Introduction
Lung cancer ranks as the most frequent type of
cancer in males worldwide with increasing incidence rates according
to the epidemiology data (1).
Although great advances have been achieved in chemotherapy and
radiotherapy, only small, incremental improvements in the outcome
of lung cancer have been realized, and the long-term survival rate
of lung cancer patients has not improved significantly. Metastatic
spread to the regional lymph nodes, liver, bone and brain, which is
characteristic of non-small cell lung cancer (NSCLC), constitutes
the primary source of morbidity and mortality in lung cancer. The
majority of patients present with locally advanced (37%) or
metastatic (38%) disease at the time of diagnosis (2,3).
Increased understanding of the molecular mechanisms and processes
underlying the metastasis of lung cancer cells is critical for
developing effective new therapies for lung cancer. Specific
receptors have been identified that are required for cancer cells
to proliferate and migrate, modulating cancer progression (4). Thus, receptors may represent
opportunistic targets for engineering vehicles that localize in
primary and distal lung tumors.
Chemokines are a superfamily of chemoattractant,
cytokine-like proteins that bind to and activate a family of
chemokine receptors. Over 50 chemokines have been identified and
can be divided into four families (CXC, CX3C, CC and C), according
to the positions of four conserved cysteine residues (5). Chemokines, which are structurally and
functionally similar to growth factors, bind to G protein-coupled
receptors on leukocytes and stem cells and process guanine
nucleotide-binding proteins to initiate intracellular signaling
cascades that promote migration towards the chemokine source
(6). Chemokine CXC motif ligand 12
(CXCL12) is a CXC chemokine that interacts with a specific
receptor, CXC chemokine receptor 4 (CXCR4). There is increasing
evidence to suggest that the CXCL12/CXCR4 axis functions as a
critical molecular determinant for events, including maintaining
embryo development, mediating immune and inflammatory reactions and
the modulation of the hematopoietic system, involving HIV infection
and angiogenesis (7). CXCR4 has
previously been highlighted for its role in cancer metastasis. The
CXCL12/CXCR4 axis is important for activating a plethora of
phenomena, including chemotaxis, invasion, tumorigenicity and
angiogenesis and proliferation in cancer, particularly in the
process of organ-selective metastasis (6,8,9),
demonstrating that tumor cells expressing a high level of CXCR4
exhibit metastasis to target tissues (lung, liver and bone). The
target tissues express high levels of CXCL12, allowing tumor cells
to directionally migrate to target organs via the CXCL12-CXCR4
chemotactic axis. CXCR4 is hypothesized to be involved in cancer
invasion and metastasis, and higher levels of this receptor are
associated with higher grades and poor prognosis of cancer
(10,11). Notably, several retrospective
studies have also examined the role of CXCR4 in NSCLC by
investigating the association between CXCR4 expression with
clinical outcome; NSCLC patients with greater CXCR4 expression on
the surface of tumor cells have been observed to be more likely to
have metastatic disease (12,13).
Few studies have investigated the effects of small interfering RNA
(siRNA)-directed inhibition of CXCR4 in NSCLC. To gain further
insight into the effect of CXCR4 in the A549 lung cancer cell line,
CXCR4 expression was selectively knocked down in the present study
using RNAi. The effect of CXCL12/CXCR4 on the metastatic potential
of NSCLC was also observed.
Materials and methods
Cell culture
The A549 human lung cancer cell line was purchased
from the Archives Center of Wuhan University (Wuhan, China) and
cultured in RPMI-1640 culture medium containing 10% fetal bovine
serum, 100 mg/l penicillin and 100 mg/l streptomycin.
Construction of
pBSilence1.1-CXCR4-siRNA
The short hairpin RNA (shRNA) sequence with short
hairpin structure was designed for the coding region sequence by
online design software, BLOCK-iT™ RNAi Designer (Invitrogen Life
Technologies, Carlsbad, CA, USA), according to the design
principles of the RNA interference (RNAi) sequence. Sequences with
nonspecific inhibition to other genes were excluded following BLAST
homology analysis. The following three pairs of shRNA were
designed: i) CXCR4-1-1.1 sense,
5′-CACCTGGGCAATGGATTGGTCATTTCAAGACGATGACCAATCCATTGCCCATTTTTTG-3′
and antisense,
5′-AGCTCAAAAAATGGGCAATGGATTGGTCATCGTCTTGAAATGACCAATCCATTGCCCA-3′;
ii) CXCR4-2-1.1 sense,
5′-CACCGTGGCAAACTGGTACTTTGTTCAAGACGCAAAGTACCAGTTTGCCACTTTTTTG-3′
and antisense,
5′-AGCTCAAAAAAGTGGCAAACTGGTACTTTGCGTCTTGAACAAAGTACCAGTTTGCCAC-3′;
and iii) CXCR4-3-1.1 sense,
5′-CACCGGCTGAAAAGGTGGTCTATTTCAAGACGATAGACCACC TTTTCAGCCTTTTTTG-3′
and antisense,
5′-AGCTCAAAAAAGGCTGAAAAGGTGGTCTATCGTCTTGAAATAGACCACCTTTTCAGCC-3′.
pBSilence1.1 plasmid was completely digested by BsaI, and 1%
agarose gel electrophoresis was used to recycle the large
fragments. Each pair of shRNA-CXCR4 provided the corresponding
sequence following annealing. T4 ligase was used to associate the
annealing products CXCR4-1, -2 and -3 with pBSilence1.1 linearized
plasmid at 16°C overnight. The positive clone was obtained by
transforming E. coli DH5α and extracting the plasmids.
Following SacI digestion, 1% agarose gel electrophoresis and
sequencing identification were performed.
Cell culture and transfection
The A549 human lung cancer cell line was cultured in
RPMI-1640 culture medium containing 10% fetal bovine serum, 100
U/ml penicillin and 100 ng/ml streptomycin. The exponentially
growing cells were seeded and cultured in six-well culture plates.
Once the cell density had increased to 60–80%, Lipofectamine™ 2000
liposome transfection kit (Invitrogen Life Technologies) was used
to transfect the plasmid into the cells according to the
manufacturer’s instructions, and the transfection solution was
discarded after 4–6 h. Following cultivation for two days, the
original culture medium was discarded and screened by RPMI-1640
culture medium containing 400 mg/l hygromycin, and the
anti-hygromycin cells were collected after two weeks.
Cells were harvested 48 h following transfection for
the evaluation of CXCR4 expression. The transfection efficiency was
monitored by measuring the percentage of fluorescent cells among
500 total cells using fluorescence microscopy (model 7900; ABI,
Vernon, CA, USA).
Detection of CXCR4 mRNA expression by
reverse-transcription polymerase chain reaction (RT-PCR)
Following transfection for 48 h, total RNA was
extracted from the lung cancer cell line A549 by TRIzol reagent and
quantified by UV spectrophotometer. In addition, purity and RNA
concentration were measured by a UV spectrophotometer (model 752;
Shimadzu Corp., Kyoto, Japan). cDNA was obtained by reverse
transcription and was used as a template for PCR amplification of
the target genes and β-actin was used as standard control. The
amplification program used was as follows: Denaturation for 4 min
at 94°C, 30 sec at 94°C, 30 sec at 52°C, 5 sec at 72°C and,
following 30 cycles, the total extension was 4 min at 72°C. The
CXCR4 primers used were 5′-CCGTGGCAAACTGGTACTTT-3′ and
5′-GACGCCAACATAGACCACCT-3′ and the length of the product was 188
bp. The β-actin primers used were upstream,
5′-CACGATGGAGGGGCCGGACTCATC-3′ and downstream,
5′-TAAAGACCTCTATGCCAACACAGT-3′ (Tm=56°C). The PCR products were
collected for electrophoresis with 5% agarose gel and imaging. The
absorbance value of each strip was measured following analysis by a
gel imaging system (Gel-Doc, Bio-Rad, Hercules, CA, USA).
Western blot analysis of CXCR4 protein
expression in A549 cells following transfection
Following transfection for 48 h, the A549 lung
cancer cell line was washed with phosphate-buffered saline and 50
μl lysis buffer solution was added. Cells were collected and
allowed to stand for 30 min at 4°C, and then centrifuged at 13,800
× g for 20 min. Next, the supernatant was collected and total
protein concentration was measured by BCA assay (P0010; Beyotime,
Shanghai, China). Protein (50 μg) was obtained for 12%
polyacrylamide gel electrophoresis and electrically transferred to
PVDF membrane, which had been soaked in Tris-Buffered Saline and
Tween 20 (TBST) containing 5% skimmed milk powder. Nonspecific
antigens were blocked for 2 h at room temperature. Mouse anti-human
CXCR4 polyclonal antibody (1:400) was added and the membranes were
incubated overnight at 4°C, before being washed with TBST
containing 5% skimmed milk powder. Horseradish peroxidase-labeled
goat anti-rabbit secondary antibody (1:50,000) was added and the
mixture was reacted at room temperature for 2 h. The
chemiluminescent substrate (NCI5079; Thermo Fisher Scientific,
Rockford, IL, USA), ECL, was added following membrane washing, and
the results were analyzed by a GIS image analysis system
(Bio-Rad).
Detection of cell proliferation by MTT
assay
The stably transfected A549 lung cancer cell line
was prepared for single cell suspension with RPMI-1640 medium
containing 10% inactivated fetal bovine serum and the cell density
was adjusted to lx108/ml. Cells were added to 96-well
plates at a density of 1×104/100 μl. The following five
groups were set up: Group A, A549 cell line plus RPMI-1640 and 10%
newborn calf serum (NBS); group B, empty vector A549 cell line plus
RPMI-1640 and 10% NBS; group C, A549 cell line plus RPMI-1640, 10%
NBS and 100 ng/ml CXCL12; group D, A549 cell line, following RNAi,
plus RPMI-1640, 10% NBS and 100 ng/ml CXCL12; and group E, A549
cell line, following RNAi, plus RPMI-1640 and 10% NBS. MTT solution
(20 μl; 5 mg/ml) was added to each well after 24, 48 and 72 h,
respectively, and incubated for 4 h. Following termination of the
culture, the culture medium in each well was removed and discarded.
Next, 150 μl DMSO was added to each well for full dissolution of
the crystals. Cell proliferation was indicated by the absorption
value (measured using BE2100 system, Bug lab, Concord, CA, USA) of
each well at a wavelength of 570 nm.
Detection of A549 cell migration
capability by Transwell migration assay
Polycarbonate microporous membranes (pore size, 8
μm) were paved between the upper and lower Transwell chambers.
Different concentrations of CXCL12 (0, 30 and 100 ng/ml, groups A,
B and C, respectively) were added to the lower section of a
Transwell chamber. Equal cell numbers of A549 were seeded in the
upper chamber in the medium without CXCL12 (200 μl A549 cell
suspension with a density of 1×105/ml). The effect of RNAi on
chemotaxis migration was assessed by another Transwell-assay.
Various concentrations of CXCL12 (100 and 0 ng/ml, groups D and E,
respectively) were added to the lower section of a Transwell
chamber. Equal cell numbers of RNA-interfered A549 were seeded in
the upper chamber in medium without CXCL12 (200 μl RNA-interfered
A549 cell suspension with a density of 1×105/ml). Cells
were cultured for 24 h in a wet incubator at 37°C with 5%
CO2, prior to the removal of the small chamber. Cells on
the membrane were removed carefully with a swab. Methanol was used
to fix migration and was adhesive to the cells of the lower
chamber. Next, conventional hematoxylin and eosin staining was
carried out. Five fields of view (up, down, left, right and center)
were selected under a light microscope (magnification, ×200; IX71,
Olympus, Japan), cells in the lower chamber were counted and the
mean value was representative of infiltration strength value. The
cell migration inhibition ratio was calculated as follows: Cell
migration inhibition ratio (%)= [number of migrating cells in the
nonsilencing double-stranded (ds) RNA group - number of migrating
cells in the siRNA group] / number of migrating cells in the
nonsilencing dsRNA group × 100. Results were analyzed
statistically.
Statistical analysis
Data are presented as the means ± standard
deviation. Statistical analysis was performed using SPSS 15.0
software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered
to indicate a statistically significant difference.
Results
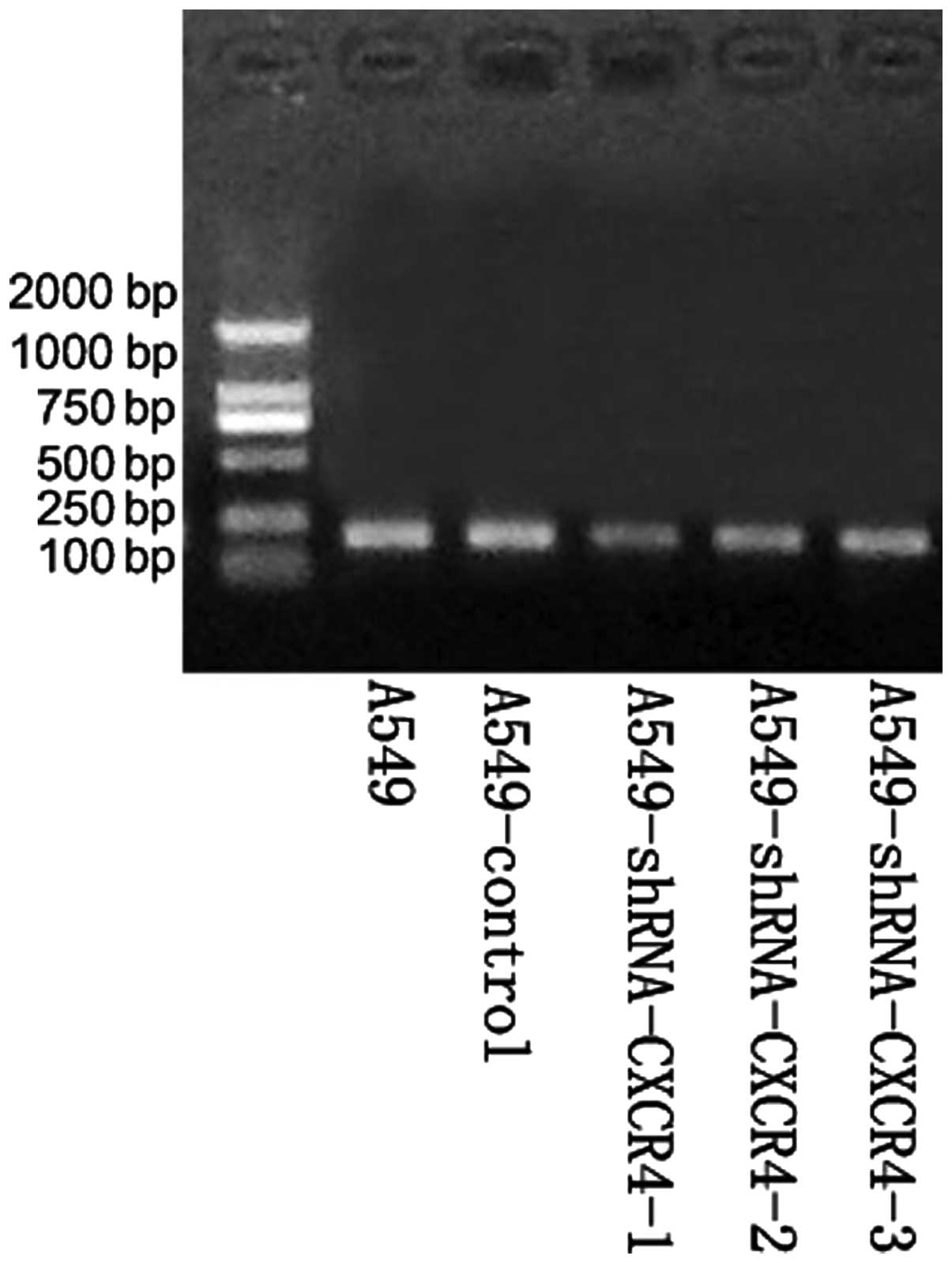
Efficacy of siRNA expression vectors in
transfection
Following PCR, recombinants were digested with the
SacI restriction enzyme. All plasmids, CXCR4-1, -2 and -3,
produced ~900-bp DNA fragments, indicating that the target fragment
had been successfully inserted into the pBSilence1.1 plasmid and in
the right direction. DNA sequencing analysis confirmed that the
sequence was consistent with the theoretical sequence (Fig. 1). In addition, restriction enzyme
digestion and sequencing analysis confirmed that the recombinant
vector, expressing three siRNA targeting the A549 CXCR4 gene in
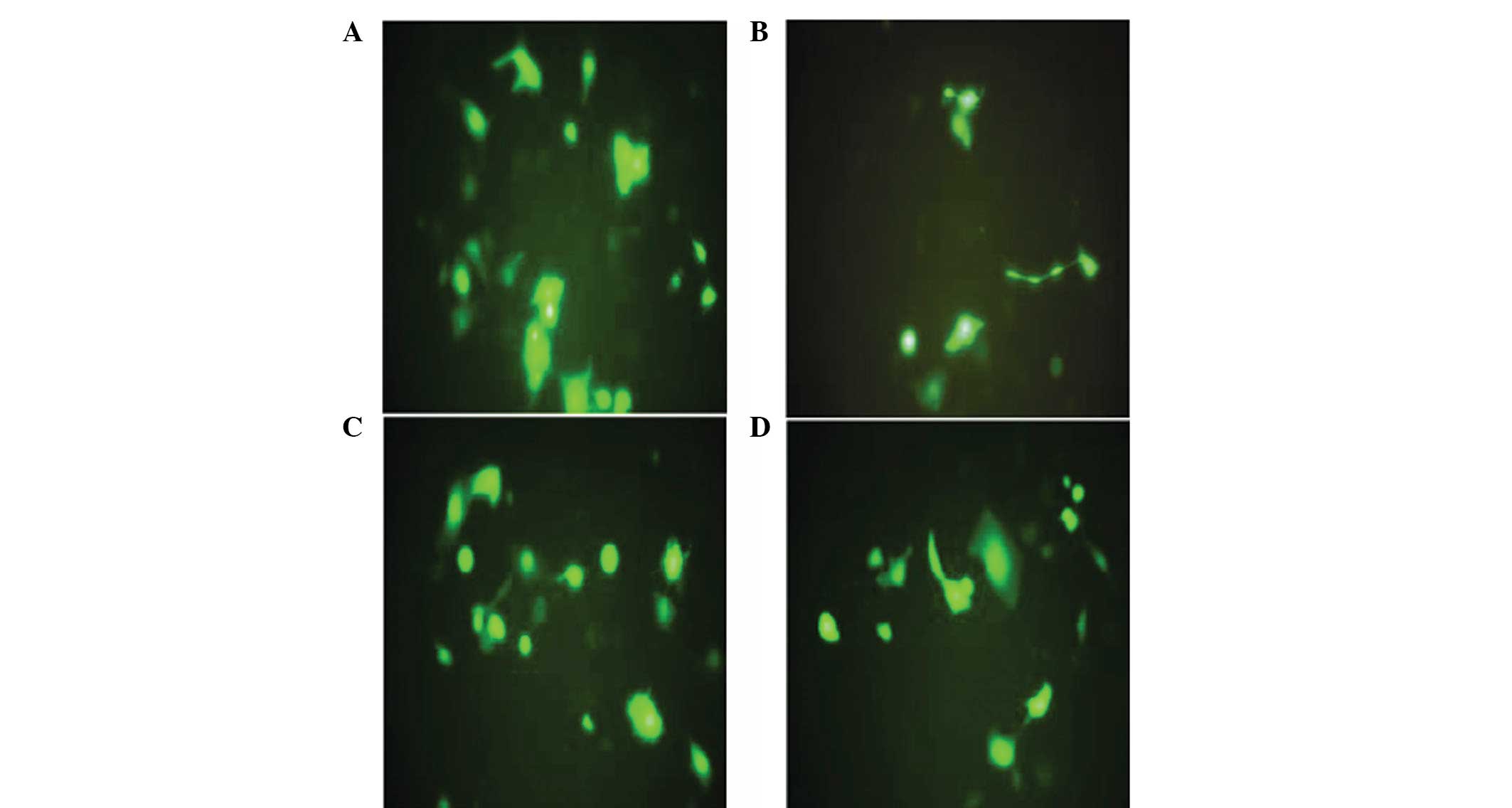
tandem, was constructed successfully. Following 48 h of
transfection, no green fluorescence was identified in the
untransfected group. By contrast, the expression of green
fluorescent protein was detected under a fluorescence microscope in
the empty vector and pBSilence1.1-CXCR4-1,-2 and -3 groups
(Fig. 2). Transfection efficiency
was determined as ~85%.
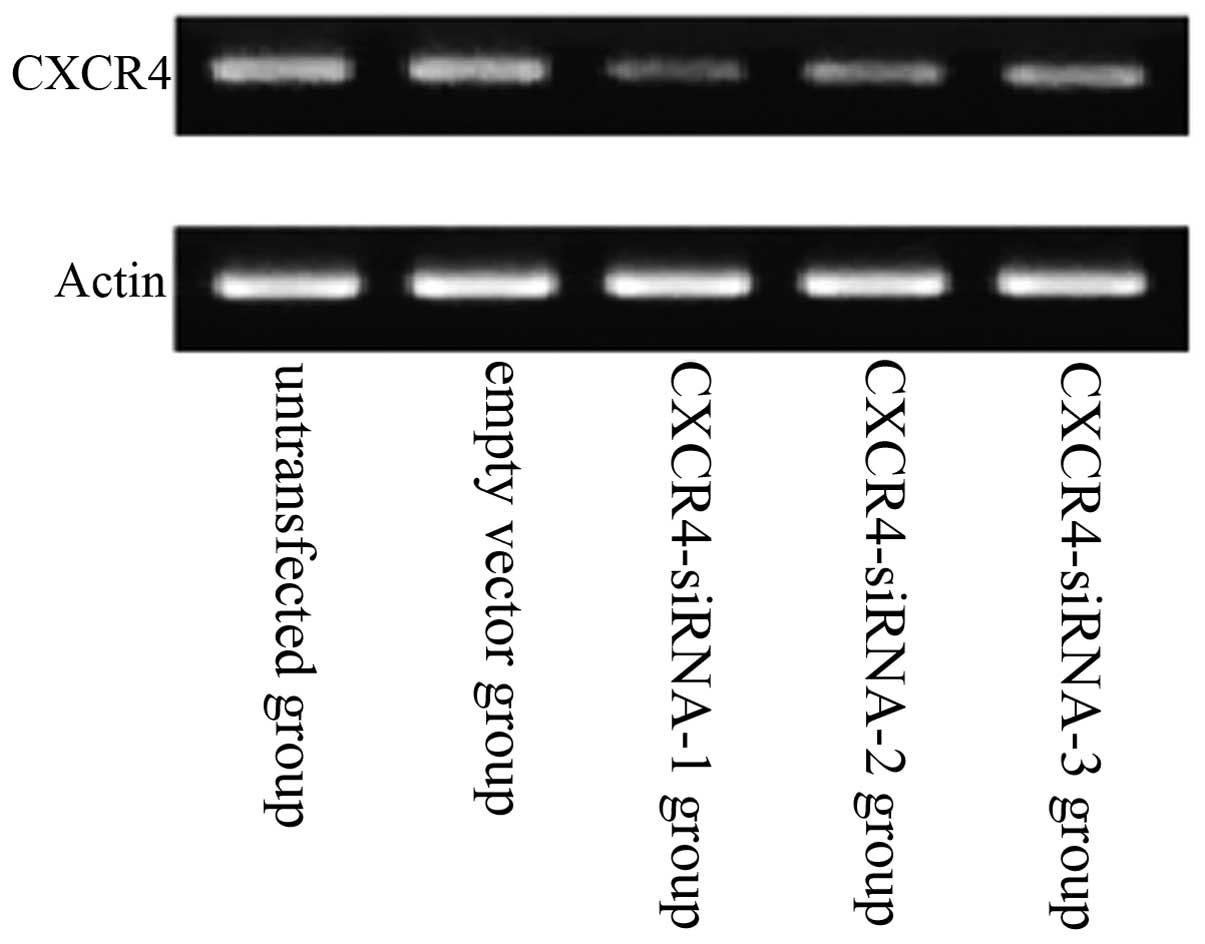
siRNA-expressing vector inhibits CXCR4
mRNA expression
Compared with the untransfected group, the mRNA
expression levels of CXCR4 in the A549 human lung carcinoma cell
line were downregulated in the CXCR4-siRNA-transfected group
(Fig. 3). Results also showed that
siRNA targeting CXCR4-1, -2 and -3 decreased CXCR4 expression
significantly at the mRNA level when compared with that of
scrambled siRNA. Of the three CXCR4 siRNAs, the strongest
interference efficiency siRNA was pBSilence1.1-CXCR4-1. However,
CXCR4-1 exhibited the most significant inhibitory effects on the
lung cancer cells (P<0.05). No significant difference was
identified between the untransfected and empty vector groups
(P>0.05).
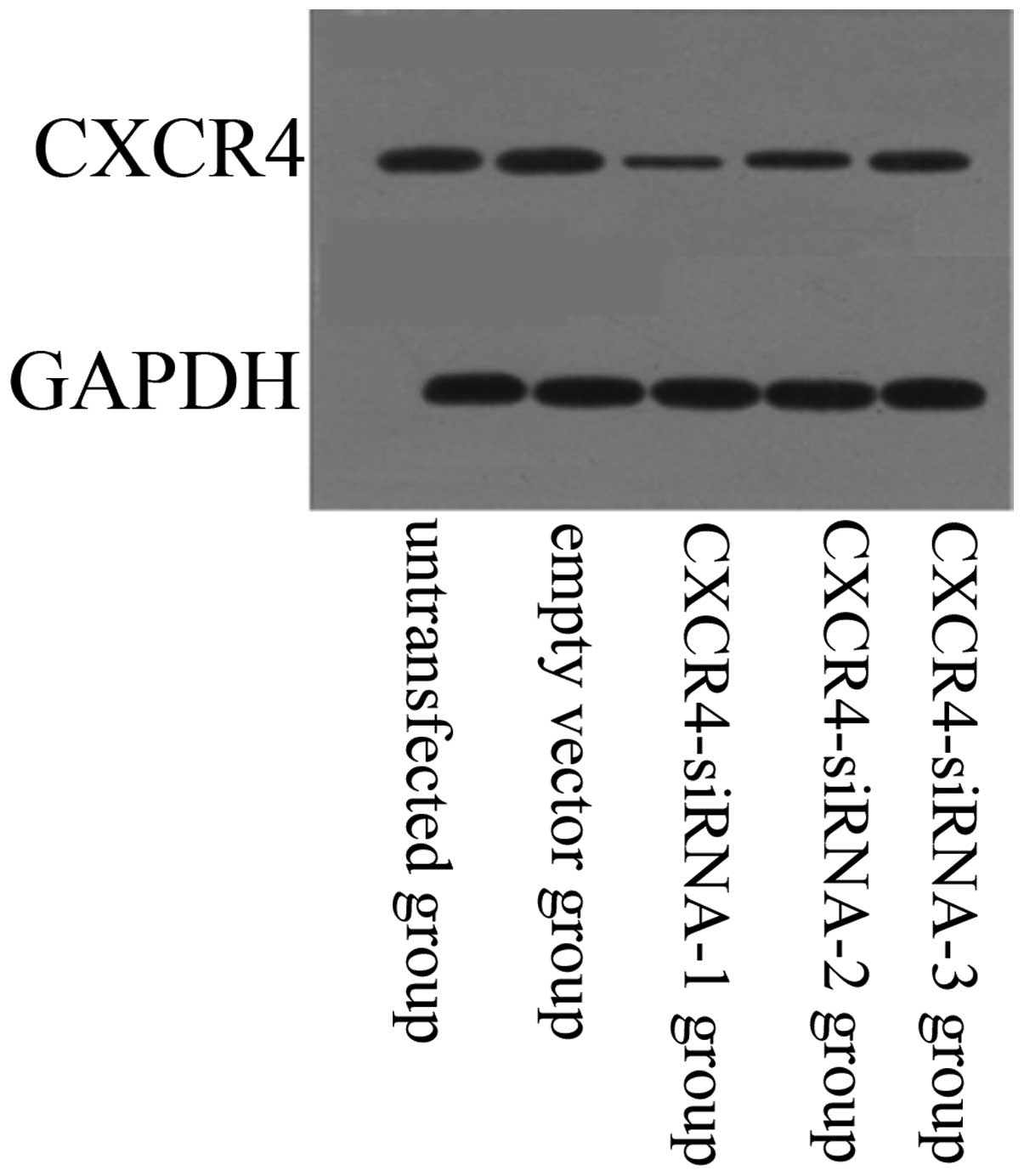
Effect of siRNA-expressing vectors on
CXCR4 protein expression
The effect of siRNA-expressing vectors on target
protein CXCR4 was examined by western blotting (Fig. 4). Compared with the untransfected
group, protein expression levels of CXCR4 were downregulated in the
CXCR4-siRNA transfected group (P<0.05). In addition, results
showed that siRNA targeting CXCR4-1, -2 and -3 decreased CXCR4
expression significantly at the protein level when compared with
that of scrambled siRNA. However, CXCR4-1 exhibited the most
significant inhibitory effects on the lung cancer cells
(P<0.05). No significant difference was identified between the
untransfected and empty vector groups (P>0.05).
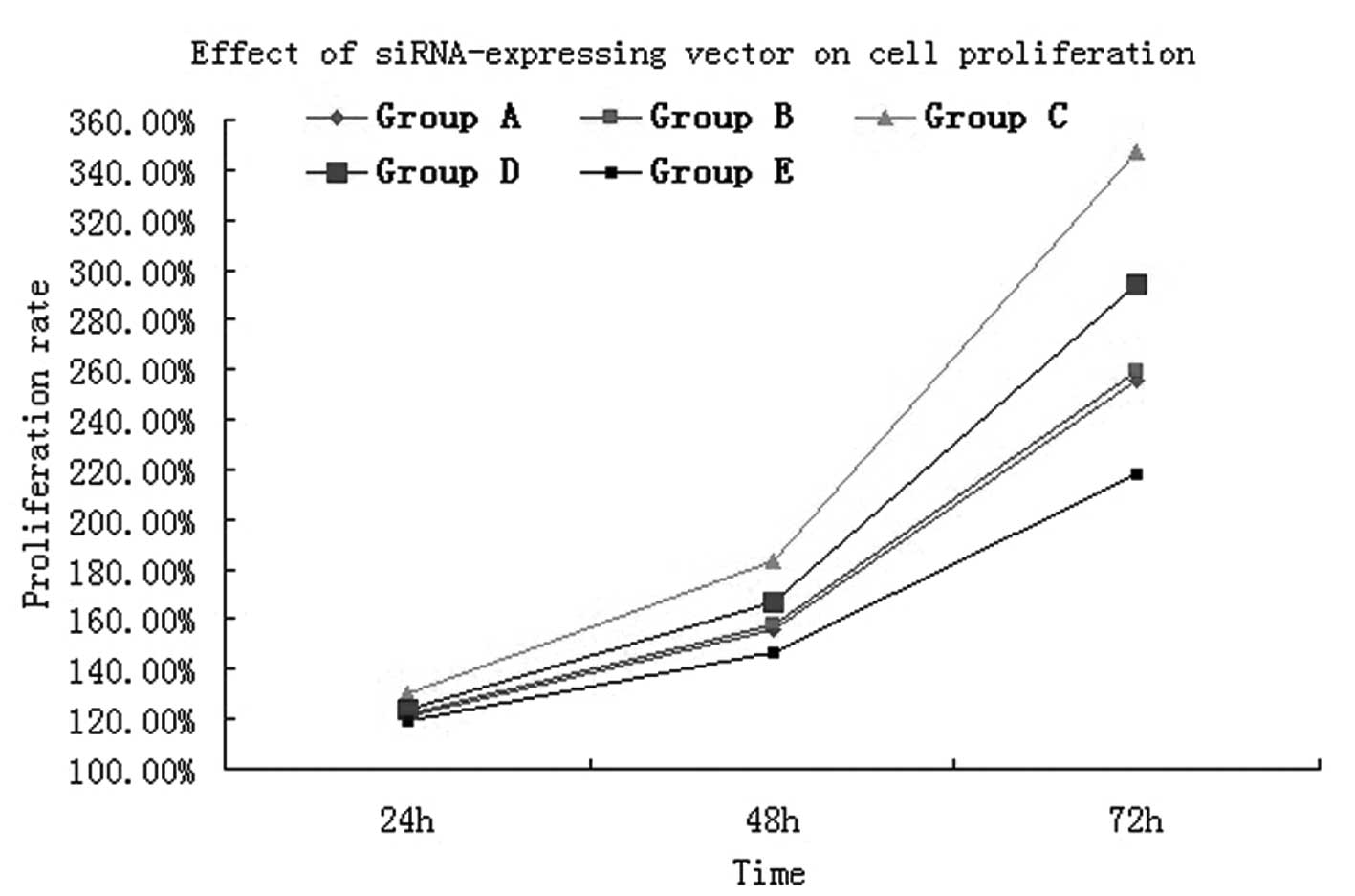
Effect of siRNA-expressing vectors on
cell proliferation
MTT assay results showed that following cultivation
for 24 h, the cell proliferative activity in groups A (normal
A549), B (empty vector), C (normal A549 and 100 ng/ml CXCL12), D
(CXCR4-siRNA A549 and 100 ng/ml CXCL12) and E (CXCR4-siRNA A549)
was 0.378±0.002, 0.380±0.002, 0.402±0.003, 0.385±0.002 and
0.373±0.002, respectively (Table
I). Following A549 cell interference with CXCR4, the
proliferative activity was significantly lower when compared with
that of the normal A549 cells (t=12.57, P<0.05). While under the
effect of chemokine CXCL12, the proliferative activity between
RNAi-treated A549 cells and normal cells was evident and, when
compared with normal A549 cells, a marked and statistically
significant difference was identified (t=5.383, P<0.05). Cell
proliferation was verified by MTT assay and for 48 and 72 h, the
results were the same (Fig. 5). The
analysis confirmed that the CXCL12/CXCR4 biological axis can induce
lung cancer cell proliferation.
 | Table IEffect of siRNA expression vector on
the cell proliferation of A549 cells following transfection. |
Table I
Effect of siRNA expression vector on
the cell proliferation of A549 cells following transfection.
| Group | OD (24 h) | OD (48 h) | OD (72 h) |
|---|
| A | 0.368±0.002 | 0.471±0.002 | 0.704±0.003 |
| B | 0.380±0.002a | 0.476±0.002a | 0.711±0.009a |
| C | 0.402±0.003b | 0.542±0.005b | 0.928±0.009b |
| D | 0.385±0.002b | 0.502±0.007b | 0.798±0.005b |
| E | 0.373±0.002a | 0.449±0.009b | 0.610±0.011b |
Effect of siRNA-expressing vectors on
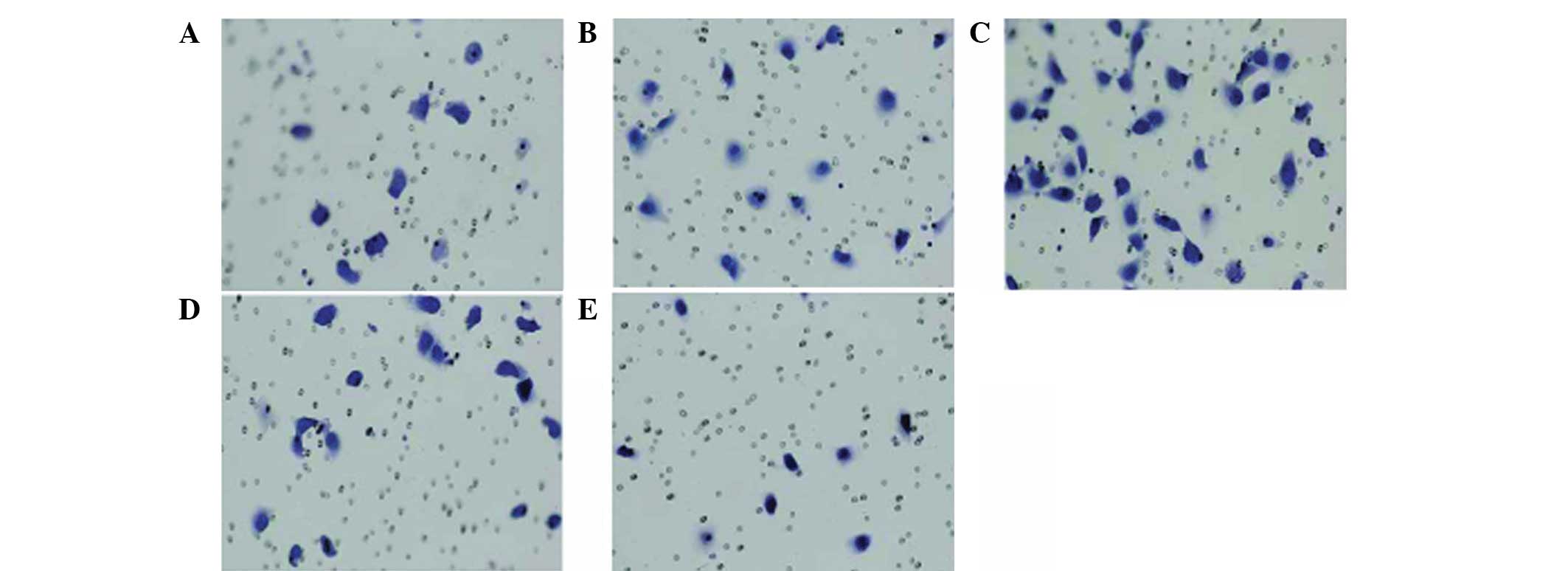
cell invasion and migration
To further clarify the impact of the CXCL12/CXCR4
interaction on the migration capacity of lung cancer cells in
vitro, chemotaxis and the chemotactic invasion response of lung
cancer cells to the CXCR4 ligand, CXCL12, were detected following
the downregulation of CXCR4 expression. The chemotactic invasion
assay was performed using a Transwell chamber with CXCL12 as a
chemoattractant, and the results showed that CXCL12 may induce
various degrees of cell chemotactic invasion. In addition, the A549
cell line was able to spontaneously pass through the microporous
membrane without CXCLl2 induction (Fig.
6) and the average transmembrane cell number was ~13.9±2.1
(Fig. 6A). Under the induction of
30 ng/ml CXCLl2, the number of A549 cells passing through the
microporous membrane was higher than that of the control group, and
the average transmembrane cell number was ~17.0±2.9 (Fig. 6B; t=2.988, P<0.05). When the
concentration of CXCLl2 was increased to 100 ng/ml, the number of
A549 cells passing through the microporous membrane increased
significantly compared with that of the 30 ng/ml group and the
average transmembrane cell number was ~30.6±7.3 (Fig. 6C; t=4.538, P<0.05). As
demonstrated above, CXCLl2 may induce and enhance the chemotactic
invasion ability of the CXCR4+ A549 cells and the
chemotactic invasion ability enhanced gradually with the increase
of CXCLl2 concentration in a concentration-dependent manner.
Following RNAi treatment, the number of A549 cells
passing through the microporous membrane was 9.7±2.7 (Fig. 6E; t=3.953, P<0.05). While under
the induction of 100 ng/ml CXCLl2, the number of RNA-interfered
A549 cells passing through the microporous membrane was ~20.1±2.4
(Fig. 6D; t=5.568, P<0.05).
Under the effect of 100 ng/ml CXCLl2, the number of A549 cells
passing through the microporous membrane markedly decreased
compared with that of the normal A549 group. Invasion analysis
found that CXCL12 is mediated by CXCR4. If the expression of CXCR4
is effectively suppressed, the binding capacity between CXCL12 and
CXCR4 is likely to be reduced and, thus, prevent CXCLl2 from
exerting its biological function. If the expression of CXCR4 is
suppressed, CXCLl2 is likely to lose specific binding sites even in
the presence of CXCLl2 induction. Therefore, the chemotactic
invasion capacity is not likely to increase, indicating that
CXCLl2-induced chemotaxis or chemotactic invasion is mediated by
CXCR4 specificity.
Discussion
There is increasing evidence to suggest that the
CXCL12/CXCR4 chemokine axis is important for the cell invasion and
migration of several types of tumor, particularly lung cancer. It
has been shown that a number of NSCLC cell lines express high
levels of CXCR4, which is associated with aggressive behavior, and
that CXCL12-activated CXCR4 promotes migration and invasion of
these cell lines in vitro (11,13).
Furthermore, preferential sites of lung cancer metastases in
vivo exhibit significantly higher levels of CXCL12 protein
expression compared with that of the primary tumor or plasma
levels, indicating that a chemotactic gradient may be established
between the site of the primary tumor and metastatic sites
(14). The results of previous
studies using various cancer cell lines have shown that inhibition
of CXCR4 reduces the frequency of metastasis, indicating that the
receptor is essential for tumor cell dissemination and invasion of
tissues (15,16). In the present study, siRNA-mediated
downregulation of CXCR4 expression in human lung cancer cells led
to a significant decrease in A549 cell proliferation and invasion.
This result is consistent with previous studies showing that CXCR4
mediates the invasive and metastatic potential of lung cancer cells
(17). The direct effect of
CXCL12/CXCR4 in tumor metastasis is that CXCL12 increases
CXCR4-mediated motility, and the cell surface expression of
integrins is mediated by the phosphorylation of extracellular
signal regulated kinase (ERK) and downstream activation of the
IKKαβ/NFκβ/RELA signaling (18). In
addition, it has previously been reported that following binding to
CXCR4, CXCL12 induces the mobilization of calcium, decreases the
levels of cyclic AMP within cells and activates multiple signal
transduction pathways, including PI3K/Akt/eNOS, which may enhance
cell proliferation, migration, survival and angiogenesis signals by
inducing eNOS activity (19).
RNAi is characterized by high efficiency, high
specificity and low toxicity of post-transcriptional gene
silencing, mediated by ds siRNAs. siRNA has become a powerful tool
for studying gene function in carcinoma and viral disease therapy
(15). Silencing is carried out by
an RNA-induced silencing complex-associated RNase III-like
endonuclease that cleaves the target homologous mRNA. The
technology of RNA silencing is likely to have a major impact on the
treatment of human diseases, particularly cancer (20,21).
In the present study, three pairs of ds siRNA oligonucleotides were
designed and constructed against CXCR4. These transcripts form a
shRNA with an inverted repeat sequence separated by a short loop
sequence. Three siRNAs targeting various sequences of human CXCR4
were cloned into a pBSilence1.1 vector for siRNA expression. The
shRNA was processed into functional siRNA to degrade target mRNA
and silence the expression. The cationic liposomal method has been
widely used due to its ease, high transfection efficacy, widespread
application and non-immunogenicity. Certain studies have achieved
particularly high transfection efficiencies by using adenoviral
vector-mediated siRNA delivery (22). Fluorescence microscopy was used to
monitor the cell plating and transfection efficacy, which for A549
lung cancer cells was >85%. In addition, the suppressed
expression of CXCR4 was confirmed by western blotting and RT-PCR at
protein and mRNA levels, respectively. The results revealed that
the RNAi constructs induced the selective degradation of CXCR4 mRNA
and thereby decreased CXCR4 protein expression levels in lung
cancer cells.
The proliferation of the A549 lung cancer cell line
in response to CXCL12 was found to be reduced by the downregulation
of CXCR4 expression by the pBSilence1.1-siRNA-CXCR4 vector, as
determined by MTT assay. This led to the examination of the effects
of CXCL12 stimulation on the A549 cell line. The results of the
in vitro proliferation assay revealed that the reduction in
cell absorbance in the CXCR4-siRNA group was greater compared with
that of the untransfected and empty vector groups at 24, 48 and 72
h following transfection, respectively. This indicated that CXCR4
functions as a positive regulator in the growth of A549 cells and,
thus, supports A549 cell proliferation. CXCL12 promoted the
colony-forming capacity of A549 cells and CXCR4-positive cells were
highly viable in response to CXCL12. By contrast, the proliferation
of A549 cells was significantly reduced by CXCR4-siRNA, indicating
that the downregulation of CXCR4 impaired the ability of the lung
cancer cells to grow. The inhibitory effect was not time dependent,
as no differences in CXCR4 inhibition were identified at 24, 48 and
72 h, respectively. This result indicated that the CXCL12/CXCR4
signaling pathway promotes tumor cell proliferation and is
consistent with previous studies showing that the CXCL12-CXCR4 axis
supports cancer cell growth (22).
The mechanisms and signaling pathways involved in CXCL12/CXCR4
activation in NSCLC reported by Lee et al indicated that ERK
activation is a key pathway in NSCLC development (23). Distant sites where CXCL12 is highly
expressed may serve as favorable niches for metastasis to occur.
The CXCL12/CXCR4 loop may stimulate tumor cell proliferation and
induce extracellular matrix rearrangement, necessary for metastasis
formation (24).
The current study also investigated the metastatic
potential of lung cancer cell line A549 in response to CXCL12,
which may be reduced by the downregulation of CXCR4 expression by
the pBSilence1.1 vector, as determined by the Transwell assay.
siRNA-CXCR4 mediated the downregulation of CXCR4 expression in
human lung cancer cells and led to a significant decrease in the
invasion and migration of A549 cells. By contrast, CXCR4-positive
cells were highly invasive in response to CXCL12 and the CXCL12
mediated chemotaxis was dose dependent, indicating that the
downregulation of CXCR4 had impaired the ability of the lung cancer
cells to migrate. This result shows that CXCR4 mediates the
invasive and metastatic potential of lung cancer cells, indicating
that CXCR4 is important for the invasion and migration of lung
cancer A549 cells toward CXCL12. Similarly, Sun et al
demonstrated that the CXCR4-CXCL12 interaction and downstream
signaling promoted the growth/survival of tumor cells, allowing
them to grow in distant and less favorable sites (25).
In conclusion, the results of the current study
indicate that CXCR4 siRNA treatment may significantly inhibit the
growth, invasion and metastasis of lung cancer cells. Thus, we
propose that CXCR4 may represent a therapeutic target for lung
cancer patients, and that RNAi with siRNA targeting CXCR4 may
establish an effective strategy for the treatment of lung
cancer.
References
|
1
|
Arriagada R, Bergman B, Dunant A, Le
Chevalier T, Pignon JP and Vansteenkiste J; International Adjuvant
Lung Cancer Trial Collaborative Group. Cisplatin-based adjuvant
chemotherapy in patients with completely resected non-small-cell
lung cancer. N Engl J Med. 350:351–360. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Murray T, Ward E, et al: Cancer
statistics. CA Cancer J Clin. 55:10–30. 2005.
|
|
3
|
Mehlen P and Puisieux A: Metastasis: a
question of life or death. Nat Rev Cancer. 6:449–458. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gangadhar T, Nandi S and Salgia R: The
role of chemokine receptor CXCR4 in lung cancer. Cancer Biol Ther.
9:409–416. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vindrieux D, Escobar P and Lazennec G:
Emerging roles of chemokines in prostate cancer. Endocr Relat
Cancer. 16:663–673. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Müller A, Homey B, Soto H, et al:
Involvement of chemokine receptors in breast cancer metastasis.
Nature. 410:50–56. 2001.PubMed/NCBI
|
|
7
|
Clapham PR and Weiss RA: Immunodeficiency
viruses. Spoilt for choice of co-receptors. Nature. 388:230–231.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liang JJ, Zhu S, Bruggeman R, et al: High
levels of expression of human stromal cell-derived factor-1 are
associated with worse prognosis in patients with stage II
pancreatic ductal adenocacinoma. Cancer Epidemiol Biomarkers Prev.
19:2598–2604. 2010. View Article : Google Scholar
|
|
9
|
Castellone MD, Guarino V, De Falco V, et
al: Functional expression of the CXCR4 chemokine receptor is
induced by RET/PTC oncogenes and is a common event in human
papillary thyroid carcinomas. Oncogene. 23:5958–5967. 2004.
View Article : Google Scholar
|
|
10
|
Torregrossa L, Giannini R, Borrelli N, et
al: CXCR4 expression correlates with the degree of tumor
infiltration and BRAF status in papillary thyroid carcinomas. Mod
Pathol. 25:46–55. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Su L, Zhang J, Xu H, et al: Differential
expression of CXCR4 is associated with the metastatic potential of
human non-small cell lung cancer cells. Clin Cancer Res.
11:8273–8280. 2005. View Article : Google Scholar
|
|
12
|
Spano JP, Andre F, Morat L, et al:
Chemokine receptor CXCR4 and early-stage non-small cell lung
cancer: pattern of expression and correlation with outcome. Ann
Oncol. 15:613–617. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Phillips RJ, Burdick MD, Lutz M, et al:
The stromal derived factor-1/CXCL12-CXC chemokine receptor 4
biological axis in non-small cell lung cancer metastases. Am J
Respir Crit Care Med. 167:1676–1686. 2003. View Article : Google Scholar
|
|
14
|
Phillips RJ, Mestas J, Gharaee-Kermani M,
et al: Epidermal growth factor and hypoxia-induced expression of
CXC chemokine receptor 4 on non-small cell lung cancer cells is
regulated by the phosphatidylinositol 3-kinase/PTEN/AKT/mammalian
target of rapamycin signaling pathway and activation of hypoxia
inducible factor-1alpha. J Biol Chem. 280:22473–22481. 2005.
|
|
15
|
Broxmeyer HE, Orschell CM, Clapp DW, et
al: Rapid mobilization of murine and human hematopoietic stem and
progenitor cells with AMD3100, a CXCR4 antagonist. J Exp Med.
201:1307–1318. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ok S, Kim SM, Kim C, et al: Emodin
inhibits invasion and migration of prostate and lung cancer cells
by downregulating the expression of chemokine receptor CXCR4.
Immunopharmacol Immunotoxicol. 34:768–778. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xie S, Zeng W, Zuo T, et al: Expression
and significance of CXCR4 chemokine receptor in non small cell lung
cancer. Chin J Gen Pract. 10:1335–1336. 2012.
|
|
18
|
Huang YC, Hsiao YC, Chen YJ, et al:
Stromal cell-derived factor-1 enhances motility and integrin
up-regulation through CXCR4, ERK and NF-kappaB-dependent pathway in
human lung cancer cells. Biochem Pharmacol. 74:1702–1712. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng H, Dai T, Zhou B, et al:
SDF-1alpha/CXCR4 decreases endothelial progenitor cells apoptosis
under serum deprivation by PI3K/Akt/eNOS pathway. Atherosclerosis.
201:36–42. 2008. View Article : Google Scholar
|
|
20
|
Abedini F, Ismail M, Hosseinkhani H, et
al: Effects of CXCR4 siRNA/dextran-spermine nanoparticles on CXCR4
expression and serum LDH levels in a mouse model of colorectal
cancer metastasis to the liver. Cancer Manag Res. 3:301–309.
2011.PubMed/NCBI
|
|
21
|
Orimo A, Gupta PB, Sgroi DC, et al:
Stromal fibroblasts present in invasive human breast carcinomas
promote tumor growth and angiogenesis through elevated SDF-1/CXCL12
secretion. Cell. 121:335–348. 2005. View Article : Google Scholar
|
|
22
|
Rubie C, Frick VO, Ghadjar P, et al: CXC
receptor-4 mRNA silencing abrogates CXCL12-induced migration of
colorectal cancer cells. J Transl Med. 9:222011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee W, Jiang Z, Liu J, Haverty PM, Guan Y,
Stinson J, et al: The mutation spectrum revealed by paired genome
sequences from a lung cancer patient. Nature. 465:473–477. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wald O, Izhar U, Amir G, et al:
Interaction between neoplastic cells and cancer-associated
fibroblasts through the CXCL12/CXCR4 axis: role in non-small cell
lung cancer tumor proliferation. J Thorac Cardiovasc Surg.
141:1503–1512. 2011. View Article : Google Scholar
|
|
25
|
Sun YX, Wang J, Shelburne CE, et al:
Expression of CXCR4 and CXCL12 (SDF-1) in human prostate cancers
(PCa) in vivo. J Cell Biochem. 89:462–473. 2003. View Article : Google Scholar : PubMed/NCBI
|