Introduction
Ubiquitination and neddylation are necessary for a
number of biological processes and have been implicated in numerous
diseases, particularly in cancer (1,2). The
process of ubiquitination has been identified as the mechanism that
labels proteins for degradation by the 26s proteasome (3). Proteins degraded by the
ubiquitin-proteasome pathway (UPS) are known to be involved in
numerous biological processes, including apoptosis (4,5), cell
cycle regulation (6) and receptor
regulation (7). Conjugation of
ubiquitin to its substrate requires three enzymes,
ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2 and
ubiquitin ligase enzyme E3.
The neural precursor cell-expressed developmentally
downregulated 8 (NEDD8) protein is 60% identical to ubiquitin and
also conjugates to target proteins (8). This process is termed neddylation and
is similar to ubiquitination. NEDD8 genes are translated into
non-conjugatable precursors, which contain additional C-terminal
residues. Ubiquitin carboxyl-terminal hydrolase isozyme L3 (UCHL3)
then cleaves the C-terminal of NEDD8, giving rise to mature NEDD8.
Subsequently, NEDD8 is bound to target proteins by a series of
enzymes. NEDD8 is activated by the E1 enzyme (APPBP1/UBA3
heterodimer), whereby a thioester bond is formed with the cysteine
residue of the UBA3 subunit. Following this, NEDD8 is transferred
to the E2 enzyme (UBC12) and, with the involvement of the E3
enzyme, NEDD8 remains bound to E2 and binds to the substrates via
its carboxy-terminal glycine (9).
Using UCHL3, NEDD8 may be hydrolyzed from the substrates.
Members of the cullin family, which serve as the
scaffold proteins of the Skp/Cul/F-box (SCF) complex, are the main
substrates of NEDD8 (10). The SCF
complex is the core subunit of E3 for ubiquitination; therefore,
neddylation may regulate the degradation of proteins by modulating
the process of ubiquitination. Ubiquitin, which labels proteins
involved in apoptosis and cell cycle regulation, has been
implicated in many cancer types (11,12).
The anticancer activity of bortezomib, which targets the UPS, has
been approved by the FDA (13,14).
However, proteasome inhibition, which irreversibly arrests the UPS
pathway, causes protein turnover to become disordered (15,16).
Study is now beginning to focus on alternative targets that
specifically inhibit the growth of tumors through UPS modulation
(16). Therefore the neddylation
pathway has attracted attention as modulation of such has a higher
safety profile which avoids removing the UPS entirely and prevents
the conversion of protein from being entirely suppressed.
Neddylation has been studied in certain cancer
cells; results have revealed that in highly proliferative cell
lines the levels of conjugated NEDD8 expression were high (17). In our previous study, it was
observed that the upregulation of neddylation was closely
associated with the proliferation capacity of melanoma cell lines
(18). In the present study,
expression levels of conjugated NEDD8 were examined in melanoma
tissues to confirm the results of our previous research. To
investigate the mechanisms by which upregulation of neddylation
affects proliferation of melanoma cell lines, the expression levels
of enzymes that regulate the process were identified and the
influence of neddylation on the cell cycle of M14 cells was
investigated. The levels of cell cycle regulators and
apoptosis-related proteins were also analyzed. To inhibit the
neddylation pathway, without altering the function of free NEDD8,
UBA3 was chosen as the target for interference (17,18).
Materials and methods
Clinical samples
Seven patient samples of melanoma tissue were
obtained from the Chinese Academy of Medical Sciences and Peking
Union Medical College (Nanjing, China). All cases of melanoma were
diagnosed pathologically, and patients had not been treated with
chemotherapy or radiation. In the seven samples of melanoma, the
majority were of acral location; however, one sample originated
from the thigh. Clark classification levels are shown in Table I. All melanoma tissues were stored
at −70°C. Patients provided informed consent and this study was
approved by the Medical Ethics Committee of the Institute of
Dermatology, Chinese Academy of Medical Sciences and Peking Union
Medical College.
 | Table IClark levels of seven cases of
melanoma. |
Table I
Clark levels of seven cases of
melanoma.
| Group | Gender | Age, years | Region | Clark level |
|---|
| 1 | Female | 50 | Big toe | 3 |
| 2 | Female | 62 | Pelma | 4 |
| 3 | Female | 40 | Pelma | 2 |
| 4 | Male | 71 | Pelma | 3 |
| 5 | Male | 85 | Thigh | 4 |
| 6 | Male | 76 | Pelma | 3 |
| 7 | Female | 66 | Heel | 4 |
Cell lines and culture conditions
The human melanoma cell line, A375, was obtained
from Xijing Hospital (Shanxi, China). M14 and MV3 human melanoma
cell lines were purchased from KeyGen Biotech Co. Ltd. (Nanjing,
China). The three cell lines, A375, M14 and MV3, were cultured in
Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine
serum (both purchased from Gibco-BRL, Eggenstein, Germany) at 37°C
with 5% CO2. The normal melanocytes were separated from
the prepuce, obtained from healthy adult males who had undergone
circumcision and had been admitted to the Institute of Dermatology,
Chinese Academy of Medical Sciences & Peking Union Medical
College, and cultured with M254 medium (Gibco-BRL) containing human
melanocyte growth supplement (Gibco-BRL) at 37°C with 5%
CO2.
Western blot assay
Following two washes with phosphate buffered-saline
(PBS), total proteins were extracted from cells and tissue masses
using lysis buffer (50 mM Tris-HCl, pH 8.0; 150 mM NaCl; 0.02%
NaN3; 0.1% SDS; 1% Nonidet P-40; 0.5% sodium
deoxycholate; 100 mg/ml phenylmethylsulfonyl fluoride; 1 mg/ml
aprotinin; 1 mg/ml leupeptin and 1 mg/ml pepstatin A; KeyGen
Biotech Co. Ltd.). Protein concentration was determined by a
spectrophotometer (UV-3540;. Seventy micrograms of protein,
separated by 10% SDS-polyacrylamide gel electrophoresis (Bio-Rad,
Hercules, CA, USA), was transferred onto a polyvinylidene flouride
(PVDF) membrane (Pall Corp., New York, NY, USA) by electroblotting.
Subsequently, the PVDF membrane was stained with polyclonal
anti-goat NEDD8 antibody and polyclonal anti-rabbit bax, p21, p27
and cyclin D antibodies (Santa Cruz Biotechnology, Inc., Santa
Cruz, CA, USA), and β-actin antibody (Santa Cruz Biotechnology,
Inc.) was used as the loading control. Peroxidase-conjugated
anti-rabbit IgG (Santa Cruz Biotechnology, Inc.) was used as a
secondary antibody and visualized using a enhanced
chemiluminescence kit (Santa Cruz Biotechnology, Inc.). Glyco
Band-Scan software 4.5 (Prozyme, San Leandro, CA, USA) was used to
quantify the relative quantity of protein.
Semi-quantitative real-time polymerase
chain reaction (PCR)
Total RNA was extracted from the A375, M14 and MV3
cell lines and the normal melanocyte cell lines, according to the
manufacturer’s protocol. Total RNA was reverse transcribed into
cDNA using the Reverse Transcription system (Promega, Madison, WI,
USA). The purity of RNA was determined by measuring the ratio of
absorbance at 260 and 280 nm, regulating the optical density
between 1.8 and 2.1 ensured a high purity. The sequences of
specific primers for each gene, synthesized by GenScript (GenScript
USA Inc., Piscataway, NJ, USA), were as follows: Forward,
5′-ACAGTGGCAAGCAGATGAATGA-3′ and reverse,
5′-ATGAGCGACAGGGTAAAGAGGT-3′ for NEDD8; forward,
5′-GCGAGGAGCCGGAGAAGAAAAG-3′ and reverse,
5′-TCGAAATCAGGGTGTGTGAAGG-3′ for UBA3; forward,
5′-GTTTTAAGGTGGGCCAGGGTTA-3′ and reverse,
5′-GGTTGGGCTCCAAGAAGAGATA-3′ for UBC12; forward,
5′-CTACATCCTAACTGGCAATTCGTT-3′ and reverse,
5′-GCGATTTTATTTTTTCTTCCTCT TCT-3′ for UCHL3; forward,
5′-CATTTTGGATTTTAGCTCG TGC-3′ and reverse,
5′-ATCTTTCTTTGCTTTTTCACGG-3′ for APPBP1; forward,
5′-GCAGAAGGAGATCACAGCCCT-3′ and reverse,
5′-GCTGATCCACATCTGCTGGAA-3′ for β-actin. The reaction system
contained: 10 μl 2X RT-PCR quick master mix (Toyobo Co. Ltd.,
Osaka, Japan), 10 pmol upstream and downstream primer and 1 μl DNA
template, made up to 20 μl with purified water. The reaction
conditions were as follows: One cycle for 5 min at 95°C; and 40
cycles of 95°C for 15 sec, 60°C for 30 sec and 72°C for 30 sec.
Size and quantity of amplified products were determined by 2%
agarose gel electrophoresis (BioSun Sci & Tech Co., Ltd.,
Shanghai, China). The ΔCt value was calculated and fluorescence was
analyzed using the thermal cycler’s software package (DA 7600, Da
An Gene Co. Ltd., Guangzhou, China). The 2-ΔΔCt value was presented
as the relative expression of each gene.
siRNA transfection
In total, three shRNA fragments against UBA3 were
designed and synthesised by GeneScript Corp. (Piscataway, NJ, USA):
shRNA-UBA3-1, 5′-GGATCCCGTTCCTCGAGCGATCTGGATTCAAGAGA
TCCAGATCGCTAGGAACTTTTTTCCAACTCGAG-3′; shRNA-UBA-3-2,
5′-GGATCCCGAACGAACAAGGCCCA AATCTTCAAGAGAGATTTGGCCTTGTTCGTTCTTTTT
TCCAACTCGAG-3′; and shRNA-UBA3-3, 5′-GGATCCCGT
GCACGCTGGAACTTTATCTTCAAGAGAGATAAAGTT CCAGCGTGCATTTTTTCCAACTCGAG-3′.
Fragments were subcloned into the pRNAT-U6.2/Lenti siRNA expression
vector (Invitrogen Life Technologies, Carlsbad, CA, USA).
Transfection was performed using FuGene® HD (Roche,
Basel, Switzerland) and Opti-DMEM (Gibco-BRL) without antibiotics.
To investigate the transfection conditions, cells were seeded in
96-well plates. The final reaction conditions were as follows: 2 μg
DNA plus 30 μl FuGene HD in a six-well plate with a volume of 2 ml
culture medium and 80–90% cell density. Following two weeks of
selection using 1000 μg/ml G418 (Invitrogen Life Technologies),
G418-resistant clones were selected and maintained with 500 μg/ml
G418 for further analysis. The optimum shRNA sequence for UBA3,
analyzed by western blotting, was determined as follows:
GGATCCCGTGCACGCTGGAACTTTATCTT CAAGAGAGATAAAGTTCCAGCGTGCATTTTT
TCCAACTCGAG (71 bp).
Cell cycle analysis
The Cell Cycle Detection kit (KeyGen Biotech Co.
Ltd.) was used to evaluate the cell cycle of non-transfected M14,
M14/shRNAT-U6.2 and M14/shRNA-UBA3 cell lines. Cells
(1.0×106) were collected, washed twice with PBS and
fixed with 70% ethanol at 4°C for 24 h. After discarding the
fixation fluid, cells were washed again and incubated with 100 μl
RnaseA for 30 min at 37°C followed by staining with 400 μl PI at
4°C for 30 min in the absence of light. The DNA content was
determined by flow cytometry (BD FACSCanto II, BD Biosciences,
Franklin Lakes, NJ, USA). Cell cycle distribution was evaluated
using ModFit software (Verity Software House, Inc., Topsham, USA).
Each experiment was repeated four times independently.
Statistical analysis
Data are expressed as the mean ± SD. SPSS 13.0
statistical software package (SPSS Inc., Chicago, IL, USA) was used
to perform the independent samples t-test on the semi-quantitative
real-time PCR analysis and cell cycle analysis data, while the
paired samples t-test was performed on the western blot analysis
data regarding melanoma tissues and surrounding normal tissues.
P<0.05 was considered to indicate a statistically significant
difference.
Results
NEDD8 conjugation in tissues
In our previous study, it was identified that NEDD8
conjugation was upregulated in melanoma cell lines (18). In the current study, conjugated
NEDD8 expression was also demonstrated to be upregulated in
melanoma tissue. In a previous study using electrophoresis, it was
observed that NEDD8 conjugation may be identified by a series of
bands including a dominant band of 90 kDa and a minor band of 66
kDa; such findings are consistent with the molecular mass of
NEDD8-cullins conjugates (17). As
a result of this, in the current study, 90- and 66-kDa bands were
observed.
In this study, NEDD8 conjugation was detected in all
seven cases of melanoma and in the normal tissues around them. As
shown in Fig. 1, the densities of
90- and 66-kDa bands in melanoma tissues were higher than these in
the corresponding normal tissues surrounding them. In Fig. 1, distinct bands of 66-kDa were
detected; however in the cell lines analyzed in our previous study
(A375, M14 and MV3), this band was absent (18).
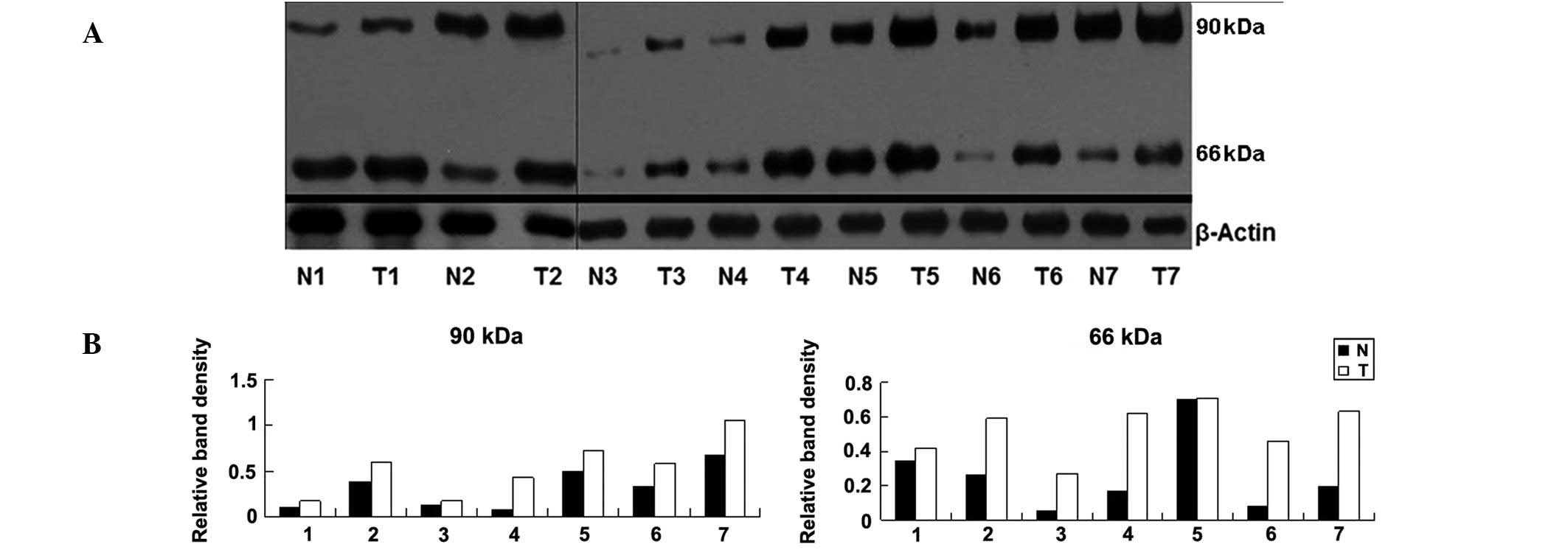 | Figure 1(A) Western blot assay of the NEDD8
conjugation levels in seven paired samples of melanoma (T) and
surrounding normal (N) tissues. (B) Column diagram of relative
density of NEDD8 conjugation in these tissues. Paired sample
t-tests were performed and statistically significant differences
were identified between the two groups (t=5.732, P=0.001 at 90 kDa;
t=4.132, P=0.006 at 66 kDa). The relative density of NEDD8 to
β-actin was as follows: (0.10, 0.17), (0.38, 0.59), (0.03, 0.18),
(0.08, 0.43), (0.5, 0.72), (0.32, 0.58), (0.68, 1.06) at 90 kDa;
(0.34, 0.42), (0.26, 0.59), (0.05, 0.27), (0.17, 0.62), (0.70,
0.71), (0.08, 0.46), (0.19, 0.63) at 66 kDa. |
Expression of NEDD8-related proteins in
melanoma cell lines and melanocytes
Gene expression of UBC12, APPBP1 and UCHL3 in three
melanoma cell lines and in melanocytes were analyzed by real-time
PCR (Fig. 2A). To observe the
associations between the NEDD8-related proteins, gene expression
data for NEDD8-related proteins, NEDD8 and UBA3, as reported in our
previous paper (Fig. 2B) (18), was combined for analysis. It was
found that the expression levels of UBC12 and APPBP1 in melanoma
cells were upregulated when compared with those in melanocytes. In
M14 cells, NEDD8 and UCHL3 expression were both downregulated, but
the UBA3 and UBC12 expression was highest compared with those in
the A375 and MV3 cell lines and in normal menalocytes.
Cell cycle arrest of M14 cells following
neddylation inhibition in vitro
It has been hypothesized that interfering with the
expression of UBA3 may inhibit NEDD8 conjugation and, subsequently,
prevent the proliferation of melanoma. However, the mechanisms of
this process are unknown. It has been reported that the degradation
of cell cycle regulators, such as cyclin D and p21, could be
regulated by the UPS (6). In the
present study, the effect of NEDD8 conjugation on the cell cycle of
M14 cells was investigated.
Percentage distribution of the cell cycle phases in
non-transfected M14, M14/shRNAT-U6.2 and M14/shRNA-UBA3 cell lines
are shown in Table II. After
transfection with shRNA-UBA3, the G0/G1 and apoptotic phases
accumulated, whereas the G2/M+S phase declined, when compared with
non-transfected M14 and M14/shRNAT-U6.2 cells. The statistical
differences between groups were identified to be significant
(P<0.05). The increase in G0/G1 phase percentage distribution
indicates the inhibition of cell division. These results suggest
that inhibition of neddylation in M14 cells may cause cell cycle
arrest at G0/G1 phase, promoting apoptosis.
 | Table IIPercentage of cells in GI, G2+S and
apoptosis phases in transfected and non-transfected M14 cells. |
Table II
Percentage of cells in GI, G2+S and
apoptosis phases in transfected and non-transfected M14 cells.
| Group | G0/G1 (%) | G2+S (%) | Apoptosis (%) |
|---|
| Non-transfected
M14 | 57.4900±4.527a | 42.4800±4.547a | 0.3050±0.187a |
| M14/shRNAT-U6.2 | 59.1325±1.019a | 40.8725±1.013a |
0.4375±0.103a |
| M14/shRNA-UBA3 | 68.3275±1.263 | 31.6675±1.265 | 1.2400±0.404 |
Effects of neddylation on proteins
involved in cell cycle or apoptosis
It was observed that the G0/G1 phase and the
apoptotic phase of the cell cycle increased in M14/shRNA-UBA3 cells
compared with M14/shRNA-U6.2 and non-transfected M14 cells. To
explore the possible molecular mechanisms by which neddylation
affects melanoma proliferation, the levels of proteins involved in
the cell cycle and apoptosis, including bax, p21, p27 and cyclin D
were analyzed.
Western blot analysis demonstrated that the levels
of bax, p21 and p27 were all upregulated, whereas those of cyclin D
were downregulated in M14/shRNA-UBA3 cells, when compared with such
levels in non-transfected M14 and M14/shRNAT-U6.2 cells (Fig. 3 and Table III).
 | Table IIIRelative density of Bax, P21, P27 and
Cyclin D in transfected and non-transfected M14 cells. |
Table III
Relative density of Bax, P21, P27 and
Cyclin D in transfected and non-transfected M14 cells.
| Group | bax | p21 | p27 | cyclin D |
|---|
| Non-transfected
M14 | 0.482 | 0.572 | 0.463 | 0.904 |
|
M14/shRNAT-U6.2 | 0.428 | 0.490 | 0.470 | 0.971 |
| M14/shRNA-UBA3 | 0.744 | 0.763 | 0.653 | 0.230 |
Discussion
Ubiquitination, a post-translational protein
modification process, mediates proteasome-dependent degradation of
numerous intracellular proteins and has been implicated in a number
of cancer types (1). However,
complete inhibition of the UPS pathway may cause protein turnover
to become disordered (15,19). Recently, there has been increased
focus on the neddylation pathway, which is more specialized and may
avoid the need to remove the UPS entirely and prevents protein
conversion from being suppressed entirely.
Neddylation plays an essential role in cellular
survival and is considered to be involved in cancer progression
(2,17). In our previous study, the
association between neddylation and the proliferation of melanoma
was investigated. The results indicated that NEDD8 conjugation was
upregulated in melanoma cell lines, and was closely associated with
the proliferation of the melanoma cell line, M14 (18).
In the current study, it was identified that there
was increased expression of NEDD8 conjugates in melanoma tissues,
which indicates the close association between the neddylation
pathway and melanoma. The results from seven samples of melanoma
showed that NEDD8 conjugation in melanoma tissues was higher than
that in the normal tissues surrounding the melanoma. However,
individual variations were evident. Certain band densities in
normal tissues were higher than the band densities exhibited in the
tumor tissues of other groups. For example, in Fig. 1, the band density at 90 kDa
exhibited by N7 is higher than that of T3. Biochemical changes may
have occurred in the tissues, which appeared normal when observed
microscopically; however, they may have contributed to the
individual variations observed.
In tissues, not only bands of 90 kDa but also 66 kDa
were detected, which is consistent with the study of Chairatvit and
Ngamkitidechakul (17), but differs
from the results of melanoma cell lines (18). In the current study, the band of 9
kDa, which represents free NEDD8, was negative in tissues, A375,
M14 and MV3 cell lines and in normal menalocytes, which was similar
to the results of a study by Chairatvit and Ngamkitidechakul
(17). These results revealed that
the upregulation of NEDD8 conjugation was not accompanied by an
increase in free NEDD8 concentration.
The neddylation process may be regulated by a number
of enzymes. Expression levels of NEDD8, UBA3 (18), APPBP1, UBC12 and UCHL3 proteins were
studied in three melanoma cell lines to determine their association
with the upregulation of NEDD8 conjugation. With the exception of
NEDD8 and UCHL3 in the M14 cell line, all the other enzymes, which
correlated closely with neddylation, were upregulated in the three
melanoma cell lines. These upregulated enzymes may be implicated in
the increased levels of NEDD8 conjugation observed. As mentioned
above, in M14 cells which exhibited the highest levels of NEDD8
conjugation, the expression levels of NEDD8 and UCHL3 were
decreased (18). The expression of
UCHL3, essential for the NEDD8 precursor to mature, was the lowest
in M14 cells, which may be one reason for the lowest levels of
NEDD8 expression in M14 cells compared with those the A375 and MV3
cells (21,22). Otherwise, UCHL3 not only makes NEDD8
mature, but also hydrolyzes the NEDD8 conjugation in order to gree
NEDD8 (20,21). The UCHL3 decline in M14 may reduce
the hydrolysis of NEDD8 conjugation. The relationships among these
enzymes are complicated and further studies are required.
In our previous study, proliferation of M14 cells
declined following neddylation pathway interference (18). In this study, to investigate the
mechanism by which neddylation may affect melanoma growth, cell
cycle progression was identified using flow cytometry. The number
of the cells in G0/G1 phase and in the apoptotic phase were
observed to increase following neddylation interference. Cells
never get into the S phase, which is safer than out of control
cells coming into the S phase which may lead to the development of
cancer. Disturbance the neddylation pathway could arrest cells at
the G0/G1 phase but they would be unable to get into the S phase,
so that inhibition of mitosis and induction of apoptosis of M14
were onset.
Inhibition of neddylation, which is coupled to the
modulation of proteins involved in cell cycle and apoptosis
regulation, is thought to induce cell cycle arrest at the G0/G1
phase, consequently promoting apoptosis. Cyclin D, an important
growth factor, is essential for cellular fission and cell
progression into S phase. Cyclin D upregulation in melanoma is
closely associated with melanoma proliferation (22). Inhibition of neddylation, coupled
with the decline in cyclin D levels causes cell cycle arrest at G1
phase, leading to apoptosis. Cell cycle progression is also
regulated by other CDK inhibitors, including p27 and p21. In the
present study, p27 and p21 were identified to be upregulated
following inhibition of the neddylation pathway. Overexpression of
p27 has been observed to inhibit the formation of the CDK
activation complex, preventing cells from entering S phase
(23). p21 may promote apoptosis
through interaction with DNA repair machinery (24). In this study, the expression of the
apoptosis promoter, bax, was also upregulated. Bax is released from
mitochondria and cytochrome c is released into the cytosol
for intrinsic cellular apoptosis (25). In our previous study, it was
observed that p53 expression increased following inhibition of
neddylation (18). Therefore, G0/G1
cell cycle arrest and apoptosis induction in M14 cells transfected
with shRNA-UBA3 may be mediated by the activation of these cell
cycle regulators and apoptosis-related proteins.
It has been demonstrated that p53 (26), p21, cyclin D (6) and bax (27) are degraded by the UPS. The
upregulation of p53, p21 and bax verified transfection in the
current study. However, cyclin D levels decreased following
transfection, and the reason for this remains unknown.
Based on the results above, it was concluded that
the neddylation pathway may be involved in the development of
melanoma. Inhibition of the neddylation pathway affects cell cycle
regulators and apoptosis promoters, leading to the depression of
melanoma growth. In this study, UBA3 was chosen for neddylation
interference. However, UBC12 and APPBP1 were also upregulated in
the three melanoma cell lines. In conclusion, proteins which
regulate the neddylation pathway may represent potential
therapeutic targets in the treatment of melanoma and other tumors
associated with neddylation promotion.
Acknowledgements
The authors thank Ming-Jun Jiang and Wu-Qing Zhou
(Central Laboratory of the Institute of Dermatology, Chinese
Academy of Medical Sciences and Peking Union Medical College) for
their technical assistance.
Abbreviations:
|
NEDD8
|
neural precursor cell-expressed
developmentally downregulated 8
|
References
|
1
|
Chen RH, Lee YR and Yuan WC: The role of
PML ubiquitination in human malignancies. J Biomed Sci. 19:812012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Duncan K, Schäfer G, Vava A, Parker MI and
Zerbini LF: Targeting neddylation in cancer therapy. Future Oncol.
8:1461–1470. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hjerpe R and Rodríguez MS: Alternative UPS
drug targets upstream the 26S proteasome. Int J Biochem Cell Biol.
40:1126–1140. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tsvetkov P, Reuven N and Shaul Y:
Ubiquitin-independent p53 proteasomal degradation. Cell Death
Differ. 17:103–108. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Allende-Vega N and Saville MK: Targeting
the ubiquitin-proteasome system to activate wild-type p53 for
cancer therapy. Semin Cancer Biol. 20:29–39. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yu ZK, Gervais JL and Zhang H: Human CUL-1
associates with the SKP1/SKP2 complex and regulates p21 (CIP1/WAF1)
and cyclin D proteins. Proc Natl Acad Sci USA. 95:11324–11329.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dace A, Zhao L, Park KS, et al: Hormone
binding induces rapid proteasome-mediated degradation of thyroid
hormone receptors. Proc Natl Acad Sci USA. 97:8985–8990. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wada H, Yeh ET and Kamitani T: A
dominant-negative UBC12 mutant sequesters NEDD8 and inhibits NEDD8
conjugation in vivo. J Biol Chem. 275:17008–17015. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rabut G and Peter M: Function and
regulation of protein neddylation. ‘Protein modifications: beyond
the usual suspects’ review series. EMBO Rep. 9:969–976. 2008.
|
|
10
|
Wu K, Chen A and Pan ZQ: Conjugation of
Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to
promote ubiquitin polymerization. J Biol Chem. 275:32317–32324.
2000. View Article : Google Scholar
|
|
11
|
Hoeller D and Dikic I: Targeting the
ubiquitin system in cancer therapy. Nature. 458:438–444. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bergink S and Jentsch S: Principles of
ubiquitin and SUMO modifications in DNA repair. Nature.
458:461–467. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Paoluzzi L and O’Connor OA: Mechanistic
rationale and clinical evidence for the efficacy of proteasome
inhibitors against indolent and mantle cell lymphomas. BioDrugs.
20:13–23. 2006. View Article : Google Scholar
|
|
14
|
O’Connor OA: Marked clinical activity of
the proteasome inhibitor bortezomib in patients with follicular and
mantle-cell lymphoma. Clin Lymphoma Myeloma. 6:191–199. 2005.
|
|
15
|
Yang Y, Kitagaki J, Dai RM, et al:
Inhibitors of ubiquitin-activating enzyme (E1), a new class of
potential cancer therapeutics. Cancer Res. 67:9472–9481. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Soucy TA, Smith PG and Rolfe M: Targeting
NEDD8-activated cullin-RING ligases for the treatment of cancer.
Clin Cancer Res. 15:3912–3916. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chairatvit K and Ngamkitidechakul C:
Control of cell proliferation via elevated NEDD8 conjugation in
oral squamous cell carcinoma. Mol Cell Biochem. 306:163–169. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cheng F, Chen H, Zhang L, Li RH, Liu Y and
Sun JF: Inhibition of the NEDD8 conjugation pathway by shRNA to
UBA3, the subunit of the NEDD8-activating enzyme, suppresses the
growth of melanoma cells. Asian Pac J Cancer Prev. 13:57–62. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Soucy TA, Smith PG, Milhollen MA, et al:
An inhibitor of NEDD8-activating enzyme as a new approach to treat
cancer. Nature. 458:732–736. 2009. View Article : Google Scholar
|
|
20
|
Gong L, Kamitani T, Millas S and Yeh ET:
Identification of a novel isopeptidase with dual specificity for
ubiquitin- and NEDD8-conjugated proteins. J Biol Chem.
275:14212–14216. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hemelaar J, Borodovsky A, Kessler BM, et
al: Specific and covalent targeting of conjugating and
deconjugating enzymes of ubiquitin-like proteins. Mol Cell Biol.
24:84–95. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Coupland SE, Bechrakis N, Schüler A,
Anagnostopoulos I, Hummel M, Bornfeld N and Stein H: Expression
patterns of cyclin D1 and related proteins regulating G1-S phase
transition in uveal melanoma and retinoblastoma. Br J Ophthalmol.
82:961–970. 1998. View Article : Google Scholar
|
|
23
|
Abukhdeir AM and Park BH: P21 and p27:
roles in carcinogenesis and drug resistance. Expert Rev Mol Med.
10:e192008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gartel AL and Tyner AL: The role of the
cyclin-dependent kinase inhibitor p21 in apoptosis. Mol Cancer
Ther. 1:639–649. 2002.PubMed/NCBI
|
|
25
|
Finucane DM, Bossy-Wetzel E, Waterhouse
NJ, Cotter TG and Green DR: Bax-induced caspase activation and
apoptosis via cytochrome c release from mitochondria is inhibitable
by Bcl-xL. J Biol Chem. 274:2225–2233. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xirodimas DP and Scheffner M: Ubiquitin
family members in the regulation of the tumor suppressor p53.
Subcell Biochem. 54:116–135. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zuo J, Bi C, Fan Y, Buac D, Nardon C,
Daniel KG and Dou QP: Cellular and computational studies of
proteasome inhibition and apoptosis induction in human cancer cells
by amino acid Schiff base-copper complexes. J Inorg Biochem.
118:83–93. 2013. View Article : Google Scholar
|

















