Introduction
Gliomas are the most common type of tumor of the
central nervous system in adults, with the glioma subtype,
glioblastoma multiforme, the most common (1). A previous genome-wide mutational
analysis of glioblastomas identified novel mutations in the
isocitrate dehydrogenase (IDH) 1 gene (2). Further studies have demonstrated that
IDH1 mutations are present in 50–90% of cases of grade II
and III astrocytoma and oligodendroglioma, but rarely present in
primary glioblastoma or pilocytic astrocytoma (3–11). In
addition to IDH1 mutations, mutations in the homologous
gene, IDH2, have also been identified in gliomas, but are
much less common than the IDH1 mutations (12,13).
Various retrospective and prospective studies have
demonstrated that the IDH mutation is associated with longer
survival in glioma patients (12–15).
However, none of these studies have analyzed the correlation
between IDH status and tumor location/magnetic resonance
imaging (MRI) characteristics. Notably, different intracranial
locations, such as functional or non-functional lobes, as well as
the cortex or deep brain, are likely to cause variable difficulty
levels for surgery, and thus, correspondingly influence the
prognosis of patients. Furthermore, different presurgical MRI
features, including well-defined or blurred interfaces, homogeneous
or heterogeneous signal intensity and contrast enhancement level,
are also likely to result in different extents of resection and
residual tumor and therefore, different prognoses.
Since IDH mutation, tumor location and MRI
features correlate with patient prognosis in glioma, the aim of the
present study was to clarify the tumor location and MRI features of
IDH-mutated gliomas to determine whether the prolonged
survival of IDH-mutated patients is associated with tumor
location and presurgical MRI features. Therefore, the radiological
and genetic features of 193 patients affected by astrocytic
neoplasms, with specific associations with IDH gene status
were analyzed to address the aforementioned issues.
Materials and methods
Tumor samples
Patients were selected from a database of glioma
patients treated at the Nanfang Hospital (Guangzhou, China) between
January 2003 and December 2007. The eligibility criteria were as
follows: Unequivocal pathological diagnosis according to the 2007
World Health Organization (WHO) criteria (16); availability of genetic analysis for
IDH1/2; MRI scan at the time of the diagnosis or during the
perioperative period; and ≥18 years old at the time of surgery.
Following approval from the institutional review board, 193
evaluable patients with astrocytic neoplasms, consisting of 111
diffuse astrocytomas (DA) and 82 anaplastic astrocytomas (AA), were
selected. The archival surgical specimens were enrolled after all
patients had provided written informed consent allowing the
molecular analysis of their tumor specimens. The pathology slides
were reviewed by two neuropathologists and all tumor samples were
confirmed by microscopic examination in which ≥70% of the visible
cells in the section were tumor cells.
Radiological assessment
All patients underwent MRI scanning prior to and
following surgery (within 72 h) to evaluate the extent of surgery.
All the MRI scans were independently reviewed by two
neuroradiologists and a neurosurgeon blinded to the genetic
alterations in the tumors. The tumor location and the following MRI
features were evaluated qualitatively: Unilateral versus bilateral
pattern of growth (tumors that traversed the corpus callosum to
involve the opposite cerebral hemisphere were determined to be
bilateral); sharp versus indistinct tumor margins; homogeneous
versus heterogeneous signal intensity; absent or slight versus
significant contrast enhancement; absent or moderate versus severe
mass effect; and absent or moderate versus severe edema.
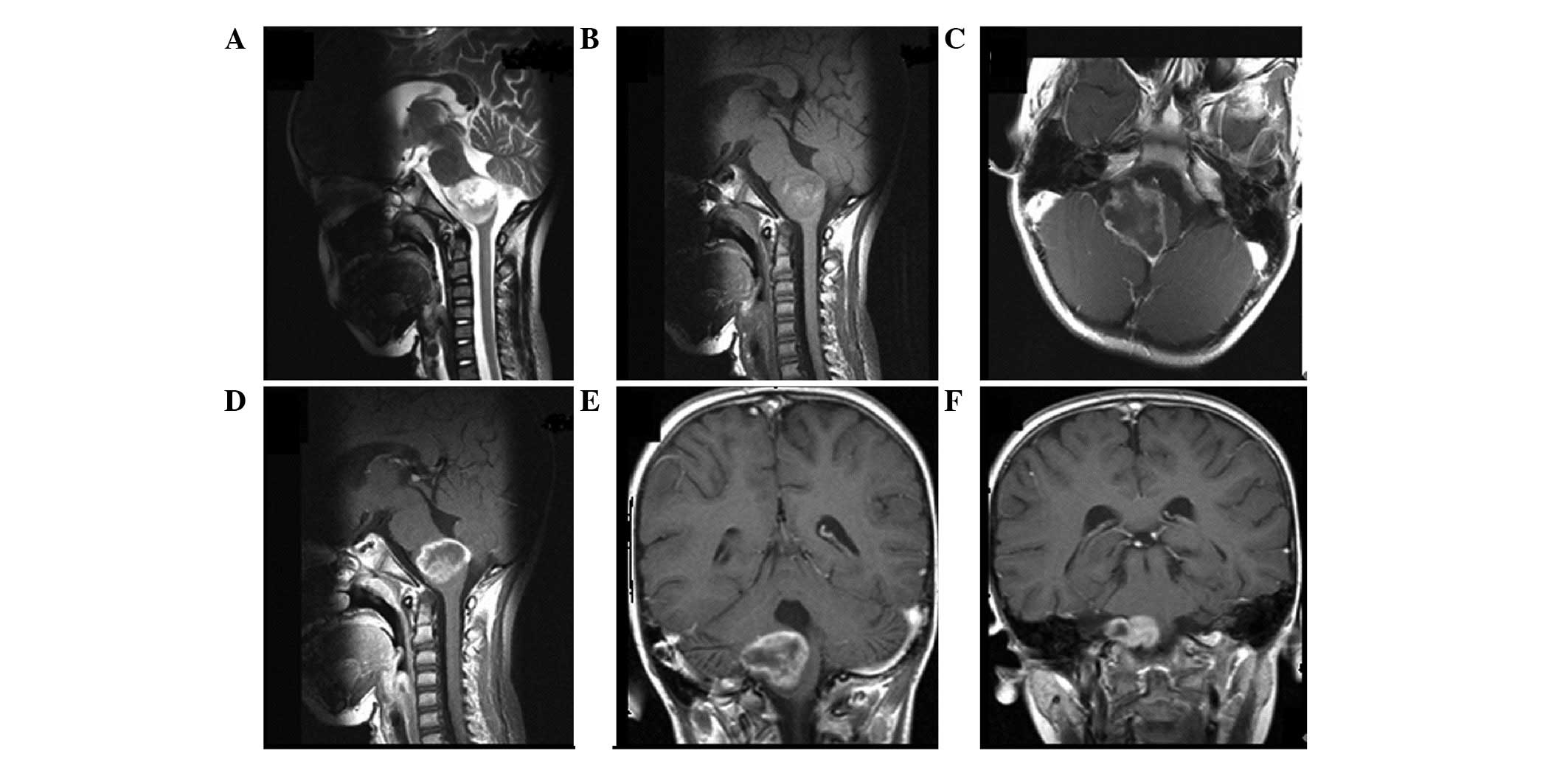
To define the tumor location, the following grouping
methods were considered. Tumor location defined using the following
prelabeled anatomical structures in the brain: i) Frontal,
temporal, parietal, occipital lobes and cerebellum; ii) insula,
diencephalon, basal ganglia; and iii) brain stem. Tumor location
divided into the following three groups according to the risk of
surgery: Group I, high-risk regions (such as the hypothalamus,
midbrain and medulla oblongata); group II, functional regions (for
example the primary sensorimotor area, supplementary motor area,
internal capsule and basal ganglia); and group III, non-functional
regions (remote from high-risk regions and functional regions) at
the time of diagnosis (Figs.
1–3). Regardless of the
grouping methods, the tumor location was typically determined by
joint analysis of sagittal, coronal and axial MRI sequences. In
addition, the site of origin and location of the epicenter of the
tumor were considered simultaneously to determine the tumor
location.
IDH1/2 mutation analysis
Genomic DNA was extracted from the formalin-fixed,
paraffin-embedded tissues using the QIAamp DNA mini kit (Qiagen,
Hilden, Germany) according to the manufacturer’s instructions.
IDH1 and IDH2 alterations characterized by mutational
hotspots at codons R132 and R172, respectively, were assessed by
high resolution melting (HRM) analysis (17) and direct sequencing, which were
performed using the ABI PRISM 3730xl DNA analyzer (Applied
Biosystems, Carlsbad, CA, USA). The polymerase chain reaction (PCR)
products generated after HRM were sequenced directly following
purification with the QIAquick PCR purification kit (Qiagen,
Valencia, CA, USA). The PCR primers for mutations were designed
using Primer Express software version 3.0 (Applied Biosystems). The
primers sequences used were as follows: Forward,
5′-CGGTCTTCAGAGAAGCCATT-3′ and reverse,
5′-GCAAAATCACATTATTGCCAAC-3′ for IDH1; and forward,
5′-CCAAGCCCATCACCATTG-3′ and reverse, 5′-ACTGGAGCTCCTCGCCTAGC-3′
for IDH2. The PCR and HRM analyses were performed in a
single run using the LightCycler 480 instrument (Roche Diagnostics
GmbH, Penzberg, Germany) in a reaction mixture to discriminate
between the wild-type and mutant DNA. Samples exhibiting
conflicting HRM and direct sequencing results were retested and
only the HRM-positive samples confirmed by direct sequencing were
considered mutated.
Statistical analysis
Statistical analyses were conducted using SPSS
version 13.0 (SPSS, Inc., Chicago, IL, USA). The χ2 test
(or Fisher’s exact test when one subgroup was n<5) was used to
determine the significance of associations. The Bonferroni test was
used for multiple comparisons and the independent samples t-test
was used to compare data acquired in each group for patients age.
Progression-free survival (PFS) and overall survival (OS) were used
to analyze the prognostic impact of IDH1/2 mutations. PFS
was calculated from the initial surgery until the first unequivocal
clinical or radiological sign of progressive disease, or the last
follow-up (for censored cases) and OS was defined as the time
between the initial surgery and mortality, or the last follow-up
(for censored cases). The survival distributions were estimated
using the Kaplan-Meier method and compared among the patient
subsets using log-rank tests. All statistical tests were two-sided
and P<0.05 was considered to indicate a statistically
significant difference. Patients who succumbed to the disease
within two months for DA and one month for AA following surgery
were excluded from the analysis to avoid the inclusion of cases in
which mortality may have been attributable to surgical
complications.
Results
IDH1 and IDH2 mutations
A total of 193 astrocytic neoplasms, which fulfilled
the inclusion criteria, were retrospectively analyzed. IDH1
and IDH2 mutations were identified in 57.5% (111/193) and
3.1% (6/193) of the patients, respectively. All IDH1
mutations were located at the amino acid residue 132, of which 102
were R132H (G395A Arg132His), six were R132C (C394T Arg132Cys) and
three were R132S (C394A Arg132Ser) mutations, whereas all
IDH2 mutations were observed at the amino acid residue 132,
of which four were R172K (G515A Arg172Lys) and two were R172M
(G515T Arg172Met) mutations. As previously reported (18,19),
these two mutations are mutually exclusive as observed in 100% of
cases in this series, suggesting that they are involved in similar
tumorigenesis pathways. Therefore, in the current statistical
analysis the IDH1 and IDH2 mutations were grouped
together. The main clinical characteristics of the patients are
summarized in Table I.
 | Table IMain clinical characteristics. |
Table I
Main clinical characteristics.
| Variables | Total
population | IDH1/2
mutations | IDH wild-type | P-value |
|---|
| n, (%) | 193 (100) | 117 (60.6) | 76 (39.4) | |
| Age, years | | | | <0.001 |
| Median | 36.5 | 32.7 | 42.5 | |
| Range | 18–72 | 19–67 | 18–72 | |
| Gender, n (%) | | | | 0.768 |
| Male | 109 (100) | 65 (59.6) | 44 (40.4) | |
| Female | 84 (100) | 52 (61.9) | 32 (38.1) | |
| KPS at diagnosis, n
(%) | | | | 0.289 |
| ≥80 | 119 (100) | 76 (63.9) | 43 (36.1) | |
| <80 | 74 (100) | 41 (55.4) | 33 (44.6) | |
| Histology (WHO
grade), n (%) | | | | 0.181 |
| DA (II) | 111 (100) | 72 (64.9) | 39( 35.1) | |
| AA (III) | 82 (100) | 45 (54.9) | 37 (45.1) | |
| Extent of surgery,
n (%) | | | | 0.002 |
| Biopsy/PR | 89 (100) | 43 (48.3) | 46 (51.7) | |
| STR/GTR | 104 (100) | 74 (71.2) | 30 (28.8) | |
| PFS, months | | | | <0.001 |
| Median | 45.8 | 56.7 | 34.4 | |
| 95% CI | 42.0–49.6 | 51.1–62.3 | 29.1–39.7 | |
| OS, months | | | | <0.001 |
| Median | 71.3 | 84.3 | 57.3 | |
| 95% CI | 66.1–76.5 | 79.0–89.6 | 46.2–68.4 | |
IDH1/2 mutations predict longer
survival
The mean follow-up of the patients was 63.3 months
(range, 15–101 months) and as indicated in Table I, a significant difference was
indicated between patients with and without the IDH1/2
mutations and PFS (P<0.001) and OS (P<0.001), independent of
WHO grade. The prognostic impact of the IDH1/2 mutations in
the DA and AA subgroups was also investigated and the IDH1/2
mutations were found to significantly correlate with increased
survival in the subgroups (data not shown).
Correlation between IDH status and tumor
location
In this series of astrocytic neoplasms, a
statistically significant correlation was identified between
IDH status (IDH-mutated versus IDH wild-type)
and tumor location. In addition, the lobar distribution of the
neoplasms was analyzed in all patients and the IDH-mutated
tumors were more frequently located in a single lobe, such as the
frontal lobe, temporal lobe or cerebellum, whereas the IDH
wild-type tumors were predominantly located in combined lobes, such
as the diencephalon or brain stem (P<0.001; χ2 test;
Table II). According to an
additional grouping method, the IDH-mutated tumors were
rarely located in the high-risk regions of the brain and more
frequently located in the non-functional and functional regions
(P<0.001; χ2 test; Table III). Although IDH-mutated
tumors invaded high-risk regions less frequently than the
non-functional and functional regions, no difference was observed
in the frequency of IDH1/2 mutations between the
non-functional and functional regions (Table III).
 | Table IIAnalyzing the frequency of
IDH1/2 mutations and tumor location according to anatomical
structures. |
Table II
Analyzing the frequency of
IDH1/2 mutations and tumor location according to anatomical
structures.
| Histology | F, n | T, n | P or O, n | Multilobes, n | I or BG, n | D or BS, n | CB, n |
|---|
| DA |
| n | 45 | 9 | 9 | 11 | 14 | 12 | 11 |
| IDH mutation | 38 | 7 | 8 | 3 | 6 | 1 | 9 |
| IDH wild-type | 7 | 2 | 1 | 8 | 8 | 11 | 2 |
| AA |
| n | 21 | 16 | 8 | 17 | 4 | 14 | 2 |
| IDH mutation | 15 | 13 | 4 | 5 | 3 | 4 | 1 |
| IDH wild-type | 6 | 3 | 4 | 12 | 1 | 10 | 1 |
| Overall |
| n | 66 | 25 | 17 | 28 | 18 | 26 | 13 |
| IDH mutation | 53 | 20 | 12 | 8 | 9 | 5 | 10 |
| IDH wild-type | 13 | 5 | 5 | 20 | 9 | 21 | 3 |
 | Table IIIAnalyzing the frequency of the
IDH1/2 mutations and tumor location according to the risk of
surgery. |
Table III
Analyzing the frequency of the
IDH1/2 mutations and tumor location according to the risk of
surgery.
| Variables | Group I vs. II | Group I vs.
III | Group II vs.
III |
|---|
| Overall, n (%) | 37 vs. 63 | 37 vs. 93 | 63 vs. 93 |
| IDH mutation | 11 (29.7) vs. 39
(61.9) | 11 (29.7) vs. 67
(72.0) | 39 (61.9) vs. 67
(72.0) |
| IDH wild-type | 26 (70.3) vs. 24
(38.1) | 26 (70.3) vs. 26
(28.0) | 24 (38.1) vs. 26
(28.0) |
| χ2
test | 9.653 | 19.746 | 1.773 |
| P-valuea | 0.003 | <0.001 | 0.222 |
Correlation between IDH status and MRI
characteristics
The correlation between the preoperative MRI
features and IDH status in all patients and the different
histological subgroups was also investigated. The histopathological
IDH status and imaging features are summarized in Table IV. As illustrated in Table IV, the gliomas with IDH
mutations were significantly more likely to exhibit a unilateral
pattern of growth, sharp tumor margins, homogeneous signal
intensity and less contrast enhancement in the DA and AA subgroups.
Analysis of all the patients also confirmed this difference,
however, no significant correlation was identified between the
remaining MRI features (mass effect and edema) and IDH
status in subgroups and all patients.
 | Table IVAnalyzing the frequency of
IDH1/2 mutations and different MRI features of gliomas. |
Table IV
Analyzing the frequency of
IDH1/2 mutations and different MRI features of gliomas.
| MRI features | Alla, n (%) | P-value | DAa, n (%) | P-value | AAa, n (%) | P-value |
|---|
| Pattern of
growth | | <0.001 | | 0.007 | | 0.001 |
| Unilateral | 116/178 (65.2) | | 71/104 (68.3) | | 45/74 (60.8) | |
| Bilateral | 1/15 (6.7) | | 1/7 (14.3) | | 0/8 (0.0) | |
| Tumor margins | | <0.001 | | 0.001 | | 0.012 |
| Sharp | 66/85 (77.6) | | 44/55 (80.0) | | 22/30 (73.3) | |
| Indistinct | 51/108 (47.2) | | 28/56 (50.0) | | 23/52 (44.2) | |
| Tumor signal
intensity | | <0.001 | | 0.003 | | <0.001 |
| Homogeneous | 70/89 (78.7) | | 45/58 (77.6) | | 25/31 (80.6) | |
| Heterogeneous | 47/104 (45.2) | | 27/53 (50.9) | | 20/51 (39.2) | |
| Contrast
enhancement | | <0.001 | | 0.001 | | 0.003 |
| Absent or
slight | 74/97 (76.3) | | 47/60 (78.3) | | 27/37 (73.0) | |
| Significant | 43/96 (44.8) | | 25/51 (49.0) | | 18/45 (40.0) | |
| Mass effect | | 0.654 | | 0.320 | | 0.216 |
| Absent or
moderate | 47/75 (62.7) | | 38/54 (70.4) | | 9/21 (42.9) | |
| Severe | 70/118 (59.3) | | 34/57 (59.6) | | 36/61 (59.0) | |
| Edema | | 0.181 | | 0.533 | | 0.375 |
| Absent or
moderate | 71/109 (65.1) | | 49/73 (67.1) | | 22/36 (61.1) | |
| Severe | 46/84 (54.8) | | 23/38 (60.5) | | 23/46 (50.0) | |
Discussion
The tumor location and MRI features of glioma are
important indicators of prognosis and the tumor location determines
the resectability of the glioma; tumors located in the critical
areas of the brain are typically non-resectable whereas those
located in the non-functional regions may undergo en bloc extended
resection to prolong survival. MRI findings which are suggestive of
high-grade gliomas, including a bilateral pattern of growth,
undefined margins, mixed signal intensity and significant
enhancement reflect increased ‘invasiveness’ and high malignancy of
the glioma, which often indicate unfavorable outcomes. Notably,
increasing age has classically been associated with a poor
prognosis in gliomas and patients with IDH wild-type tumors
are significantly older than those with IDH-mutated tumors,
which is consistent with the results of the present study (Table I) (2,3,5,7).
However, few studies have analyzed the correlation between
IDH status and tumor location/MRI characteristics.
Recently, Metellus et al (20) reported that IDH wild-type WHO
grade II gliomas are preferentially located in the
fronto-temporo-insular region and exhibit a greater volume and
therefore, require a reduced extent of surgery and demonstrate an
infiltrative pattern on MRI. This is the only study to identify a
statistically significant correlation between IDH status and
specific brain subregions of tumor locations. By contrast, patients
with oligodendroglial tumors were excluded in the current study as
astrocytoma and oligodendroglioma exhibit different biological
behaviors and clinical features (21–23).
In addition, the correlation between IDH status and tumor
location was analyzed in the present study based on a larger sample
size and more comprehensive grouping methods, determined not only
by prelabeled anatomical structures but also by the risk of
surgery. The IDH-mutated gliomas were rarely found to locate
in the high-risk regions of brain, such as the diencephalon or
brain stem, where surgical resection is limited and exhibits a high
mortality rate intraoperatively and postoperatively. The
IDH-mutated gliomas were instead preferentially found to
locate in the functional or non-functional regions, particularly
the frontal and temporal lobes, where tumors can be removed easily.
These results were supported by the observation that the extent of
surgery (gross total resection and subtotal resection) was
significantly higher in IDH-mutated gliomas compared with
IDH wild-type gliomas (P=0.002; Table I).
This correlation between genotype and tumor site has
also been reported in oligodendroglial neoplasms and glioblastoma.
Furthermore, in a pioneering study by Zlatescu et al
(24), the anaplastic
oligodendrogliomas located in the frontal, parietal and occipital
lobes were significantly more likely to harbor the 1p19q codeletion
than tumors arising in the temporal lobe, insula and diencephalon.
More recent studies have identified a significant correlation
between the 1p19q codeletion and tumor location in
oligodendrogliomas or oligoastrocytomas (25–29).
However, conflicting results have been reported regarding the
correlation between O6-methylguanine DNA methyltransferase (MGMT)
promoter methylation status and tumor location in glioblastoma.
Eoli et al (30) also
demonstrated that tumors with MGMT promoter methylation were more
frequently located in the parietal and occipital lobes, whereas
tumors without were frequently located in the temporal lobes,
however, other studies have not identified such a correlation
(31). In addition, astrocytomas
have not been found to exhibit different frequencies of the 1p19q
codeletion or tumor protein p53 mutations with respect to tumor
location (25,26).
Significant associations have also been reported
between MRI characteristics and genotype in oligodendrogliomas,
oligoastrocytomas or glioblastomas (20,29,31–37),
however, IDH mutation status was not addressed in these
studies, with the exception of that by Metellus et al.
Consistent with the results reported by Metellus et al
(20), the present study also
revealed that IDH-mutated gliomas are significantly more
likely to exhibit sharp tumor margins. Furthermore, it was observed
that IDH-mutated tumors tend to exhibit a higher incidence
of unilateral pattern of growth, homogeneous signal intensity and
less contrast enhancement. Therefore, the IDH-mutated tumors
exhibit less invasiveness when compared with IDH wild-type
tumors in MRI.
It is evident that the presence of IDH
mutations is of major prognostic significance for patient outcome
in gliomas and the current study found a marked correlation between
the IDH mutations and OS (independent of WHO grade) in this
series of patients. However, at present, the underlying mechanism
of IDH mutations in tumorigenesis and prognostic
significance remains unclear. A prospective randomized European
Organization for Research and Treatment of Cancer study 26951
reported by van den Bent et al (38), revealed no indication that the
presence of the IDH1 mutation predicts the outcome to
adjuvant procarbazine, 1-(2-chloroethyl)-3-cyclohexyl-nitrosourea,
and vincristine chemotherapy. In an additional retrospective report
on temozolomide chemotherapy in progressive low-grade astrocytoma,
no correlation was identified between outcome and the IDH
mutations (39). These results
suggested that the improved survival observed in
IDH1-mutated tumors is primarily due to a less aggressive
biological behavior and not due to an improved outcome for
chemotherapy treatment (38).
In conclusion, the current study investigated the
correlation between IDH status and tumor location, as well
as MRI characteristics in astrocytic neoplasms and revealed that
the prolonged survival of patients with IDH1-mutated tumors
is primarily due to a less aggressive biological behavior from the
perspective of tumor site and MRI features.
Acknowledgments
The authors would like to thank the patients
involved in the study for their participation. This study was
supported by the Nanfang Glioma Centre of Nanfang Hospital (grant
no. 2013B020), the Departments of Radiology and Pathology of the
Nanfang Hospital, and Helixgen (Guangzhou) Co., Ltd (Guangzhou,
China).
References
|
1
|
Xu BJ, An QA, Srinivasa GS, et al:
Identification of blood protein biomarkers that aid in the clinical
assessment of patients with malignant glioma. Int J Oncol.
40:1995–2003. 2012.
|
|
2
|
Parsons DW, Jones S, Zhang X, et al: An
integrated genomic analysis of human glioblastoma multiforme.
Science. 321:1807–1812. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Balss J, Meyer J, Mueller W, Korshunov A,
Hartmann C and von Deimling A: Analysis of the IDH1 codon 132
mutation in brain tumors. Acta Neuropathol. 116:597–602. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hartmann C, Meyer J, Balss J, et al: Type
and frequency of IDH1 and IDH2 mutations are related to astrocytic
and oligodendroglial differentiation and age: a study of 1,010
diffuse gliomas. Acta Neuropathol. 118:469–474. 2009. View Article : Google Scholar
|
|
5
|
Ichimura K, Pearson DM, Kocialkowski S, et
al: IDH1 mutations are present in the majority of common adult
gliomas but rare in primary glioblastomas. Neuro Oncol. 11:341–347.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Watanabe T, Nobusawa S, Kleihues P and
Ohgaki H: IDH1 mutations are early events in the development of
astrocytomas and oligodendrogliomas. Am J Pathol. 174:1149–1153.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yan H, Parsons DW, Jin G, et al: IDH1 and
IDH2 mutations in gliomas. N Engl J Med. 360:765–773. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nobusawa S, Watanabe T, Kleihues P and
Ohgaki H: IDH1 mutations as molecular signature and predictive
factor of secondary glioblastomas. Clin Cancer Res. 15:6002–6007.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sanson M, Marie Y, Paris S, et al:
Isocitrate dehydrogenase 1 codon 132 mutation is an important
prognostic biomarker in gliomas. J Clin Oncol. 27:4150–4154. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Toedt G, Barbus S, Wolter M, et al:
Molecular signatures classify astrocytic gliomas by IDH1 mutation
status. Int J Cancer. 128:1095–1103. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qi ST, Yu L, Lu YT, et al: IDH mutations
occur frequently in Chinese glioma patients and predict longer
survival but not response to concomitant chemoradiotherapy in
anaplastic gliomas. Oncol Rep. 26:1479–1485. 2011.
|
|
12
|
Reitman ZJ and Yan H: Isocitrate
dehydrogenase 1 and 2 mutations in cancer: alterations at a
crossroads of cellular metabolism. J Natl Cancer Inst. 102:932–941.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kloosterhof NK, Bralten LB, Dubbink HJ,
French PJ and van den Bent MJ: Isocitrate dehydrogenase-1
mutations: a fundamentally new understanding of diffuse glioma?
Lancet Oncol. 12:83–91. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Labussiere M, Sanson M, Idbaih A and
Delattre JY: IDH1 gene mutations: a new paradigm in glioma
prognosis and therapy? Oncologist. 15:196–199. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zou P, Xu H, Chen P, et al: IDH1/IDH2
mutations define the prognosis and molecular profiles of patients
with gliomas: a meta-analysis. PLoS One. 8:e687822013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Louis DN, Ohgaki H, Wiestler OD, et al:
The 2007 WHO classification of tumours of the central nervous
system. Acta Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Reed GH, Kent JO and Wittwer CT:
High-resolution DNA melting analysis for simple and efficient
molecular diagnostics. Pharmacogenomics. 8:597–608. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dang L, Jin S and Su SM: IDH mutations in
glioma and acute myeloid leukemia. Trends Mol Med. 16:387–397.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yen KE, Bittinger MA, Su SM and Fantin VR:
Cancer-associated IDH mutations: biomarker and therapeutic
opportunities. Oncogene. 29:6409–6417. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Metellus P, Coulibaly B, Colin C, et al:
Absence of IDH mutation identifies a novel radiologic and molecular
subtype of WHO grade II gliomas with dismal prognosis. Acta
Neuropathol. 120:719–729. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Persson AI, Petritsch C, Swartling FJ, et
al: Non-stem cell origin for oligodendroglioma. Cancer Cell.
18:669–682. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ohgaki H and Kleihues P: Genetic
alterations and signaling pathways in the evolution of gliomas.
Cancer Sci. 100:2235–2241. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zlatescu MC, TehraniYazdi A, Sasaki H, et
al: Tumor location and growth pattern correlate with genetic
signature in oligodendroglial neoplasms. Cancer Res. 61:6713–6715.
2001.PubMed/NCBI
|
|
25
|
Mueller W, Hartmann C, Hoffmann A, et al:
Genetic signature of oligoastrocytomas correlates with tumor
location and denotes distinct molecular subsets. Am J Pathol.
161:313–319. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang L, Jiang T, Yuan F, Li GL, Liu EZ
and Wang ZC: Correlations between molecular profile and tumor
location in Chinese patients with oligodendroglial tumors. Clin
Neurol Neurosurg. 110:1020–1024. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Laigle-Donadey F, Martin-Duverneuil N,
Lejeune J, et al: Correlations between molecular profile and
radiologic pattern in oligodendroglial tumors. Neurology.
63:2360–2362. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gozé C, Rigau V, Gibert L, Maudelonde T
and Duffau H: Lack of complete 1p19q deletion in a consecutive
series of 12 WHO grade II gliomas involving the insula: a marker of
worse prognosis? J Neurooncol. 91:1–5. 2009.PubMed/NCBI
|
|
29
|
Eoli M, Bissola L, Bruzzone MG, et al:
Reclassification of oligoastrocytomas by loss of heterozygosity
studies. Int J Cancer. 119:84–90. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Eoli M, Menghi F, Bruzzone MG, et al:
Methylation of O6-methylguanine DNA methyltransferase and loss of
heterozygosity on 19q and/or 17p are overlapping features of
secondary glioblastomas with prolonged survival. Clin Cancer Res.
13:2606–2613. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Drabycz S, Roldán G, de Robles P, et al:
An analysis of image texture, tumor location, and MGMT promoter
methylation in glioblastoma using magnetic resonance imaging.
Neuroimage. 49:1398–1405. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Megyesi JF, Kachur E, Lee DH, et al:
Imaging correlates of molecular signatures in oligodendrogliomas.
Clin Cancer Res. 10:4303–4306. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jenkinson MD, du Plessis DG, Smith TS,
Joyce KA, Warnke PC and Walker C: Histological growth patterns and
genotype in oligodendroglial tumours: correlation with MRI
features. Brain. 129:1884–1891. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jenkinson MD, Smith TS, Brodbelt AR, Joyce
KA, Warnke PC and Walker C: Apparent diffusion coefficients in
oligodendroglial tumors characterized by genotype. J Magn Reson
Imaging. 26:1405–1412. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jenkinson MD, Smith TS, Joyce KA, et al:
Cerebral blood volume, genotype and chemosensitivity in
oligodendroglial tumours. Neuroradiology. 48:703–713.
2006.PubMed/NCBI
|
|
36
|
Walker C, du Plessis DG, Fildes D, et al:
Correlation of molecular genetics with molecular and morphological
imaging in gliomas with an oligodendroglial component. Clin Cancer
Res. 10:7182–7191. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Aghi M, Gaviani P, Henson JW, Batchelor
TT, Louis DN and Barker FN II: Magnetic resonance imaging
characteristics predict epidermal growth factor receptor
amplification status in glioblastoma. Clin Cancer Res.
11:8600–8605. 2005. View Article : Google Scholar
|
|
38
|
van den Bent MJ, Dubbink HJ, Marie Y, et
al: IDH1 and IDH2 mutations are prognostic but not predictive for
outcome in anaplastic oligodendroglial tumors: a report of the
European Organization for Research and Treatment of Cancer Brain
Tumor Group. Clin Cancer Res. 16:1597–1604. 2010.
|
|
39
|
Dubbink HJ, Taal W, van Marion R, et al:
IDH1 mutations in low-grade astrocytomas predict survival but not
response to temozolomide. Neurology. 73:1792–1795. 2009. View Article : Google Scholar : PubMed/NCBI
|