Introduction
Triple-negative breast cancer (TNBC) is recognized
as a subtype of breast cancer in which three receptors [estrogen
receptor (ER), progesterone receptor and human epidermal growth
factor receptor 2] are negatively expressed on the surface of
breast cancer cells (1,2). TNBC is an invasive tumor with a high
degree of malignancy, and accounts for approximately 12–20% of
breast cancer cases (2–5). Compared with other breast cancer
subtypes, the overall prognosis for TNBC is poor, as it is prone to
brain metastasis and bone metastasis. Currently, according to
genomic differences, TNBC is divided into three categories with a
total of six subtypes: i) basal-like (BL), including BL1, BL2 and
immunomodulatory (IM) subtypes; ii) mesenchymal-like (ML),
including mesenchymal (M) and mesenchymal stem-like (MSL) subtypes;
and iii) luminal androgen receptor (LAR) subtype (4,6–8). The
genes involved in steroid synthesis and androgen metabolism are
highly expressed in the LAR subtype of TNBC.
There are no uniform treatment guidelines and no
standard chemotherapy program for TNBC to date (4,9–11).
Chemotherapy is the only option for TNBC treatment and has a good
prognosis for those TNBC patients who are sensitive to
chemotherapy. Results from clinical observations have shown that
the pathological complete remission rate of TNBC patients is higher
than that in ER+ breast cancer patients after standard
chemotherapy (treatment with anthracycline and taxane drugs)
(4,9,12–16).
However, therapeutic drugs are limited for chemotherapy-resistant
TNBC patients, as they are resistant to common chemotherapy drugs
for breast cancer, such as anthracycline and taxane (4,9,10).
Capecitabine and ixabepilone have been used for the treatment of
anthracycline and taxane chemotherapy-resistant TNBC patients.
Preclinical studies have shown that capecitabine with ixabepilone
has synergistic antitumor activity (17). As monotherapy with ixabepilone has a
good human tolerance and synergistic antitumor activity without
overlapping toxicities with capecitabine, ixabepilone monotherapy
or combination therapy with capecitabine has become an effective
treatment strategy for TNBC patients (4,9,10,18–25).
MicroRNAs (miRNAs) are a class of endogenous
non-coding small RNAs that can inhibit gene expression at the
post-transcriptional level by inhibiting mRNA translation and
promoting mRNA degradation, and are involved in many key processes
of cell activity, such as development, differentiation, metabolism,
apoptosis and proliferation (26).
An increasing number of studies have revealed that miRNA is
involved in tumorigenesis, differentiation and metastasis, and that
anticancer drugs can change the miRNA expression profiles of tumor
cells, indicating that the change in miRNA expression profiles may
account for the mechanism behind the antitumor effect of these
chemotherapy drugs (27). Moreover,
it has been reported that the chemotherapy drug-tolerance of tumors
is also associated with miRNA expression (28). Therefore, miRNA has been used in
studies on early tumor diagnosis, classification, prognosis, drug
sensitivity prediction and exploration of drug mechanisms (26,28–36).
To date, the abnormal expression of miRNA profiles has been found
to exist in a variety of breast cancer types. It has been reported
that miR-21, miR-155, miR-210, miR-29c, miR-196a, miR-213, miR-191,
miR-203, miR-29b and miR-93 are highly expressed in breast cancer,
while miR-125b, miR-145, miR-100, miR-10b, miR-125-b2, miR-497 and
miR143 are minimally expressed, producing an anti-apoptotic effect,
and promoting proliferation, metastasis and invasion by modulating
the target gene expression (13,26,29–33).
In Radojicic et al’s study on miRNA expression in TNBC, a
significantly higher expression of miR-21, miR-210 and miR-221 and
a notably lower expression of miR-10b, miR-145, miR-205 and
miR-122a were observed compared with that in normal tissues.
However, there were no statistically significant differences in
miR-222 and miR-296 expression between TNBC and normal tissues
(31). miRNA expression profiles
have been recognized as a potential diagnostic method for tumors.
However, the literature documenting the miRNA expression profiles
of pathological specimens of TNBC subtypes has been limited to
date.
The TNBC subtype cell line MDA-MB-231 is the most
commonly used cell line in previous studies. However, these studies
have rarely focused on miRNA expression profiles (34–36).
There are very few reported studies on miRNA expression profiles
for other TNBC subtypes (31).
Capecitabine and ixabepilone can be used for the treatment of
metastatic breast cancer (including TNBC), but no studies have
stated the impact of these two drugs on miRNA expression profiles
in TNBC. miRNA plays an essential role in early tumor diagnosis,
classification, prognosis, anti-drug sensitivity forecast, drug
action mechanism and cancer treatment; therefore, it is important
to explore the miRNA profile expression of different TNBC subtypes
and the changes in miRNA profile expression when various TNBC
subtype cells are treated with chemotherapy drugs.
This study aimed to explore the miRNA profile
expression differences between the LAR-type TNBC MDA-MB-453 cell
line and normal breast cells, including six carcinogenic miRNAs
(miR-296, miR-222, miR-221, miR-210, miR-10b and miR-21) and three
antitumor miRNAs (miR-145, miR-205 and miR-122a). The effect of the
capecitabine active metabolite 5-fluorouracil (5-FU), ixabepilone
and 5-FU + ixabepilone on the miRNA profile expression of
MDA-MB-453 was also explored, aiming to further clarify the
association between miRNAs and LAR-type TNBC. In addition, we aimed
to discover potential new mechanisms of chemotherapy with these
drugs in LAR-type TNBC treatment.
Materials and methods
Cell culture
The human LAR-type TNBC MDA-MB-453 cell line and the
normal breast CRL-2713 (MDA-kb2) cell line were obtained from ATCC
Company (Manassas, VA, USA). MDA-MB-453 cells were cultured in
RPMI-1640 (SH30809.01B; Hyclone Laboratories, Inc., Logan, UT, USA)
and incubated at 37ºC with humidified air containing 5%
CO2, while MDA-kb2 cells were cultured in Leibovitz’s
L-15 medium (SH30525.01; Hyclone Laboratories, Inc.) with
atmospheric air at 37ºC. Both types of medium were supplemented
with 10% FBS (SV30087.02; Hyclone Laboratories, Inc.), penicillin
(100 U/ml) and streptomycin (0.1 mg/ml).
Drug treatment and MTT assay
MDA-MB-453 cells cultured in 96-well plates (3599;
Corning, Tewksbury, MA, USA) were treated with either a gradient
concentration of 5-FU (F6627; Sigma-Aldrich, St. Louis, MO, USA)
ranging from 0.0001–10,000 μM or ixabepilone (Hubei Honch
Pharmaceutical Co., Ltd., Wuhan, China) ranging from 0.0001–10,000
nM. The IC10 value of these drugs, the concentration
that can induce 10% cell inhibition, was analyzed by an MTT assay
and determined for further analysis. For the MTT assay, MDA-MB-453
cells were seeded in quadruplicate on 96-well plates at a density
of 3×105 cells per well in 100 μl RPMI-1640 medium, and
incubated in an air-humidified incubator at 37ºC with 5%
CO2 for 24 h. The cells were then treated with a
gradient concentration of drugs and cultured for another 24 h.
Subsequently, 20 μl of 5 mg/ml MTT (AR1156; Wuhan Boster Biological
Technology, Wuhan, China) dissolved in PBS (BD-1070; Hubei Biossci,
Wuhan, China) were added to each well and the cells were incubated
for 4 h, followed by the addition of 100 μl of formazan and
incubation for 30 min on a flat shaker at room temperature in the
dark to completely dissolve the crystals. The optical density was
determined by a microplate reader (RT-6100; Rayto Life and
Analytical Sciences Co., Ltd, Shenzhen, China) using a 560-nM
filter. The results were statistically analyzed and the cell
inhibition rate was determined with the following formula: cell
inhibition % = 100% −
(OD2-OD0)/(OD1-OD0) ×
100%. OD0, culture medium alone; OD1, cells
untreated; OD2, cells treated with drugs.
RNA extraction and stem-loop RT-PCR
Total RNA was extracted from MDA-MB-453 or MDA-kb2
cell lines using TRIzol reagent (15596026; Invitrogen Life
Technologies, Carlsbad, CA, USA) according to the manufacturer’s
instructions. Stem-loop RT-PCR assay was performed using an
All-in-One™ First Strand cDNA Synthesis kit (AORT-100; GeneCopoeia,
Inc., Rockville, MD, USA) as described previously (37). A total of 13 μl of reaction system
was prepared containing extracted RNA (10 ng-1 μg), U6 primer and
miRNA stem-loop primer, incubated at 65ºC for 10 min, and then
incubated on ice for 2 min. The 13 μl of RNA-primer mixture was
then mixed with 5 μl of 5X reaction buffer, 1 μl of dNTP (25 mM), 1
μl of Rnase inhibitor (15 U/μl) and 1 μl of M-MLV RTase (200 U/μl),
and incubated at 42ºC for 60 min prior to heat-inactivation at 85ºC
for 5 min. The product was stored at −20ºC for subsequent usage.
The sequences of miRNA stem-loop primers are shown in Table I.
 | Table ISequences of microRNA stem-loop
primers. |
Table I
Sequences of microRNA stem-loop
primers.
| microRNA | microRNA
sequence | microRNA stem-loop
primer |
|---|
| 296 |
AGGGCCCCCCCTCAATCCTGT |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACacagga |
| 222 |
AGCTACATCTGGCTACTGGGT |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACacccag |
| 221 |
AGCTACATTGTCTGCTGGGTTTC |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACgaaacc |
| 210 |
CTGTGCGTGTGACAGCGGCTGA |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACtcagcc |
| 10b |
TACCCTGTAGAACCGAATTTGTG |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACcacaaa |
| 21 |
TAGCTTATCAGACTGATGTTGA |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACtcaaca |
| 145 |
GTCCAGTTTTCCCAGGAATCCCT |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACagggat |
| 205 |
TCCTTCATTCCACCGGAGTCTG |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACcagact |
| 122a |
TGGAGTGTGACAATGGTGTTTG |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACcaaaca |
qPCR
qPCR was conducted with a real-time PCR system (ABI
Step One Plus; Applied Biosystems, Inc., Foster City, CA, USA) in
20 μl of reaction mixture containing 5 μl of cDNA, 10 μl of 2X
All-in-One qPCR mix (AORT-1200; GeneCopoeia, Inc.), 0.4 μl of each
forward and reverse primer and 50X ROX reference dye buffer. The
PCR conditions were 3 min at 95ºC, and 40 cycles consisting of 15
sec at 95ºC, 20 sec at 56ºC and 20 sec at 72ºC. The internal
control primer was designed with Primer 5.0 software (PREMIER
Biosoft, Palo Alto, CA, USA) and the sequence of the common reverse
primer for qPCR was 5′-GTGCAGGGTCCGAGGT-3′, while the sequences of
the forward primers for the detection of miRNAs are shown in
Table II.
 | Table IISequences of forward primers for the
detection of microRNAs. |
Table II
Sequences of forward primers for the
detection of microRNAs.
| microRNA | Forward primers of
microRNA |
|---|
| 296 |
AGGGCCCCCCCTCAA |
| 222 |
CTGGGTGTCGTATCCAGTGC |
| 221 |
TTGTCTGCTGGGTTTCGTCG |
| 210 |
GTGTGACAGCGGCTGAGT |
| 10b |
AGAACCGAATTTGTGGTCGT |
| 21 |
TCAGACTGATGTTGAGTCGT |
| 145 |
TTCCCAGGAATCCCTGTCGT |
| 205 |
TCCACCGGAGTCTGGTCGTAT |
| 122a |
TGTGACAATGGTGTTTGGTCG |
Statistical analysis
The data from the MTT assay were statistically
analyzed by SPSS 11.5 software (SPSS, Inc., Chicago, IL, USA), and
the data from the miRNA expression level detection were
statistically analyzed by GraphPad Prism 5 software (GraphPad, San
Diego, CA, USA). Comparisons between the two groups were performed
by Student’s t test, while comparisons among multiple groups were
performed by one-way analysis of variance. P<0.05 was considered
to indicate a statistically significant difference.
Results
Effect of 5-FU and ixabepilone on the
human LAR-type TNBC cell line, MDA-MB-453
As 5-FU and ixabepilone have been implicated in the
treatment of TNBC, the sensitivity of the LAR-type TNBC cell line,
MDA-MB-453, to 5-FU and ixabepilone was determined and a
dose-response curve was generated by detecting the cell viability
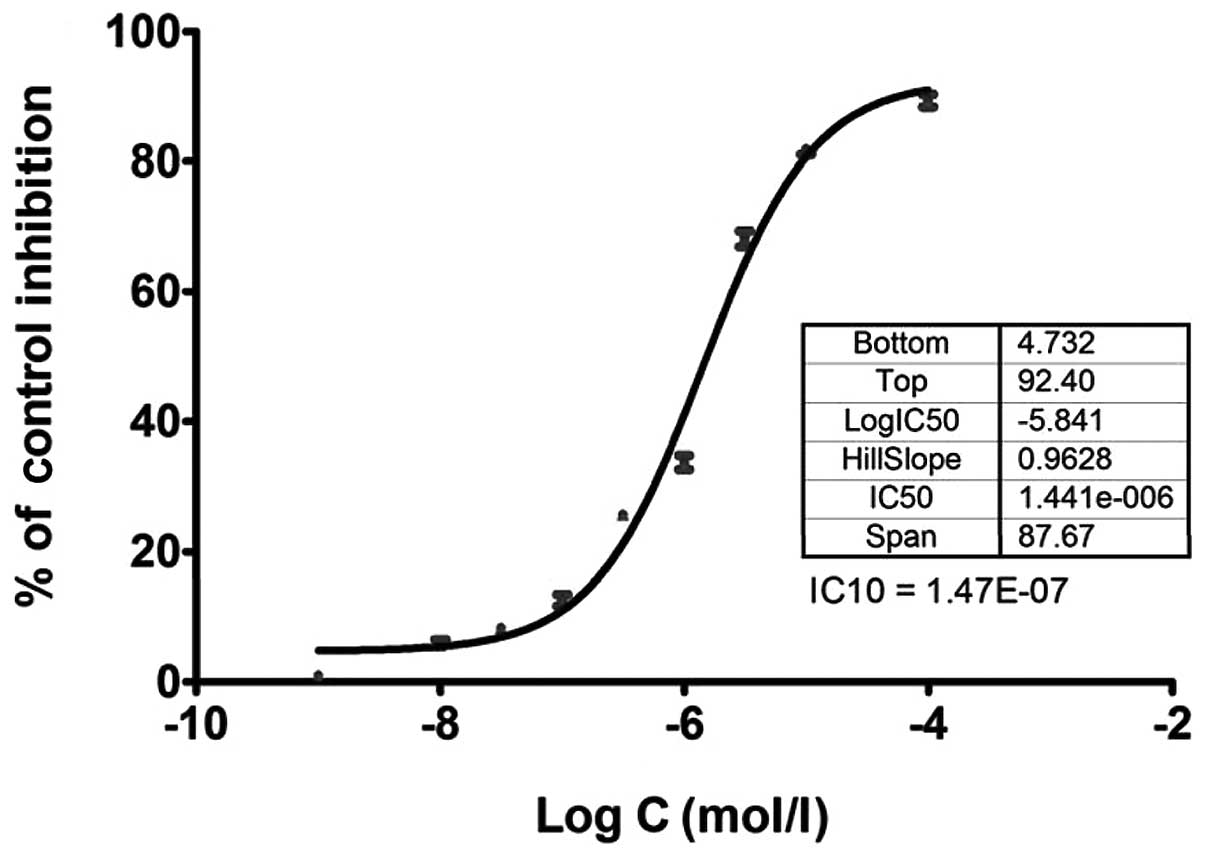
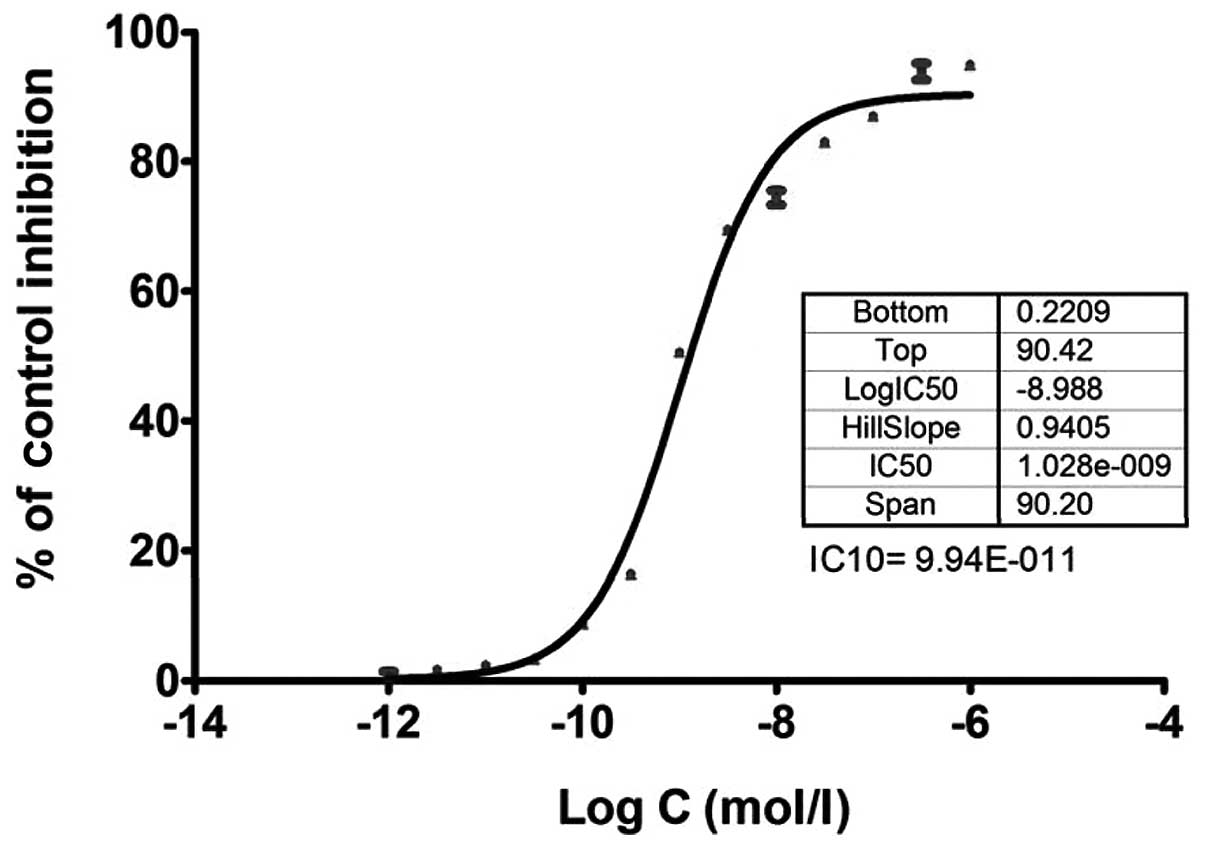
with an MTT assay as shown in Figs.
1 and 2. The low concentration
of 10% cell inhibition (IC10 value) by 5-FU and
ixabepilone drugs was selected to further examine the effects of
these two drugs on miRNA expression in MDA-MB-453 cells, as a high
concentration of drugs may lead to cellular changes rather than
genetic changes (27). The
IC10 value of 5-FU was 1.47E-07 mol/l (Fig. 1), while that of ixabepilone was
9.94E-011 mol/l (Fig. 2).
Alternation of three anti-tumor miRNA
expression levels in MDA-MB-453 cells after 5-FU and ixabepilone
treatment
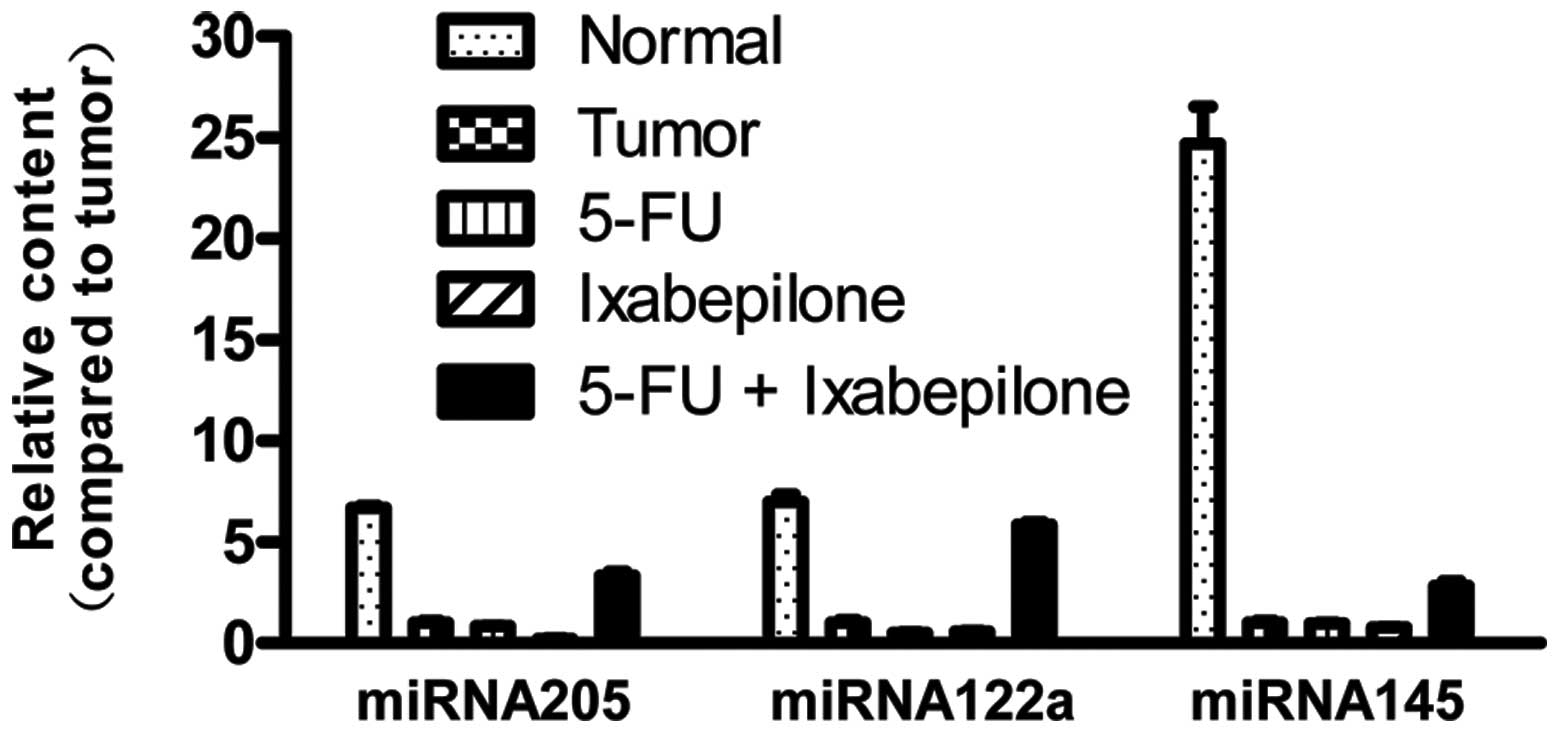
A previous study demonstrated that expression levels
of miR-122a, miR-145 and miR-205 in TNBC are significantly lower
than those in normal tissues, and that these three miRNAs have
antitumor effects (31). In this
study, we analyzed the expression levels of miR-122a, miR-145 and
miR-205 in normal breast MDA-kb2 cells and MDA-MB-453 cells prior
to and after 5-FU and ixabepilone treatment. As shown in Fig. 3, the expression levels of miR-122a,
miR-145 and miR-205 in MDA-MB-453 cells were significantly lower
than those in normal breast cells (P<0.05, Table III). When MDA-MB-453 cells were
treated with 5-FU or ixabepilone, the expression levels of these
three miRNAs marginally decreased (Fig.
3, Table III). However, when
tumor cells were treated with 5-FU together with ixabepilone, the
expression levels increased significantly (P<0.05, Fig. 3, Table
III).
 | Table IIIP-values of comparisons of three
antitumor microRNA expression levels between the tumor group and
normal or drug-treated groups. |
Table III
P-values of comparisons of three
antitumor microRNA expression levels between the tumor group and
normal or drug-treated groups.
| P-value |
|---|
|
|
|---|
| microRNA | Normal vs.
tumor | 5-FU vs. tumor | Ixabepilone vs.
tumor | 5-FU+ ixabepilone
vs. tumor |
|---|
| miRNA122a | <0.0001 | 0.0171 | 0.0305 | <0.0001 |
| miRNA145 | 0.0002 | 0.7914 | 0.0853 | 0.0020 |
| miRNA205 | <0.0001 | 0.2423 | 0.0022 | 0.0004 |
Alternation of six carcinogenic miRNA
expression levels in MDA-MB-453 cells after 5-FU and ixabepilone
treatment
It has been reported that miR-296, miR-222, miR-221,
miR-210, miR-10b and miR-21 are abnormally expressed in breast
cancer, and are recognized as carcinogenic miRNAs. In the present
study, we examined expression levels of these six miRNAs in normal
breast MDA-kb2 cells and MDA-MB-453 cells. In Fig. 4, we can observe that the expression
of miR-210 in MDA-MB-453 cells was at an undetected level while
that of miR-222 was marginally higher than in normal cells.
Expression levels of miR-296, miR-221, miR-21 and miR-10b in
MDA-MB-453 cells were notably lower than those in normal cells
(P<0.05, Fig. 4, Table IV). When MDA-MB-453 cells were
treated with 5-FU or ixabepilone, expression levels of all six
miRNAs were marginally decreased compared with those in untreated
tumor cells (Fig. 4, Table IV). However, when cells were
treated with 5-FU + ixabepilone, expression levels of miR-221,
miR-210, miR-21 and miR-10b increased significantly, while
expression levels of miR-296 showed a significant decrease
(P<0.05, Fig. 4, Table IV). No significant difference was
observed between expression levels of miR-222 after treatment with
5-FU + ixabepilone compared with those in the untreated tumor cells
(P>0.05, Table IV).
 | Table IVP-values of comparisons of six
carcinogenic microRNA expression levels between the tumor group and
normal or drug-treated groups. |
Table IV
P-values of comparisons of six
carcinogenic microRNA expression levels between the tumor group and
normal or drug-treated groups.
| P-value |
|---|
|
|
|---|
| microRNA | Normal vs.
tumor | 5-FU vs. tumor | Ixabepilone vs.
tumor | 5-FU+ ixabepilone
vs. tumor |
|---|
| miRNA10b | <0.0001 | 0.9165 | 0.0039 | 0.0022 |
| miRNA21 | <0.0001 | 0.0097 | 0.0009 | 0.0015 |
| miRNA210 | - | <0.0001 | 0.4806 | 0.0016 |
| miRNA221 | 0.0006 | 0.1021 | 0.0090 | 0.0013 |
| miRNA222 | 0.0141 | 0.0027 | 0.0006 | 0.6478 |
| miRNA296 | 0.0007 | 0.0016 | 0.0182 | 0.0005 |
Discussion
Breast cancer is the most common type of malignant
tumor in women worldwide. Approximately 1.2 million women suffer
from breast cancer each year, of whom around 500,000 die, and the
incidence is growing at a rate of 2% per annum (38). As a systemic disease, the
therapeutic effect of breast cancer depends on early diagnosis, the
degree of metastasis and treatment options (3,4,12,13).
Generally, the early stage of breast cancer is estrogen receptor
(ERα)-positive and non-metastatic, so an early diagnosis is vital
for effective treatment of breast cancer. However, breast cancer
patients in the earlier stages usually lack clear clinical
characteristics and there are currently no clinically effective
auxiliary diagnostic methods for early breast cancer detection
(3,29). Therefore, the majority of cases are
diagnosed as advanced tumors with ineffective treatment options and
a poor prognosis (16). Although
certain diagnostic tools and biomarkers are currently being used
for the clinical diagnosis of breast cancer, there are many
shortcomings: Ionizing radiation and a high false-positive rate
exist in breast X-ray examination techniques, and tumor markers
such as ERα lack specificity (3).
Therefore, detecting the early signs of breast cancer and
developing early diagnostic reagents are essential for the primary
prevention of breast cancer.
Increasing evidence has revealed that miRNA
expression profiles can be used to identify the tissue samples that
are difficult to be determined by histology, since miRNA expression
profiles may represent the degree of differentiation of tissues
(3). Therefore, the establishment
of miRNA expression profiles in normal tissue and tumor tissue is
crucial for the accurate and efficient diagnosis of the disease.
The first detailed study of the correlation between miRNA and
breast cancer was reported by Croce’s group (39). In their study, they detected 76
types of miRNA expression in breast cancer by miRNA microarray
analysis. The results showed that, compared with normal tissue, the
expression levels of 29 miRNAs significantly changed in breast
cancer. They found that five miRNAs are required for the 100%
successful identification of normal tissue and cancerous tissue:
miR10b, miR-125b, miR-145, miR-21 and miR-155. Moreover, the
expression levels of miR-125b and miR-145 decreased markedly while
the other three miRNAs showed a clear increase (39). Currently, there is increasing
evidence to suggest that microRNA analysis may be implemented in
breast cancer diagnosis, prognosis and treatment (40–42).
However, few studies have revealed the miRNA
expression profiles of pathological specimens of TNBC subtypes
(31). Studies on TNBC have often
not carefully distinguished the TNBC subtypes, resulting in a
certain degree of confusion when studying TNBC. In the present
study, we examined miRNA expression in the LAR-type TNBC cell line,
MDA-MB-453, and explored the variations in this expression compared
with the normal breast cell line, MDA-kb2. Six carcinogenic miRNAs
(miR-296, miR-222, miR-221, miR-210, miR-10b and miR-21) and three
antitumor miRNAs (miR-145, miR-205 and miR-122a) were analyzed. The
results showed that the expression levels of miR-122a, miR-145 and
miR-205 in MDA-MB-453 cells were significantly lower than those in
normal breast cells, as has been observed in other subtypes of TNBC
(31). However, the expression
levels of miR-296, miR-221, miR-21 and miR-10b in MDA-MB-453 cells
were also notably lower than those in normal cells, which is in
contrast to previous studies on miRNA expression profiles in other
subtypes of TNBC (31). This
indicates that different TNBC subtypes exhibit different microRNA
expression features, which can be used for early diagnosis and
classification.
Capecitabine is an oral nucleoside metabolic
inhibitor that can be metabolized to 5-FU in tissue to block DNA
synthesis (43). Preclinical
studies have shown that capecitabine combined with ixabepilone has
synergistic antitumor activity, and that the effect of this
combination therapy on metastatic TNBC is superior to a single drug
application (4,9,10,18–25).
However, the mechanisms underlying this effect remain unclear. We
hypothesized that combination therapy with these two drugs may
change miRNA expression profiles in tumor cells, thereby
efficiently inhibiting tumor growth via modulation of target gene
expression. It has been reported that antitumor drugs can change
miRNA expression profiles in tumor cells, and that chemotherapy
drug-resistant tumors are also correlated with miRNA expression
level. Therefore, the variation in miRNA expression profiles may be
one mechanism by which drug treatment inhibits tumors (27,28).
Studies by Shah et al have found that variations in miRNA
expression profiles occur in 5-FU- treated breast cancer MCF-7
cells (27), prompting us to
explore the miRNA expression profiles in LAR-type TNBC MDA-MB-453
cells that have been treated with capecitabine and ixabepilone,
alone or in combination. In the present study, we examined the
expression levels of nine miRNAs and found that when MDA-MB-453
cells were treated with 5-FU or ixabepilone alone, the expression
of almost all miRNAs decreased marginally. By contrast, following
combined treatment with 5-FU and ixabepilone, the expression of
seven miRNAs increased significantly, the expression levels of
miR-296 decreased and those of miR-222 showed no notable change.
Although the expression levels of three antitumor miRNAs (miR-145,
miR-205 and miR-122a) increased significantly when tumor cells were
treated with 5-FU + ixabepilone, those of other carcinogenic miRNAs
(miR-221, miR-210, miR-10b and miR-21) also increased. The
treatment effect of 5-FU + ixabepilone on LAR-type TNBC therefore
requires further study. The abovementioned results indicate that
joint treatment with 5-FU and ixabepilone had a more marked effect
on LAR-type TNBC MDA-MB-453 cells compared with treatment with 5-FU
or ixabepilone alone. Moreover, it significantly altered the miRNA
expression profiles in tumor cells, indicating that the examination
of miRNA expression profiles could be used for prognosis.
TNBC has a variety of subtypes. In this study, we
examined the miRNA expression profiles of LAR-type TNBC MDA-MB-453
cells, and analyzed the correlation between miRNA expression
profiles in tumor cells and in normal breast MDA-kb2 cells. We
found that the expression levels of nine microRNAs in LAR-type TNBC
were unlike those previously reported in other subtypes of TNBC.
Therefore, it is necessary to distinguish the subtypes of TNBC
during treatment. We also explored the variation in miRNA
expression profiles in TNBC cells when treated with the
chemotherapy drugs capecitabine and ixabepilone, alone or in
combination, in order to clarify the potential mechanisms of
chemotherapy in LAR-type TNBC. Our study provides a theoretical
basis for the future clinical application of miRNA expression
profiles in the early diagnosis, classification and prognosis of
breast cancer.
Acknowledgements
This study was financially supported by the Hubei
Provincial Department of Education Natural Science Foundation
(B20121304).
References
|
1
|
Brenton JD, Carey LA, Ahmed AA and Caldas
C: Molecular classification and molecular forecasting of breast
cancer: ready for clinical application? J Clin Oncol. 23:7350–7360.
2005. View Article : Google Scholar
|
|
2
|
de Ruijter TC, Veeck J, de Hoon JP, van
Engeland M and Tjan-Heijnen VC: Characteristics of triple-negative
breast cancer. J Cancer Res Clin Oncol. 137:183–192.
2011.PubMed/NCBI
|
|
3
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. New Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Peddi PF, Ellis MJ and Ma C: Molecular
basis of triple negative breast cancer and implications for
therapy. Int J Breast Cancer. 2012:2171852012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yao-Lung K, Dar-Ren C and Tsai-Wang C:
Clinicopathological features of triple-negative breast cancer in
Taiwanese women. Int J Clin Oncol. 16:500–505. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hall RE, Birrell SN, Tilley WD and
Sutherland RL: MDA-MB-453, an androgen-responsive human breast
carcinoma cell line with high level androgen receptor expression.
Eur J Cancer. 30A:484–490. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lehmann BD, Bauer JA, Chen X, et al:
Identification of human triple-negative breast cancer subtypes and
preclinical models for selection of targeted therapies. J Clin
Invest. 121:2750–2767. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Singh G, Odriozola L, Guan H, Kennedy CR
and Chan AM: Characterization of a novel PTEN mutation in
MDA-MB-453 breast carcinoma cell line. BMC Cancer. 11:4902011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fornier M and Fumoleau P: The paradox of
triple negative breast cancer: novel approaches to treatment.
Breast J. 18:41–51. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gucalp A and Traina TA: Triple-negative
breast cancer: adjuvant therapeutic options. Chemother Res Pract.
2011:6962082011.PubMed/NCBI
|
|
11
|
Yagata H, Kajiura Y and Yamauchi H:
Current strategy for triple-negative breast cancer: appropriate
combination of surgery, radiation, and chemotherapy. Breast Cancer.
18:165–173. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Minami CA, Chung DU and Chang HR:
Management options in triple-negative breast cancer. Breast Cancer
(Auckl). 5:175–199. 2011.PubMed/NCBI
|
|
13
|
Rakha EA and Chan S: Metastatic
triple-negative breast cancer. Clin Oncol (R Coll Radiol).
23:587–600. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Steponaviciene L, Lachej-Mikeroviene N,
Smailyte G, Aleknavicius E, Meskauskas R and Didziapetriene J:
Triple negative breast cancer: adjuvant chemotherapy effect on
survival. Adv Med Sci. 56:285–290. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu J, Li S, Jia W and Su F: Response and
prognosis of taxanes and anthracyclines neoadjuvant chemotherapy in
patients with triple-negative breast cancer. J Cancer Res Clin
Oncol. 137:1505–1510. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brady-West DC and McGrowder DA: Triple
negative breast cancer: therapeutic and prognostic implications.
Asian Pac J Cancer Prev. 12:2139–2143. 2011.PubMed/NCBI
|
|
17
|
Lee F, Jure-Kunkel MN and Salvati ME:
Synergistic activity of ixabepilone plus other anticancer agents:
preclinical and clinical evidence. Ther Adv Med Oncol. 3:11–25.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fornier M: Ixabepilone plus capecitabine
for breast cancer patients with an early metastatic relapse after
adjuvant chemotherapy: two clinical trials. Clin Breast Cancer.
10:352–358. 2010. View Article : Google Scholar
|
|
19
|
Perez EA, Patel T and Moreno-Aspitia A:
Efficacy of ixabepilone in ER/PR/HER2-negative (triple-negative)
breast cancer. Breast cancer Res Treat. 121:261–271. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang J, Fan Y and Xu B: Ixabepilone plus
capecitabine for Chinese patients with metastatic breast cancer
progressing after anthracycline and taxane treatment. Cancer
Chemother Pharmacol. 66:597–603. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jassem J, Fein L, Karwal M, et al:
Ixabepilone plus capecitabine in advanced breast cancer patients
with early relapse after adjuvant anthracyclines and taxanes: a
pooled subset analysis of two phase III studies. Breast. 21:89–94.
2012. View Article : Google Scholar
|
|
22
|
Li L, Li J, Yang K, et al: Ixabepilone
plus capecitabine with capecitabine alone for metastatic breast
cancer. Future Oncol. 6:201–207. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Roché H, Conte P, Perez EA, et al:
Ixabepilone plus capecitabine in metastatic breast cancer patients
with reduced performance status previously treated with
anthracyclines and taxanes: a pooled analysis by performance status
of efficacy and safety data from 2 phase III studies. Breast Cancer
Res Treat. 125:755–765. 2011.
|
|
24
|
Denduluri N and Swain S: Ixabepilone:
clinical role in metastatic breast cancer. Clin Breast Cancer.
11:139–145. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sparano JA, Vrdoljak E, Rixe O, et al:
Randomized phase III trial of ixabepilone plus capecitabine versus
capecitabine in patients with metastatic breast cancer previously
treated with an anthracycline and a taxane. J Clin Oncol.
28:3256–3263. 2010. View Article : Google Scholar
|
|
26
|
Yu Z, Baserga R, Chen L, Wang C, Lisanti
MP and Pestell RG: microRNA, cell cycle, and human breast cancer.
Am J Pathol. 176:1058–1064. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shah MY, Pan X, Fix LN, Farwell MA and
Zhang B: 5-Fluorouracil drug alters the microRNA expression
profiles in MCF-7 breast cancer cells. J Cell Physiol.
226:1868–1878. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kutanzi KR, Yurchenko OV, Beland FA,
Checkhun VF and Pogribny IP: MicroRNA-mediated drug resistance in
breast cancer. Clin Epigenetics. 2:171–185. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Andorfer CA, Necela BM, Thompson EA and
Perez EA: MicroRNA signatures: clinical biomarkers for the
diagnosis and treatment of breast cancer. Trends in Mol Med.
17:313–319. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang L and Wang J: MicroRNA-mediated
breast cancer metastasis: from primary site to distant organs.
Oncogene. 31:2499–2511. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Radojicic J, Zaravinos A, Vrekoussis T,
Kafousi M, Spandidos DA and Stathopoulos EN: MicroRNA expression
analysis in triple-negative (ER, PR and Her2/neu) breast cancer.
Cell Cycle. 10:507–517. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rothé F, Ignatiadis M, Chaboteaux C, et
al: Global microRNA expression profiling identifies MiR-210
associated with tumor proliferation, invasion and poor clinical
outcome in breast cancer. PloS One. 6:e209802011.
|
|
33
|
Song B, Wang C, Liu J, et al: MicroRNA-21
regulates breast cancer invasion partly by targeting tissue
inhibitor of metalloproteinase 3 expression. J Exp Clin Cancer Res.
29:292010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang L, Wei L, Zhao W, et al:
Down-regulation of osteopontin expression by RNA interference
affects cell proliferation and chemotherapy sensitivity of breast
cancer MDA-MB-231 cells. Mol Med Rep. 5:373–376. 2012.PubMed/NCBI
|
|
35
|
Liu H, Cao YD, Ye WX and Sun YY: Effect of
microRNA-206 on cytoskeleton remodelling by downregulating Cdc42 in
MDA-MB-231 cells. Tumori. 96:751–755. 2010.PubMed/NCBI
|
|
36
|
Wang L, Shan BE, Sang MX, Lian YS, Wang B
and Ding CY: Effect of microRNA-mediated p65 gene silencing on the
proliferation and apoptosis of human breast cancer MDA-MB-231
cells. Nan Fang Yi Ke Da Xue Xue Bao. 31:1742–1747. 2011.(In
Chinese).
|
|
37
|
Varkonyi-Gasic E and Hellens RP:
Quantitative stem-loop RT-PCR for detection of microRNAs. Methods
Mol Biol. 744:145–157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Scully OJ, Bay BH, Yip G and Yu Y: Breast
cancer metastasis. Cancer Genomics Proteomics. 9:311–320. 2012.
|
|
39
|
Iorio MV, Ferracin M, Liu CG, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Calin GA and Croce CM: MicroRNA-cancer
connection: the beginning of a new tale. Cancer Res. 66:7390–7394.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jeyaseelan K, Herath WB and Armugam A:
MicroRNAs as therapeutic targets in human diseases. Expert Opin
Ther Targets. 11:1119–1129. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Walko CM and Lindley C: Capecitabine: a
review. Clin Ther. 27:23–44. 2005. View Article : Google Scholar
|