 1 min).
Subsequent to normalization with ribosomal protein L15 (RPLO)
expression levels for each cDNA, relative quantification of target
cDNA was performed using 2−ΔΔct values.
Western blot analysis
The cells were cultured in 6-well plates with normal
culture medium until near confluency, after which they were rinsed
with sterile phosphate-buffered saline (PBS) and cultured further
for the indicated times in serum-free culture medium. At the
desired time-points, the culture medium was discarded and the cells
were quickly harvested in lysis buffer (Cell Signaling Technology,
Inc., Danvers, MA, USA) and clarified by centrifugation, as
previously described (8).
Subsequent to boiling the supernatants in reducing sodium dodecyl
sulphate (SDS) sample buffer, equal amounts of protein (~100 μg)
were loaded per lane and the samples were electrophoresed into 10
or 4–20% gradient polyacrylamide SDS gels (Bio-Rad Laboratories,
Inc., Hercules, CA, USA), then transferred to a nitrocellulose
membrane. To detect TLR9, the blots were incubated overnight at 4°C
with anti-TLR9 antibodies (IMG-431; Imgenex, San Diego, CA, USA),
diluted 1:500 in Tris-buffered saline with 0.1% (v/v) Tween-20
(TBST). Equal loading was confirmed with polyclonal rabbit
anti-actin (Sigma; A-2066, used at 1:1,000 dilution). Secondary
detection was performed with horseradish peroxidase-linked
secondary antibodies (GE Healthcare, Piscataway, NJ, USA). The
protein bands were visualized by chemiluminescence using an ECL kit
(Pierce Biotechnology, Inc., Rockford, IL, USA).
Cell viability assays
The cells were plated into 96-well plates (20,000
cells per 100 μl per well) in normal growth medium. The viability
of the cells was measured with the CellTiter 96 Aqueous One
Solution Cell Proliferation assay (Promega Corporation, Madison,
WI, USA), according to the manufacturer’s recommendations. In
another set of experiments, the cells were plated into 24-well
plates and after the indicated time, the cells were trypsinized and
the viable cells were counted following trypan blue staining using
a TC10™ automated cell counter (Bio-Rad Laboratories).
Zymography
The cells were incubated for 24–48 h in serum-free
media. The supernatants were collected and concentrated using a
centrifugal filter device (Millipore, Billerica, MA, USA; cut-off
size 3 kDa, cat no. UFC5-003-24). Equal amounts of protein (~20 μg)
were loaded per lane of zymogram gels (10% gelatin, Bio-Rad
Laboratories). The gels were then run, renaturated, developed and
stained using Bio-Rad zymogram buffers, according to the
manufacturer’s recommendations.
Animal studies
Control and TLR9 siRNA MDA-MB-231 cells
(5×105 cells in 100 μl) were inoculated into the mammary
fat pads of four-week-old, immune-deficient mice (athymic nude/nu
Foxn1; Harlan Sprague Dawley, Inc., Indianapolis, IN, USA).
Treatments were started seven days after tumor cell inoculation.
The mice were treated daily either with intraperitoneal (i.p.)
chloroquine (80 mg/kg) or vehicle (PBS). The animals were monitored
daily for clinical signs. Tumor measurements were performed twice a
week and tumor volume was calculated according to the formula V =
(π / 6) (d1 × d2)3/2, where
d1 and d2 are perpendicular tumor diameters
(9). The tumors were allowed to
grow for 22 days, at which point the mice were sacrificed and the
tumors were dissected for a final measurement. Throughout the
experiments, the animals were maintained under controlled
pathogen-free environmental conditions (20–21ºC, 30–60% relative
humidity and a 12-h lighting cycle). The mice were fed with
small-animal food pellets (Harlan Sprague Dawley) and supplied with
sterile water ad libitum. The experimental procedures were
reviewed and approved by the University of Alabama at Birmingham
Institutional Animal Care and Use Committee.
Statistical analysis
The results are presented as the mean ± SD or mean ±
SEM, as stated. Unpaired Student’s t-tests were used to calculate
statistically significant differences between the various study
groups in the in vitro and pre-clinical in vivo
experiments.
Results
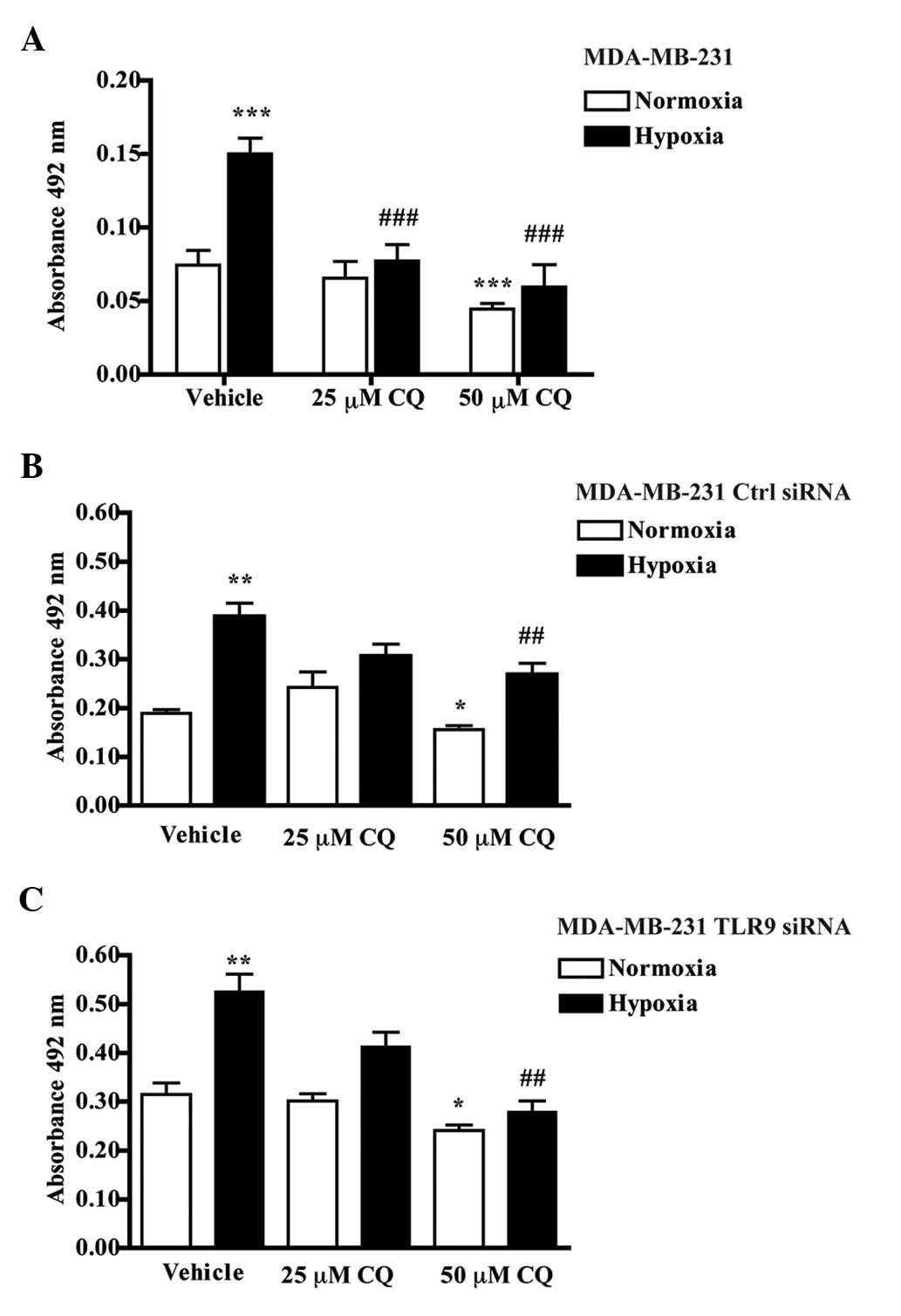
Effects of chloroquine on cellular
viability of parental MDA-MB-231 cells
Since the behavior of TNBC cells is significantly
affected by hypoxia (9,12), all experiments were conducted in
normoxic (pO2 21%) and hypoxic (pO2 5%)
culture conditions. First, the effects of chloroquine on the
cellular viability of the parental MDA-MB-231 cells were
investigated. In agreement with our previous observations (9), hypoxic culture conditions induced a
significant increase in parental MDA-MB-231 cell viability compared
with cultures that were kept in normoxia (9). The addition of 25 μM chloroquine did
not affect MDA-MB-231 viability in normoxia, whereas 50 μM
chloroquine had a slight but significant inhibitory effect. Neither
dose of chloroquine, however, completely blocked the
hypoxia-induced increase in viability (Fig. 1A). Similar studies were also
conducted with MDA-MB-231 cells that were stably transfected with
control siRNA- or TLR9 siRNA-encoding plasmids. Chloroquine also
inhibited the hypoxia-induced increase in viability in these two
cell lines (Fig. 1B and C). Taken
together, these results suggest that chloroquine dose-dependently
inhibits the hypoxia-induced viability of MDA-MB-231 cells and that
these effects are independent of the TLR9 expression status of the
cells.
Effects of chloroquine on hypoxia-induced
TLR9 expression
Next, the effects of chloroquine on hypoxia-induced
TLR9 expression were studied. As also previously detected, hypoxia
induced a significant increase in TLR9 mRNA expression in the
parental MDA-MB-231 cells (Fig.
2A). This effect was significantly enhanced by chloroquine in
the normoxic and hypoxic culture conditions. In hypoxia, the effect
of chloroquine was, however, significantly reduced (Fig. 2B). Similar effects on TLR9 mRNA
expression by chloroquine were also detected in the D54MG and
U373MG brain cancer cell lines (Fig. 2C
and D). Furthermore, a similar trend in TLR9 mRNA expression
was also detected in the Caco-2 and AGS human colorectal and
gastric adenocarcinoma cell lines, respectively (Fig. 2E). At the protein level, however,
chloroquine decreased MDA-MB-231 TLR9 protein expression, in
normoxia and hypoxia (Fig. 2F).
Similar effects on TLR9 protein were also detected in the control
siRNA and TLR9 siRNA cells (Fig.
3). Taken together, these studies suggest that chloroquine has
opposing effects on TLR9 mRNA and protein expression.
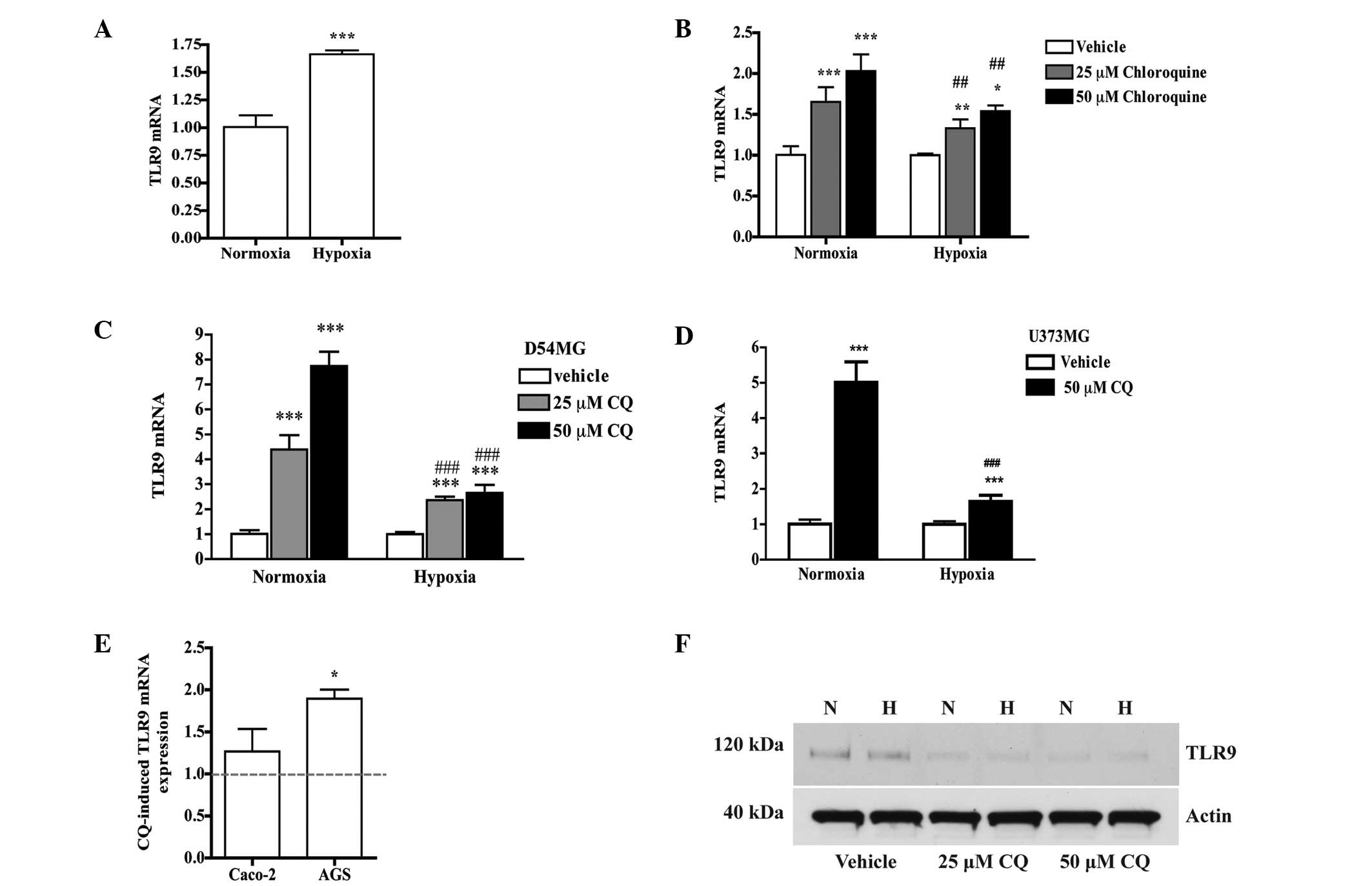 | Figure 2(A) Parental MDA-MB-231 cells were
cultured for 24 h under hypoxia and normoxia. Expression of TLR9
mRNA was measured with qPCR; mean ± SEM, n=6.
***P<0.001 vs. normoxia. (B) Expression of TLR9 mRNA
in parental MDA-MB-231 cells. Bars represent chloroquine-induced
changes in TLR9 mRNA expression relative to vehicle treatment in
normoxia and hypoxia; mean ± SD, n=6. *P<0.05,
**P<0.01 and ***P<0.001 vs. the
corresponding vehicle; ##P<0.01 vs. corresponding
chloroquine concentration in normoxia. (C) D54MG and (D) U373MG
cells were cultured in normoxia and hypoxia in the presence of
vehicle or 25 or 50 μM chloroquine for 24 h, and TLR9 mRNA was
measured with qPCR. Data are expressed as fold-change in TLR9 mRNA
expression vs. corresponding vehicle. Mean ± SEM, n=6.
***P<0.001 vs. vehicle and ###P<0.001
vs. corresponding chloroquine-treatment in normoxia. (E) Caco-2 and
AGS cells were cultured with 50 μM chloroquine in normoxia; mean ±
SEM, n=4. *P<0.05 vs. vehicle (vehicle is set to 1
and represented by the dotted line). (F) Western blot analysis of
TLR9 protein in parental MDA-MB-231 cells after culture for 24 h in
normoxia (N) and hypoxia (H), in the presence of vehicle or 25 or
50 μM chloroquine. Actin band of the same stripped blot is shown to
indicate equal loading. Mean ± SEM, n=6. qPCR, quantitative PCR;
CQ, chloroquine; TLR9, toll-like receptor-9. |
Effects of chloroquine on MMP-2, MMP-9
and MMP-13 mRNA expression and proteolytic activity of TNBC cells
with high and low TLR9 expression
TLR9 ligand-induced invasion has been shown to be
associated with the activation of MMP-13 (6–8). Since
chloroquine inhibits TLR9-ligand-induced invasion in normoxia in
vitro, the effects of chloroquine on MMP-2, MMP-9 and MMP-13
mRNA expression, as well as the proteolytic activity of TNBC cells
with high and low TLR9 expression were investigated. Chloroquine
had similar, suppressive effects on MMP-2 mRNA expression in
normoxia and hypoxia in all the studied cells (Fig. 4A). The effects on MMP-9 mRNA
expression were more dose- and oxygen-status dependent. While 25 μM
chloroquine suppressed MMP-9 mRNA expression in normoxia and
hypoxia, the 50-μM dose was less suppressive in normoxia and did
not suppress MMP-9 mRNA expression in parental MDA-MB-231 cells
under hypoxia. Similar effects were observed in the control and
TLR9 siRNA MDA-MB-231 cells, with the exception that, in the TLR9
siRNA cells compared with vehicle-treatment, 50 μM chloroquine
induced significant suppression of MMP-9 mRNA expression in
normoxia and hypoxia (Fig. 4B).
Chloroquine also had a dual, dose-dependent effect on MMP-13 mRNA
expression. In normoxia and hypoxia, the 25-μM dose induced no
change or slightly suppressed MMP-13 mRNA expression in all the
studied cells. The higher chloroquine concentration (50 μM),
however, induced a significant increase of MMP-13 mRNA expression
in normoxia in all the cells. This induction of MMP-13 mRNA was
further significantly enhanced by hypoxia in the control siRNA
cells, but decreased in the TLR9 siRNA cells (Fig. 4C). Similar dose- and oxygen
status-dependent effects on MMP-13 mRNA expression were also
detected in the human D54MG and U373MG glioblastoma cell lines. The
smaller dose had no or only a slightly suppressive effect on MMP-13
mRNA expression, while the higher dose induced MMP-13 mRNA in an
oxygen level-dependent fashion (Fig.
4D). The Caco-2 and AGS cells were studied only in normoxia. In
the Caco-2 cells, 50 μM chloroquine had no effect on MMP-9 mRNA,
but suppressed MMP-2 mRNA and significantly induced MMP-13 mRNA
expression. Similarly, 50 μM chloroquine also induced MMP-13 mRNA
expression in the AGS cells (Fig.
4E). Taken together, these studies suggest that chloroquine has
cell-, dose- and hypoxia-dependent effects on MMP-2, MMP-9 and
MMP-13 mRNA expression. Most notably, higher doses of chloroquine
appear to induce more MMP-13 mRNA expression, suppress less MMP-9
mRNA expression and, in hypoxia, these effect appear to be
TLR9-dependent.
 | Figure 4Expression of (A) MMP-2, (B) MMP-9 and
(C) MMP-13 mRNA in parental MDA-MB-231 cells, control siRNA or TLR9
siRNA cells in normoxia and hypoxia, as measured with qPCR. The
bars represent chloroquine-induced changes in mRNA expression,
relative to vehicle-treatment (dotted line) in normoxia and
hypoxia; mean ± SEM, n=3–6. *P<0.05,
**P<0.01 and ***P<0.001 vs. the
corresponding vehicle; #P<0.05 vs. corresponding
chloroquine in normoxia; ^^^P<0.001 vs. corresponding
control siRNA. (D) D54MG and U373MG cells were cultured in normoxia
and hypoxia in the presence of vehicle or 25 or 50 μM chloroquine
for 24 h, and MMP-13 mRNA expression was measured with qPCR. Data
is expressed as fold-change in MMP-13 mRNA expression vs.
corresponding vehicle (represented by the dotted line). Mean ± SEM,
n=6. *P<0.05 vs. vehicle in normoxia,
#P<0.05 vs. vehicle in hypoxia,
**P<0.01 vs. corresponding vehicle in normoxia and
##P<0.01 vs. corresponding chloroquine-treatment in
normoxia. (E) Caco-2 and AGS cells were cultured with 50 μM
chloroquine in normoxia. Mean ± SEM, n=4. *P<0.05,
**P<0.01 vs. corresponding vehicle. CQ, chloroquine;
qPCR, quantitative PCR; MMP, matrix metalloproteinase; TLR9,
toll-like receptor-9. |
Effects of chloroquine on MMP at the
functional protein level
To investigate whether chloroquine’s effects on MMP
mRNAs are translated to the functional protein level, zymograms
were performed using the cell supernatants following the various
treatments. Subsequent to 24 h of treatment, the pro-MMP-9 and
pro-MMP-2 proteolytic bands were clearly visible (13), but no clear differences were
detected in proteolytic activities between the various treatments
of the studied cells (Fig. 5A).
However, after 48 h, while MMP-2 and MMP-9 activities were
suppressed by chloroquine, MMP-13 proteolytic activity began to
emerge in the same specimens (Fig.
5B). Data is shown only for TLR9 siRNA cells in normoxia,
although similar results were detected for all studied cells in
normoxia and hypoxia.
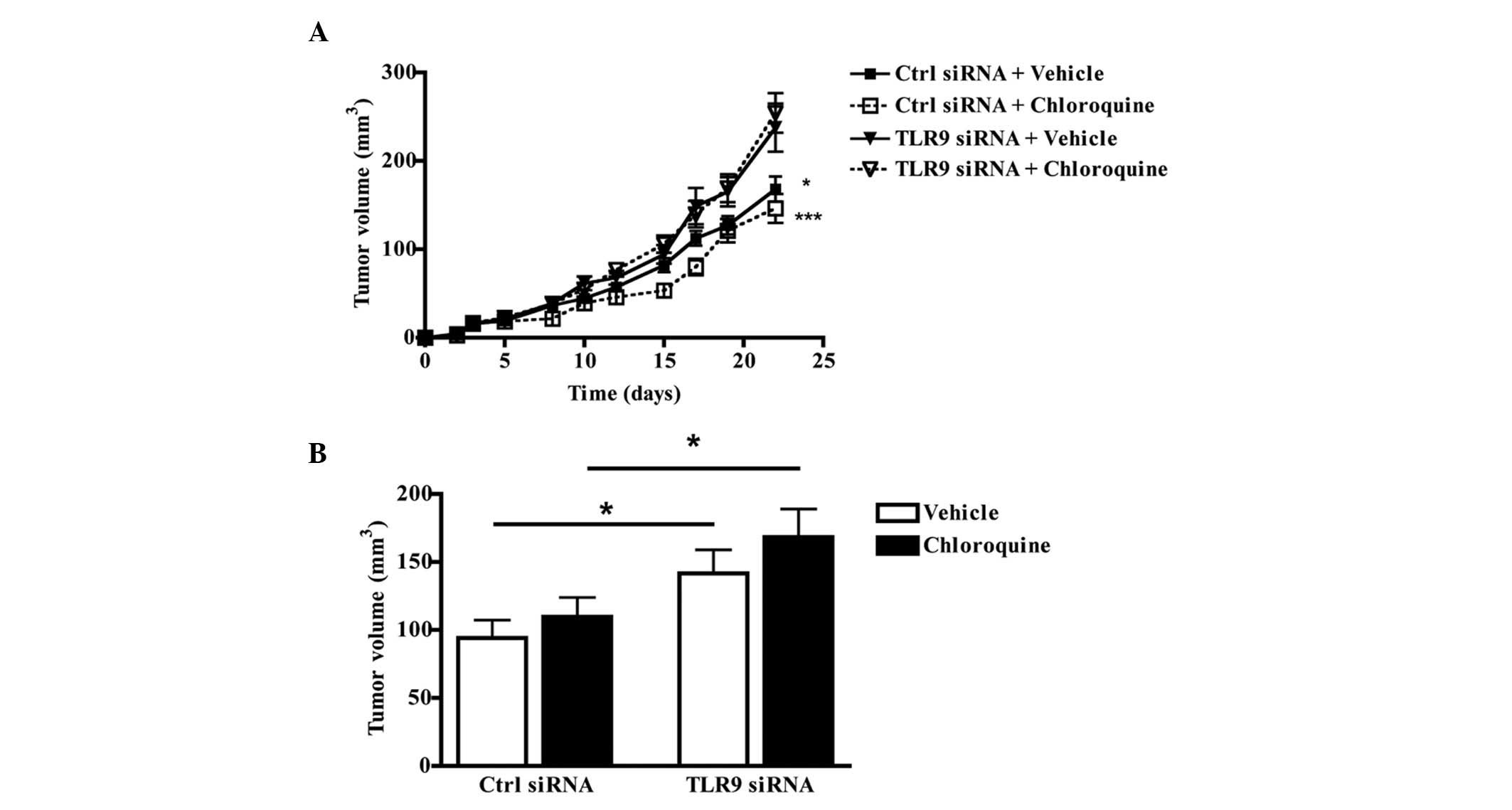
Anti-tumor efficacy of chloroquine in an
orthotopic mouse model
The anti-tumor efficacy of chloroquine was studied
in an orthotopic mouse model, using control siRNA and TLR9 siRNA
MDA-MB-231 cells. Subsequent to tumor cell inoculation and the
establishment of tumors seven days later, the mice were treated
daily with i.p. chloroquine (80 mg/kg). As expected, the TLR9 siRNA
cells formed significantly larger tumors than the control siRNA
cells during the experiment. Chloroquine treatment did not inhibit
tumor growth in either the control siRNA or TLR9 siRNA groups
(Fig. 6A and B). Taken together,
despite the favorable antitumor and anti-invasive effects that
chloroquine exhibits against the tested breast cancer cells in
vitro, the results suggest that chloroquine does not prevent
the growth of these cells at the orthotopic site in
vivo.
Discussion
In the current era of escalating cancer care costs,
there is emerging interest in identifying new uses for old drugs
(14,15). For example, chloroquine has
demonstrated promising effects as an anti-cancer agent,
particularly in breast cancers (16–19).
Chloroquine has been shown to inhibit breast cancer growth in
vitro, and low doses of chloroquine have induced resistance to
mammary carcinogenesis in a rat model of chemically-induced breast
cancers (20,21). Since our previous in vitro
data suggested that chloroquine inhibits the invasive capacity of
TNBC cells with the highly aggressive low TLR9 expression phenotype
(7,8), the present study aimed to investigate
the anti-tumor effects of this widely used, anti-malarial and
rheumatology drug in a mouse model that mimics the aggressive human
disease in vivo. According to our preliminary data, such
patients with low TLR9-TNBC and poor prognoses may represent up to
10% of all breast cancer patients (9).
The present results demonstrated that, despite the
promising hypoxia-associated growth inhibitory effects in
vitro, chloroquine does not inhibit the local growth of tumors
formed by the same cells in vivo. The reason for the
discrepancy between the in vitro and in vivo findings
is currently unclear and requires further characterization. Local
tumor growth is the sum of cell proliferation and local invasion.
Thus the lack of inhibition of tumor growth may, at least
partially, be explained by the pro-invasive effects of chloroquine,
such as increased MMP-13 activity, which at the protein level
manifests later than the anti-invasive and growth-inhibitory
effects and which may be more pronounced in hypoxic conditions. The
present results are the opposite of those published by Jiang et
al(22), who observed that
chloroquine inhibits the growth of subcutaneous (s.c.) 4T1 breast
tumors and lung metastases in vivo. Chloroquine, alone or in
combination with the mTOR inhibitor RAD001, has also been shown to
inhibit the in vivo growth of orthotopic MCF-7 tumors
(21). The differences in the
results may be explained by the different cell lines used and the
drug dosage; it is possible that, for example, the
MMP-13-activating effects of chloroquine manifest only with the
higher chloroquine doses, similar to those used in our studies. A
part of the differences in chloroquine responses may also be
explained by the p53 status of the cell lines used. Chloroquine is
known to induce cell cycle arrest through the activation of the p53
tumor suppressor, which is mutated in MDA-MB-231 cells (20). The present results, which showed
that chloroquine inhibits the hypoxia-induced increased viability
of these cells, suggest that in hypoxia, chloroquine may induce
other pathways of cell death or growth arrest, independent of p53.
This is supported by the fact that the 4T1 cells have been shown to
be p53 null (23).
TLR9 is a cellular DNA receptor which, based on our
observations, appears to regulate cancer cell invasion in the
absence of exogenously added DNA ligands. Chloroquine has been
shown to inhibit TLR9 ligand-induced inflammatory reactions in
cells and this effect has been attributed to the inhibition of
endosomal acidification and more recently, to direct binding of
chloroquine to nucleic acids, thus masking their TLR9-binding
epitopes (24). Notably, the
present study revealed that chloroquine treatment upregulates TLR9
mRNA expression in cancer cells. This effect on mRNA was slightly
reduced in hypoxia, but did not translate into increased TLR9
protein levels, even in oxygen replete conditions. By contrast,
chloroquine treatment actually resulted in decreased TLR9 protein
expression. The present results agree with those of Zhu et
al(25) who demonstrated that
chloroquine inhibits TLR9 expression in dendritic cells. The reason
for these findings is unclear, but it may be that a low pH is
required for the proper folding of the TLR9 protein or the
involvement of specific microRNAs that would inhibit TLR9
expression. By blocking the acidification of the endosomal
organelles where TLR9 resides, chloroquine may also actually hasten
the degradation of the TLR9 protein. Although Kuznik et
al(24) demonstrated that
chloroquine does not increase the pH of endosomes, the
concentration of chloroquine these authors used was significantly
smaller compared with the present experiments (4 vs. 25–50 μM).
These issues require further biochemical characterization at the
cellular level. Another possible explanation for why chloroquine
does not prevent tumor growth in this model is that reducing TLR9
expression may promote the highly aggressive low TLR9 expression
phenotype of the TNBC cells (9),
thus allowing the activation of the presently unknown pathway of
aggressive growth and invasion.
In conclusion, despite the promising TLR9
status-independent growth inhibitory and anti-invasive in
vitro effects against TNBC cells in normoxia and hypoxia,
chloroquine does not inhibit the growth of orthotopic TNBC tumors
in vivo. Furthermore, by promoting MMP-13 activation and
suppressing TLR9 expression under such conditions, chloroquine may
be a particularly poor choice for tumors that are hypoxic.
Chloroquine may, however, have growth inhibitory and
anti-metastatic effects against other types of breast or other
cancers.
Acknowledgements
This study was funded by grants from the Department
of Defense (W81XWH-10-1-0308, K.S.S.), Lapland Cultural Foundation
(K.S.S.), Elsa U. Pardee Foundation (K.S.S.), Maud Kuistila
Memorial Foundation (J.T.), Finnish Cultural Foundation (J.S.),
Emil Aaltonen Foundation (J.H.K.), Cancer Foundation of Northern
Ostrobotnia (J.H.K.), Oulu University Research Foundation (J.H.K.),
Georg C. and Mary Ehrnroot Foundation (J.H.K.), Orion-Farmos
Research Foundation (J.H.K.) and the Finnish Medical Foundation
(J.H.K.). Christine Pressey is acknowledged for providing
assistance with the qPCR assays.
References
|
1
|
Elias AD: Triple-negative breast cancer: a
short review. Am J Clin Oncol. 33:637–645. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Carotenuto P, Roma C, Rachiglio AM, Botti
G, D’Alessio A and Normanno N: Triple negative breast cancer: from
molecular portrait to therapeutic intervention. Crit Rev Eukaryot
Gene Expr. 20:17–34. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hemmi H, Takeuchi O, Kawai T, Kaisho T,
Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K and
Akira S: A Toll-like receptor recognizes bacterial DNA. Nature.
408:740–745. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Di JM, Pang J, Pu XY, Zhang Y, Liu XP,
Fang YQ, Ruan XX and Gao X: Toll-like receptor 9 agonists promote
IL-8 and TGF-beta1 production via activation of nuclear factor
kappaB in PC-3 cells. Cancer Genet Cytogenet. 192:60–67. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Assaf A, Esteves H, Curnow SJ and Browning
MJ: A threshold level of TLR9 mRNA predicts cellular responsiveness
to CpG-ODN in haematological and non-haematological tumour cell
lines. Cell Immunol. 259:90–99. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ilvesaro JM, Merrell MA, Li L, Wakchoure
S, Graves D, Brooks S, Rahko E, Jukkola-Vuorinen A, Vuopala KS,
Harris KW, et al: Toll-like receptor 9 mediates CpG
oligonucleotide-induced cellular invasion. Mol Cancer Res.
6:1534–1543. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ilvesaro JM, Merrell MA, Swain TM,
Davidson J, Zayzafoon M, Harris KW and Selander KS: Toll like
receptor-9 agonists stimulate prostate cancer invasion in vitro.
Prostate. 67:774–781. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Merrell MA, Ilvesaro JM, Lehtonen N, Sorsa
T, Gehrs B, Rosenthal E, Chen D, Shackley B, Harris KW and Selander
KS: Toll-like receptor 9 agonists promote cellular invasion by
increasing matrix metalloproteinase activity. Mol Cancer Res.
4:437–447. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tuomela J, Sandholm J, Karihtala P,
Ilvesaro J, Vuopala KS, Kauppila JH, Kauppila S, Chen D, Pressey C,
Härkönen P, et al: Low TLR9 expression defines an aggressive
subtype of triple-negative breast cancer. Breast Cancer Res Treat.
135:481–493. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Neve RM, Chin K, Fridlyand J, Yeh J,
Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al: A
collection of breast cancer cell lines for the study of
functionally distinct cancer subtypes. Cancer Cell. 10:515–527.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sandholm J, Kauppila JH, Pressey C,
Tuomela J, Jukkola-Vuorinen A, Vaarala M, Johnson MR, Harris KW and
Selander KS: Estrogen receptor-α and sex steroid hormones regulate
Toll-like receptor-9 expression and invasive function in human
breast cancer cells. Breast Cancer Res Treat. 132:411–419.
2012.
|
|
12
|
Chaudary N and Hill RP: Hypoxia and
metastasis in breast cancer. Breast Dis. 26:55–64. 2007.
|
|
13
|
Ramos-DeSimone N, Hahn-Dantona E, Sipley
J, Nagase H, French DL and Quigley JP: Activation of matrix
metalloproteinase-9 (MMP-9) via a converging plasmin/stromelysin-1
cascade enhances tumor cell invasion. J Biol Chem. 274:13066–13076.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yeomans ND: Aspirin: old drug, new uses
and challenges. J Gastroenterol Hepatol. 26:426–431. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vazquez-Martin A, López-Bonetc E, Cufi S,
Oliveras-Ferraros C, Del Barco S, Martin-Castillo B and Menendez
JA: Repositioning chloroquine and metformin to eliminate cancer
stem cell traits in pre-malignant lesions. Drug Resist Updat.
14:212–223. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Solomon VR, Hu C and Lee H: Design and
synthesis of chloroquine analogs with anti-breast cancer property.
Eur J Med Chem. 45:3916–3923. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Espina V, Mariani BD, Gallagher RI, Tran
K, Banks S, Wiedemann J, Huryk H, Mueller C, Adamo L, Deng J, et
al: Malignant precursor cells pre-exist in human breast DCIS and
require autophagy for survival. PLoS One. 5:e102402010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu C, Raja Solomon V, Cano P and Lee H: A
4-aminoquinoline derivative that markedly sensitizes tumor cell
killing by Akt inhibitors with a minimum cytotoxicity to non-cancer
cells. Eur J Med Chem. 45:705–709. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rahim R and Strobl JS: Hydroxychloroquine,
chloroquine, and all-trans retinoic acid regulate growth, survival,
and histone acetylation in breast cancer cells. Anticancer Drugs.
20:736–745. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Loehberg CR, Thompson T, Kastan MB,
Maclean KH, Edwards DG, Kittrell FS, Medina D, Conneely OM and
O’Malley BW: Ataxia telangiectasia-mutated and p53 are potential
mediators of chloroquine-induced resistance to mammary
carcinogenesis. Cancer Res. 67:12026–12033. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Loehberg CR, Strissel PL, Dittrich R,
Strick R, Dittmer J, Dittmer A, Fabry B, Kalender WA, Koch T,
Wachter DL, et al: Akt and p53 are potential mediators of reduced
mammary tumor growth by Chloroquine and the mTOR inhibitor RAD001.
Biochem Pharmacol. 83:480–488. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jiang PD, Zhao YL, Deng XQ, Mao YQ, Shi W,
Tang QQ, Li ZG, Zheng YZ, Yang SY and Wei YQ: Antitumor and
antimetastatic activities of chloroquine diphosphate in a murine
model of breast cancer. Biomed Pharmacother. 64:609–614. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yerlikaya A, Okur E and Ulukaya E: The
p53-independent induction of apoptosis in breast cancer cells in
response to proteasome inhibitor bortezomib. Tumour Biol.
33:1385–1392. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kuznik A, Bencina M, Svajger U, Jeras M,
Rozman B and Jerala R: Mechanism of endosomal TLR inhibition by
antimalarial drugs and imidazoquinolines. J Immunol. 186:4794–4804.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu X, Pan Y, Li Y, Jiang Y, Shang H,
Gowda DC, Cui L and Cao Y: Targeting Toll-like receptors by
chloroquine protects mice from experimental cerebral malaria. Int
Immunopharmacol. 13:392–397. 2012. View Article : Google Scholar : PubMed/NCBI
|