Introduction
Hepatocellular carcinoma (HCC), the most common
primary liver cancer, is a highly lethal disease that causes
~700,000 mortalities worldwide each year (1). It has been reported that as many as
80% of HCC cases can be attributed to chronic hepatitis B virus
(HBV) infection (2). Currently,
there are no biomarkers for the early detection of HCC, and the
majority of patients with HCC are diagnosed at advanced stages,
which are associated with a poor prognosis and low survival rates
due to a lack of curative treatment options. A common approach used
for screening HCC in a high risk-population is to examine serum
tumor markers, such as α-fetoprotein (AFP). However, the
sensitivity and specificity of serum AFP levels for HCC have been
reported to range from 39–64% and 76–91%, respectively, indicating
that elevated serum AFP levels are not a sufficient indicator of
HCC (3,4). Thus, it is critical to identify novel
biochemical markers for the early detection of HCC.
microRNAs (miRNAs) are small non-coding RNAs that
post-transcriptionally regulate gene expression, predominantly
through imperfect base pairing with the 3′-untranslated region of
target mRNAs (5). miRNAs affect a
broad range of biological functions including development,
apoptosis, proliferation and differentiation (6–8).
Dysregulation of miRNAs is also implicated in various diseases,
including cancer. There is evidence that clearly demonstrates that,
apart from genetic and epigenetic abnormalities, the dysregulation
of miRNAs may also contribute to the aberrant activation of
oncogenes and the inactivation of tumor suppressor genes in human
carcinogenesis (9,10). Aberrant expression of miRNAs has
been widely reported in human cancers with both up- and
downregulation detected in HCC tumor tissue relative to the
corresponding normal tissue (11–13).
miRNAs are notably stable in blood, and their expression patterns
appear to be tissue-specific. These characteristics make
circulating miRNAs good candidates for noninvasive testing for
cancer. Circulating miRNAs have been suggested as diagnostic
markers for various types of cancer (14–19).
Previously, we reported that miRNA-101 (miR-101) is downregulated
by the HBV X protein (20).
However, the applicability of circulating miR-101 in the diagnosis
of HBV-related HCC has not been explored.
Materials and methods
Serum and tissue specimens
Serum and tissues (paired tissue specimens from
HBV-related HCC tissues and adjacent noncancerous hepatic tissues)
were obtained from patients undergoing surgical HCC resection. The
specimens were collected at the Hepatobiliary Surgery Department of
the First Affiliated Hospital of Chongqing Medical University,
Chongqing, China. As a control, serum was also collected from
healthy volunteers. Volunteers had not been diagnosed previously
with any type of cancer, based on self-reporting. All participants
signed informed consent for the use of their blood samples prior to
recruitment. This study was approved by the Ethics Committee of The
First Affiliated Hospital of Chongqing Medical University.
RNA extraction
Total RNA was extracted from serum using TRIzol
reagent (Invitrogen, Carlsbad, CA, USA) according to the
instructions provided by the manufacturer. The purity of the
isolated RNA was determined by OD260/280-reading using a
NanodropND-2000 spectrophotometer (Thermo Scientific, Worcester,
MA, USA).
Real-time quantitative reverse
transcription-polymerase chain reaction (qPCR) for miRNA
expression
Reverse transcription was performed using the M-MLV
Reverse Transcription system (Promega Corporation, Madison, WI,
USA). U6 RNA was used as an internal control for the miRNA. The
primers used for stem-loop reverse-transcription PCR for miR-101
were purchased from RiboBio Co., Ltd. (Guangzhou, China). qPCR was
performed using a standard SYBR-Green PCR kit protocol for a
StepOne Plus system (Applied Biosystems, Foster City, CA, USA).
Finally, relative expression was calculated using the comparative
Ct method and normalized to the U6RNA internal control.
Statistical analysis
Statistical analysis was performed using SPSS 16.0
software (SPSS, Inc., Chicago, IL, USA). Correlations were analyzed
using Pearson’s correlation. The statistical significance of the
differences in data was determined using the Student’s t-test, and
data are expressed as the mean ± standard deviation from at least
three independent experiments. Differences were considered
statistically significant when P<0.05.
Results
Characteristics of study subjects
The demographics of the study subjects are
summarized in Table I. The gender
distribution between the two groups of study subjects was similar.
However, healthy controls were on average younger than patients
with HCC. Twenty of the 25 patients with HCC were hepatitis B
surface antigen-positive, indicating concurrent HBV infection,
whereas none of the healthy controls carried HBV.
 | Table IDemographics of healthy controls and
patients with primary HCC. |
Table I
Demographics of healthy controls and
patients with primary HCC.
| Healthy controls
(n=20) | Patients with HCC
(n=25) |
|---|
|
|
|
|---|
| No. | (%) | No. | (%) |
|---|
| Gender |
| Male | 13 | 65.0 | 19 | 76.0 |
| Female | 7 | 35.0 | 6 | 24.0 |
| Age (years) |
| ≤40 | 17 | 85.0 | 4 | 16.0 |
| 41–50 | 2 | 10.0 | 8 | 32.0 |
| 51–60 | 1 | 5.0 | 10 | 40.0 |
| >60 | 0 | 0.0 | 3 | 12.0 |
| HBV status |
|
HBsAg+ | 0 | 0.0 | 20 | 80.0 |
|
HBsAg− | 20 | 100.0 | 5 | 20.0 |
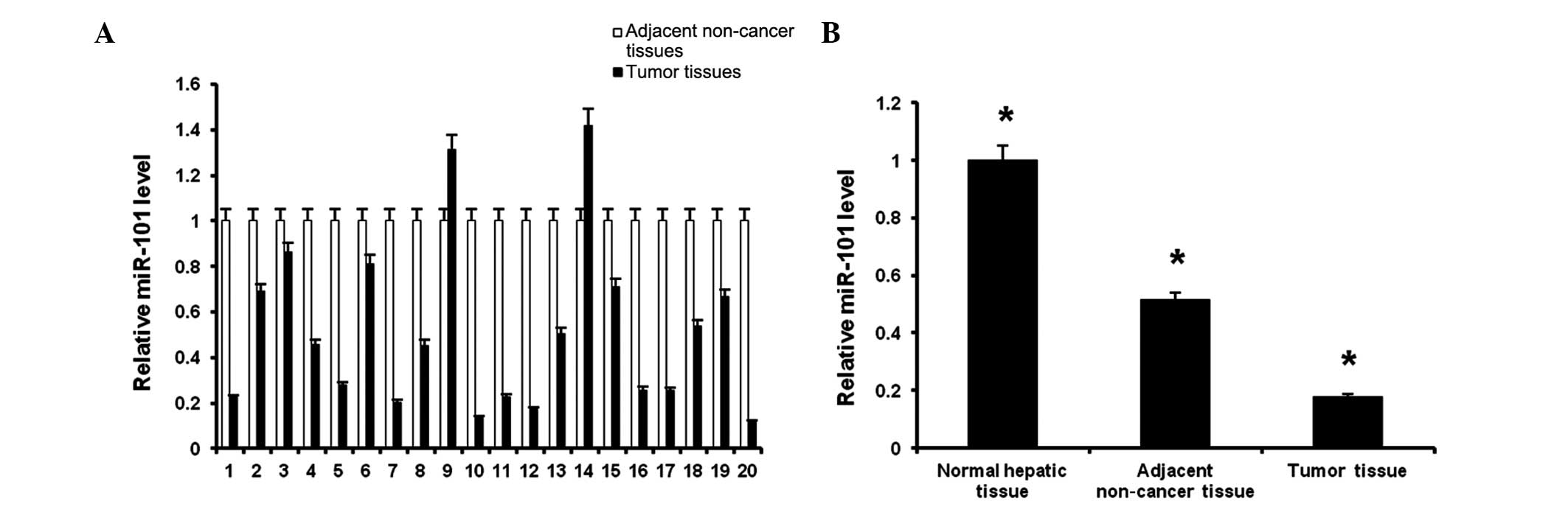
miR-101 is downregulated in human
HBV-related HCC tissues
To verify whether miR-101 is differentially
expressed in HBV-related HCC tumors, we measured miR-101 expression
levels in the 20 human HBV-related HCC tissues and adjacent
noncancerous hepatic tissues by qPCR. Among the 20 matched samples,
miR-101 expression within the same patient was significantly
decreased in 90% of the HCC samples compared with adjacent
noncancerous hepatic tissues (Fig.
1A), verifying our previous findings (20) that transformed HBV-infected HCC
cells have downregulated miR-101 expression.
On average, the miR-101 expression levels in
HBV-related HCC tissue were ~50% lower compared with those in
normal adjacent tissue (Fig. 1B,
first two bars). In addition, the levels of miR-101 in both the
tumor and adjacent normal tissues from human HBV-related HCC
patients were downregulated compared with those in tissue from the
healthy controls (Fig. 1B, third
bar). These results confirm the findings in Fig. 1A and indicate that reduced tissue
miR-101 levels are observed in all hepatic tissues from HCC
patients, but are more marked in tumor tissue.
miR-101 is upregulated in human
HBV-related HCC serum
Circulating miRNAs have emerged as candidate
diagnostic markers for various types of cancer (14–19).
We hypothesized that dysregulated miR-101 in serum may be suitable
for use as a biomarker for HCC diagnosis. To test this, we assessed
whether serum miR-101 levels show a similar decrease to tissue
miR-101 levels in HCC patients. Notably, although miR-101 tissue
levels were downregulated approximately two- to five-fold in
HBV-related HCC patients in this study (Fig. 1) and our previous study (20), the expression of miR-101 in
HBV-related HCC serum increased by approximately 10-fold compared
with the healthy controls (Fig.
2A). This discrepancy between the trends of downregulation in
tissue from HCC patients versus upregulation in serum from HCC
patients may be explained by the increased amount of overall
hepatic tissue in HCC patients with tumor masses. The miR-101
levels in the tissue samples were standardized to the U6 RNA levels
within the tissue, and therefore should account for cell number;
however, the serum levels assess absolute circulating miR-101
levels and are standardized to the higher level of U6 RNA in serum,
which may arise from a variety of cell sources. An alternative
explanation for this discrepancy is that different mechanisms may
regulate the production of miRNA within the cell and its release.
Regardless of the explanation, the marked increase in serum levels
of miR-101 may be suitable for use as a diagnostic biomarker for
HBV-related HCC.
Inverse correlation between serum miR-101
levels and HBV-related HCC tissue miR-101 levels
To verify the disparate results for tissue and serum
miR-101 expression trends, we assessed whether serum miR-101 levels
have a positive correlation with miR-101 levels in HBV-related HCC
tissue. miR-101 mRNA expression levels in HBV-related HCC tumor
tissue and serum were compared using the qPCR data for all 20 HCC
patients. Our results show that serum miR-101 levels have an
inverse correlation with the levels in tumor tissue (Fig. 2B). This verifies that different
mechanisms underlie the trends for the downregulation of miR-101 in
tissue and the upregulation of miR-101 in serum of HCC
patients.
miR-101 expression and
clinicopathological characteristics
To determine whether circulating miR-101 levels are
indicative of the state of HCC progression, we studied whether
miR-101 serum expression correlated with the clinicopathological
characteristics of HCC. miR-101 levels were not statistically
associated with patient age or gender, or several of the classic
markers for HCC progression, including AFP levels, alanine
transaminase, aspartate aminotransferase, cirrhosis and
tumor-node-metastasis staging; however, a statistically significant
correlation was observed between miR-101 expression and HBsAg, HBV
DNA level and tumor size (Table
II). This significant correlation may indicate that miR-101
expression is important in HBV-related HCC tumorigenesis and may be
a potential tumor marker for this type of cancer when it is
associated with HBV positivity, as is the case for ~80% of all HCCs
(2).
 | Table IIClinicopathological features and
miR-101 expression in HCC. |
Table II
Clinicopathological features and
miR-101 expression in HCC.
| Clinicopathological
features | No. (n=25) | miR-101 expression
(−ΔCt) | P-value |
|---|
|
|---|
| <10.13 | ≥10.13 |
|---|
| Gender |
| Male | 19 | 10 | 9 | 0.409 |
| Female | 6 | 2 | 4 | |
| Age (years) |
| <40 | 4 | 1 | 3 | 0.315 |
| ≥40 | 21 | 11 | 10 | |
| HBsAg |
| + | 19 | 12 | 7 | 0.047 |
| − | 6 | 1 | 5 | |
| HBV DNA |
|
<1.0×10e3 | 10 | 9 | 1 | 0.001 |
|
≥1.0×10e3 | 15 | 3 | 12 | |
| AFP (μg/l) |
| <400 | 18 | 8 | 10 | 0.568 |
| ≥400 | 7 | 4 | 3 | |
| ALT (U/l) |
| <40 | 9 | 2 | 7 | 0.053 |
| ≥40 | 16 | 10 | 6 | |
| AST (U/l) |
| <40 | 11 | 5 | 6 | 0.821 |
| ≥40 | 14 | 7 | 7 | |
| Tumor size
(cm) |
| <5 | 16 | 11 | 5 | 0.006 |
| ≥5 | 9 | 1 | 8 | |
| Cirrhosis |
| + | 18 | 9 | 9 | 0.748 |
| − | 7 | 3 | 4 | |
| TNM staging |
| I | 6 | 5 | 1 | 0.137 |
| II | 14 | 5 | 9 | |
| III | 5 | 3 | 2 | |
Discussion
HCC, which is the fifth most common cause of cancer
worldwide, has an extremely poor prognosis. The early diagnosis of
HCC is of great clinical importance and may improve the prognosis
of HCC if patients were to receive surgical treatment early.
Despite noteworthy advances in the effort to develop noninvasive
serum biomarkers for the diagnosis of HCC, the reliability of
biomarkers, such as AFP, remains debatable. Indeed, the specificity
of AFP is low, particularly in the context of chronic liver disease
(21). Accordingly, novel
biomarkers for early HCC diagnosis are urgently required.
miRNAs have been identified in many body fluids,
including serum and plasma (22),
and studies have indicated circulating miRNAs as potential
biomarkers for several disease conditions, including human cancer
(23–25). Circulating miRNAs can therefore be
considered representative of certain pathological conditions.
Moreover, their accessibility and high stability in the circulatory
system (15) make them ideal
biomarkers, particularly for the surveillance of early-stage,
pre-symptomatic diseases in at-risk patients (26). Studies have shown that miRNAs can
function as oncogenes or tumor suppressors to promote or prevent
HCC development (27,28). Therefore, it is anticipated that
circulating miRNAs are also affected during HCC progression. A
previous study reported altered levels of circulating miRNAs in
association with HCC. For instance, the serum level of miR-122 was
shown to be higher in HCC patients than in healthy controls and to
be reduced in post-operative serum samples (29). Although the clinical significance of
these findings has not been elucidated in detail, these findings
demonstrate that circulating miRNAs may be noninvasive diagnostic
or prognostic markers for HCC.
In this study, we demonstrated that miR-101 was
downregulated in 90% of HBV-positive HCC tumor tissues compared
with adjacent noncancerous tissue. Furthermore, these miR-101
levels were decreased compared with those in healthy controls.
Apparently contradictory to these findings, we found that the serum
miR-101 expression in patients with HBV-related HCC was
significantly higher than that in the healthy controls. Serum
miR-101 levels correlated with HBsAg, HBV DNA level and tumor size,
indicating that serum miR-101 may be used as a potential predictor
of HCC prognosis.
Although our results gave a high positive predictive
value for our HCC cohort, there are several limitations to this
study. For example, the post-surgical serum samples were small in
size and varied in time point. It is imperative to test more
longitudinal samples in order to justify the specific time or
period at which the circulating miRNAs return to basal levels.
Substantial evidence has implicated that serum-based
miRNAs are useful as noninvasive biomarkers for different types of
cancer (30–33); however, little is known regarding
the source of circulating miRNAs and the mechanisms that control
their biogenesis. It is speculated that miRNAs may enter the
circulation via secretion from blood cells or tissues/cells that
are affected by disease (15). At
present, aberrant serum miRNA levels in cancer are considered to be
due to excessive secretion by primary cancer cells (34–36).
Further studies are required to determine the exact time during
cancer progression at which circulating miRNAs become detectable in
the bloodstream.
In conclusion, our findings indicate that the
fluctuation in circulating miRNAs during HCC provides an innovative
approach that offers a sensitive and convenient means for the early
detection of HBV-related HCC carcinogenesis. Serum miR-101
expression, which was closely associated with tumoral size in this
study, provides a promising biochemical marker of HBV-related
HCC.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (no. 81171562) and the Chongqing Health
Bureau grant (no. 2011-1-010).
Abbreviations:
|
miR-101
|
microRNA-101
|
|
HBV
|
hepatitis B virus
|
|
HBsAg
|
hepatitis B surface antigen
|
|
HCC
|
hepatocellular carcinoma
|
|
AFP
|
α-fetoprotein
|
|
AST
|
aspartate aminotransferase
|
|
ALT
|
alanine transaminase
|
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Arbuthnot P and Kew M: Hepatitis B virus
and hepatocellular carcinoma. Int J Exp Pathol. 82:77–100. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Oka H, Tamori A, Kuroki T, Kobayashi K and
Yamamoto S: Prospective study of alpha-fetoprotein in cirrhotic
patients monitored for development of hepatocellular carcinoma.
Hepatology. 19:61–66. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Marrero JA and Lok AS: Newer markers for
hepatocellular carcinoma. Gastroenterology. 127:S113–S119. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stefani G and Slack FJ: Small non-coding
RNAs in animal development. Nat Rev Mol Cell Biol. 9:219–230. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mott JL: MicroRNAs involved in tumor
suppressor and oncogene pathways: implications for hepatobiliary
neoplasia. Hepatology. 50:630–637. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Visone R, Petrocca F and Croce CM:
Micro-RNAs in gastrointestinal and liver disease. Gastroenterology.
135:1866–1869. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jiang J, Gusev Y, Aderca I, Mettler TA,
Nagorney DM, Brackett DJ, et al: Association of MicroRNA expression
in hepatocellular carcinomas with hepatitis infection, cirrhosis,
and patient survival. Clin Cancer Res. 14:419–427. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fornari F, Gramantieri L, Ferracin M,
Veronese A, Sabbioni S, Calin GA, et al: miR-221 controls
CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular
carcinoma. Oncogene. 27:5651–5661. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gramantieri L, Fornari F, Callegari E,
Sabbioni S, Lanza G, Croce CM, et al: MicroRNA involvement in
hepatocellular carcinoma. J Cell Mol Med. 12:2189–2204. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cortez MA and Calin GA: MicroRNA
identification in plasma and serum: a new tool to diagnose and
monitor diseases. Expert Opin Biol Ther. 9:703–711. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, et al: Circulating microRNAs as
stable blood-based markers for cancer detection. Proc Natl Acad Sci
USA. 105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu W, Qin W, Atasoy U and Sauter ER:
Circulating microRNAs in breast cancer and healthy subjects. BMC
Res Notes. 2:892009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pain SJ, Barber RW, Ballinger JR, Solanki
CK, Mortimer PS, Purushotham AD and Peters AM: Local vascular
access of radioprotein injected subcutaneously in healthy subjects
and patients with breast cancer-related lymphedema. J Nucl Med.
45:789–796. 2004.
|
|
18
|
Croci S, Pedrazzi G, Passeri G, Delsignore
R and Ortalli I: Red cell Hb oxidation of healthy subjects compared
to breast cancer patients. Anticancer Res. 22:2903–2906.
2002.PubMed/NCBI
|
|
19
|
Würz H, Lüben G and Bohn H: Serum levels
of placental protein 10 (PP10) in women with breast cancer and
genital carcinoma and in healthy male and female subjects. Arch
Gynecol. 233:267–274. 1983.PubMed/NCBI
|
|
20
|
Wei X, Xiang T, Ren G, Tan C, Liu R, Xu X
and Wu Z: miR-101 is down-regulated by the hepatitis B virus x
protein and induces aberrant DNA methylation by targeting DNA
methyltransferase 3A. Cell Signal. 25:439–446. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zinkin NT, Grall F, Bhaskar K, Otu HH,
Spentzos D, Kalmowitz B, et al: Serum proteomics and biomarkers in
hepatocellular carcinoma and chronic liver disease. Clin Cancer
Res. 14:470–477. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Weber JA, Baxter DH, Zhang S, Huang DY,
Huang KH, Lee MJ, et al: The microRNA spectrum in 12 body fluids.
Clin Chem. 56:1733–1741. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ng EK, Chong WW, Jin H, Lam EK, Shin VY,
Yu J, et al: Differential expression of microRNAs in plasma of
patients with colorectal cancer: a potential marker for colorectal
cancer screening. Gut. 58:1375–1381. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ji X, Takahashi R, Hiura Y, Hirokawa G,
Fukushima Y and Iwai N: Plasma miR-208 as a biomarker of myocardial
injury. Clin Chem. 55:1944–1949. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang K, Zhang S, Marzolf B, Troisch P,
Brightman A, Hu Z, et al: Circulating microRNAs, potential
biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA.
106:4402–4407. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bianchi F, Nicassio F, Marzi M, Belloni E,
Dall’olio V, Bernard L, et al: A serum circulating miRNA diagnostic
test to identify asymptomatic high-risk individuals with early
stage lung cancer. EMBO Mol Med. 3:495–503. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ladeiro Y, Couchy G, Balabaud C,
Bioulac-Sage P, Pelletier L, et al: MicroRNA profiling in
hepatocellular tumors is associated with clinical features and
oncogene/tumor suppressor gene mutations. Hepatology. 47:1955–1963.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu S, Guo W, Shi J, Li N, Yu X, Xue J, et
al: MicroRNA-135a contributes to the development of portal vein
tumor thrombus by promoting metastasis in hepatocellular carcinoma.
J Hepatol. 56:389–396. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Qi P, Cheng SQ, Wang H, Li N, Chen YF and
Gao CF: Serum microRNAs as biomarkers for hepatocellular carcinoma
in Chinese patients with chronic hepatitis B virus infection. PLoS
One. 6:e284862011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Brase JC, Wuttig D, Kuner R and Sültmann
H: Serum microRNAs as non-invasive biomarkers for cancer. Mol
Cancer. 9:306 View Article : Google Scholar : 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wittmann J and Jäck HM: Serum microRNAs as
powerful cancer biomarkers. Biochim Biophys Acta. 1806:200–207.
2010.PubMed/NCBI
|
|
32
|
Jackson DB: Serum-based microRNAs: are we
blinded by potential? Proc Natl Acad Sci USA. 106:E52009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qu KZ, Zhang K, Li H, Afdhal NH and
Albitar M: Circulating microRNAs as biomarkers for hepatocellular
carcinoma. J Clin Gastroenterol. 45:355–360. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Skog J, Würdinger T, van Rijn S, Meijer
DH, Gainche L, Sena-Esteves M, et al: Glioblastoma microvesicles
transport RNA and proteins that promote tumour growth and provide
diagnostic biomarkers. Nat Cell Biol. 10:1470–1476. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cortez MA and Calin GA: MicroRNA
identification in plasma and serum: a new tool to diagnose and
monitor diseases. Expert Opin Biol Ther. 9:703–711. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pegtel DM, Cosmopoulos K, Thorley-Lawson
DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, et
al: Functional delivery of viral miRNAs via exosomes. Proc Natl
Acad Sci USA. 107:6328–6333. 2010. View Article : Google Scholar : PubMed/NCBI
|