Introduction
Cancer of the head and neck is associated with high
morbidity and mortality rates of patients worldwide, including
Brazil, and is a global health concern (1,2).
Studies have been conducted to determine the genes and
environmental factors that are involved in the etiology and
modulation of the presence and severity of head and neck cancer
(3–8).
The v-Ki-ras2 Kirsten rat sarcoma viral oncogene
homolog (KRAS)-LCS6 (G>T) polymorphism is located in the
KRAS2 gene (region 12p12.1, 6 exons; exon 5 has an
alternative splicing site and results in the B isoform,
KRASB) (9) and appears to
reduce the lifespan of head and neck cancer patients. Therefore,
this variant may be an indicator for the phenotype or therapeutic
response (10).
In 2005, three complementary RAS genes,
HRAS, KRAS and NRAS, were reported in the
3′untranslated region (UTR) region with the expression regulated by
the let-7 microRNA (miRNA). The let-7 miRNA expression was reduced
in lung tumor tissue compared with normal tissue, however, the RAS
protein expression was greater in lung tumors, which indicates a
potential underlying mechanism of let-7 miRNA in cancer (11). Furthermore, miRNAs are significant
in gene regulation and altered gene expression in tumors (12). The let-7 miRNA is complementary to
the KRAS 3′UTR polymorphism as a possible risk factor for
cancer (13) and the let-7 family
may be involved in KRAS gene regulation (14).
In small cell lung cancer, smokers aged >40 years
with the KRAS let-7 (KRAS-LCS6) variant possessed an
increased risk of disease. Furthermore, the KRAS-LCS6 variant
allele is associated with a high KRAS expression and
decreased levels of let-7 (13). In
this context, the interaction between the let-7 miRNA and
KRAS gene as a control mechanism via the KRAS variants has
been described. Following growth factor binding, cascade activation
occurs, which can be blocked via binding to let-7 miRNA. Thus, when
the binding between the KRAS gene and let-7 miRNA occurs,
the cascade is interrupted and the translation of the genes, which
are responsible for cell multiplication, remains blocked. When the
let-7 miRNA concentration decreases, the cascade is released and
translation occurs. However, the KRAS-LCS6 allele and let-7 miRNA
binding is hampered, therefore the cascade is released, and
translation is facilitated. This observation may be in response to
the reduced survival rate of individuals with head and neck cancer,
and the KRAS-LCS6 allele variant (10). In addition, the low level of
let-7a-2 is associated with a poor survival rate of patients with
lung adenocarcinoma (15).
Another polymorphism is the solute carrier family 23
member 2 (SLC23A2)-05 (variation G to C 8 in intron 8, position
48,263), which is localized in the SLC23A2 (nucleobase transporter)
member 2, region 20p13-p12. The sodium-dependent SLC23A2 protein is
responsible for the transport of vitamin C (16) and is considered to be a risk factor
of head and neck cancer in individuals carrying human
papillomavirus 16 (HPV16) (17).
The SLC23A2-05 polymorphism has been associated with squamous cell
carcinoma of the head and neck. However, the association between
the SLC23A2-05 variation and positive HPV, as a risk factor for
squamous cell carcinoma of the head and neck, is ambiguous.
Patients with positive HPV have an increased risk, while those with
negative HPV have a reduced risk for cancer (18).
In this context, the objective of the present study
was to evaluate the prevalence of the KRAS-LCS6 and SLC23A2-05
polymorphisms in patients with squamous cell carcinoma of the head
and neck, and to verify the significance of the polymorphisms on
the involvement and severity of the disease via an association
study.
Materials and methods
Patients with squamous cell carcinoma of
the head and neck
A cross-sectional study was performed according to a
case-control model. The study was conducted by comparing the case
(squamous cell carcinoma of the head and neck diagnosed by clinical
and laboratory variables) and control (subjects were paired by
gender and age to the case subjects) groups. A negative family
history of cancer was considered to be an inclusion criterion.
There were 165 males diagnosed with squamous cell
carcinoma of the head and neck that were treated at the Clinical
Oncology and Otorhinolaryngology, Faculty of Medical Sciences,
State University of Campinas (Campinas, São Paulo, Brazil). The DNA
samples were stored at the Laboratory of Cancer Genetics (State
University of Campinas, Campinas, Brazil). The control group
comprised of 230 healthy male subjects.
The present study was approved by the Ethics
committee of the State University of Campinas (no. 873/2007;
Campinas, Brazil) and written informed consent for participation in
the study was obtained from the subjects.
DNA extraction from blood
The extraction of DNA from peripheral blood
leukocytes was performed according to standard protocol for the
phenol-chloroform method (19),
with specific modifications. The DNA concentration that was used
for analysis was 50 ng/ml, evaluated using a NanoVue™
Spectrophotometer (GE Healthcare Biosciences, Pittsburgh, PA,
USA).
Clinical variables
The clinical variables analyzed in the samples were
as follows: Cancer presence (control or case group), ethnicity
(Caucasian or not), fatality, smoking habit (smoker, non-smoker or
ex-smoker; smokers were defined as individuals who smoked until the
point of diagnosis and ex-smokers were those that had stopped
smoking ≤5 years prior to diagnosis), alcohol consumption
(alcoholic, teetotal or ex-alcoholic; an alcoholic was defined as a
patient who had used alcohol until the point of diagnosis, teetotal
were patients who had never consumed alcohol and ex-alcoholics had
stopped prior to the diagnosis), age at diagnosis,
tumor-node-metastasis (TNM) staging system (I, II, III and IV),
degree of cancer differentiation (poor, moderate or well) and
localization. For the control group, the variables considered were
smoker/alcoholic (smoked/consumed alcohol up to the time of last
visit), ex-smoker/ex-alcoholic (stopped smoking/consuming alcohol
prior to the last visit) and non-smoker/teetotal.
Medical specialists assessed all of the variables,
with special consideration given to TNM stage and the
adenocarcinoma differentiation degree. The evaluated variables were
normalized according to the literature (20).
SLC23A2-05 (rs4987219) and KRAS-LCS6
(rs61764370) polymorphism analysis
The polymorphism analysis was by polymerase chain
reaction (PCR) and enzymatic digestion. To determine the KRAS-LCS6
and SLC23A2-05 polymorphism genotypes the following PCR conditions
were used: Bidistilled water, 10× Taq Buffer with KCl,
MgCl2 (25 mM), dNTP (25 mM for each nitrogenous base),
primers (0.2 pmol of sense and antisense primers), Taq polymerase
(5 units) and genomic DNA (50 ng/ml). The primers used were as
follows: Sense, 5′-GGT GTC AGA GTC TCG CTC TT-3′ and antisense,
5′-GGG TCG TAT ACC AAA GGC CTT AG-3′ for the KRAS-LCS6 polymorphism
(to amplify a fragment of 420 bp); sense, 5′-AAA TGC TCT GGG CAA
CCT TA-3′ and antisense, 5′-CCC CCA GGA CAT CGA CAA-3′ for the
SLC23A2-05 polymorphism (to amplify a fragment of 385 bp).
The annealing temperature was 64.6 and 64.9°C for
the KRAS-LCS6 and SLC23A2-05 polymorphisms, respectively, for one
minute. The initial denaturation was at 95°C for 5 min, followed by
35 cycles of 94°C for 1 min (with an annealing temperature specific
to each polymorphism) and 72°C for 2 min; the final amplification
was at 72°C for 7 min.
Enzymatic digestion
Following PCR, the enzymatic digestion was performed
with the Hinf I enzyme (New England BioLabs, Inc., Ipswich,
MA, USA) for the KRAS-LCS6 polymorphism and with Tai I (New
England BioLabs, Inc.,) for the SLC23A2-05 polymorphism, at 37°C
for 14 h according to the manufacturer’s instructions.
The reaction was analyzed on a polyacrylamide gel
(12%) with a voltage of 180 V for 4 h. The gel was stained in
ethidium bromide solution and visualized on a Typhoon™ scanner (GE
Healthcare, Wisconsin, WI, USA).
According to the fragments observed, the genotypes
were identified as follows: i) KRAS-LCS6 polymorphism; TT
(8+80+135+197 bp), TG (8+80+135+197+332 bp), and GG (8+80+332 bp)
and ii) SLC23A2-05 polymorphism; CC (385 bp), CG (128+270 bp) and
GG (128+270+385 bp).
Statistical analyses
The mean age difference between the case and control
groups was evaluated by Student’s t-test using the R program
(21). The Hardy-Weinberg
equilibrium for genotypes was evaluated by the Haploview program
(22), and P<0.05 was considered
to indicate a statistically significant difference.
Genotypic and allelic association calculations were
performed by logistic regression using the logistf function
environment in the R program (21),
considering each polymorphism individually as a risk factor for a
more severe TNM staging as well as for the presence of cancer.
To associate cancer presence with smoking habit,
alcohol consumption and ethnicity as risk factors, the Statistical
Package for the Social Sciences version 17 (SPSS Inc., Chicago, IL,
USA) (23) was used, incorporating
Fisher’s exact test and the χ2 test. Using the same
program and tests, the association between the polymorphisms, and
tumor localization and differentiation degree was assessed. A final
analysis was performed concerning the combination of the two
polymorphisms, KRAS-LCS6 and SLC23A2-05, and the presence of
cancer.
Extra data was obtained using the multi-factor
dimensionality reduction (MDR) version 2.0 and MDR permutation test
(PT) version 0.4.7 programs. The two programs were used in order to
evaluate the genetic interaction between the polymorphisms in the
samples. The MDR model is non-parametric and genetic model-free
data mining for the identification of non-linear interactions among
genetic and environmental attributes (24–26).
To adjust the results for multiple comparisons, an MDRPT, which
calculated 100,000 permutations was performed in the samples.
The statistical power of the samples was estimated
using GPower 3.1.6 (27), to detect
associations between the phenotype and polymorphisms. The
statistical power of the samples in the present study was
86.6%.
Results
Population characteristics
In the case group (mean age, 58.53±9.82 years) 100
patients were diagnosed with laryngeal cancer, and 65 were
diagnosed with throat (larynx and pharynx) cancer. With regard to
smoking habit; 76.98, 21.21 and 1.81% of the subjects were smokers,
ex-smokers and non-smokers, respectively, and for alcohol
consumption; 55.76, 30.91 and 13.33% were alcoholics, ex-alcoholics
or teetotal, respectively.
In the control group (mean age, 57.12±10.27 years),
the smoking habit group comprised of 21.61, 41.71 and 36.68%
smokers, ex-smokers and non-smokers, respectively, and for alcohol
consumption, 9.55, 27.64 and 62.81% were alcoholics, ex-alcoholics
or teetotal, respectively. Alcohol consumption was analyzed in 199
control subjects.
Analysis of the KRAS-LCS6 and SLC23A2-05
polymorphisms
In the KRAS-LCS6 polymorphism case group, 135
(81.82%) patients exhibited the TT genotype, 29 (17.58%) exhibited
the TG genotype and 1 (0.61%) exhibited the GG genotype. In the
control group, 187 (81.30%) exhibited the TT genotype, 41 (17.83%)
exhibited the GT genotype and 2 (0.87%) exhibited the GG genotype.
In the case group the allele frequency was 0.91 and 0.09, for the T
and G allele, respectively. In the control group the allele
frequency was 0.90 and 0.10, for the T and G allele,
respectively.
In the case group for the SLC23A2-05 polymorphism,
35 (21.21%) patients exhibited the CC genotype, 84 (50.91%) CG and
46 (27.88%) GG. In the control group, 43 (18.69%) exhibited the CC
genotype, 132 (57.39%) CG and 55 (23.91%) GG. In the case and
control groups the allele frequency was 0.47 and 0.53, for the C
and G allele respectively.
The KRAS-LCS6 polymorphism was in Hardy-Weinberg
equilibrium in the case and control groups: P=0.9899 and P=0.9999,
respectively; as were the SLC23A2-05 polymorphisms in the case and
control groups; P=0.9232 and P=0.0332, respectively (data not
shown).
In Table I the
association of alcohol consumption (P<0.001), smoking habit
(P<0.001) and ethnicity (P=0.1337) with cancer diagnosis is
described. In Table II, the
association between the KRAS-LCS6 and SLC23A2-05 polymorphisms, and
the degree of cancer differentiation (KRAS-LCS6, P=0.140; and
SLC23A2-05, P=0.186) and cancer location (KRAS-LCS6, P=0.640; and
SLC23A2-05, P=0.447) is shown. The association of the KRAS-LCS6 and
SLC23A2-05 polymorphisms with cancer presence is demonstrated in
Table III and in Table IV, the correlation between the two
polymorphs and TNM staging is shown; P>0.05 for all values.
 | Table IAssociation between the cancer and
control groups with environmental factors; smoking, alcohol
consumption and ethnicity. |
Table I
Association between the cancer and
control groups with environmental factors; smoking, alcohol
consumption and ethnicity.
| Group | | | | |
|---|
|
| | | | |
|---|
| Environmental
factor | Cancer | Control | Total | P-value | OR | CI (5–95%) |
|---|
| Smokera |
| EXS | 35 | 83 | 118 | <0.0001b | 0.377b | 0.334–0.600b |
| NS | 3 | 73 | 76 | | 0.032b | 0.008–0.093b |
| S | 127 | 43 | 170 | | 12.02b | 7.374–19.93b |
| Total | 165 | 199 | 364 | | | |
| Alcohol
consumptiona |
| EXA | 49 | 55 | 104 | <0.0001b | 1.115 | 0.705–1.763 |
| NA | 24 | 125 | 149 | | 0.102b | 0.06–0.1702b |
| A | 91 | 19 | 110 | | 11.72b | 6.748–21.030b |
| Total | 164 | 199 | 363 | | | |
| Ethnicityc |
| C | 143 | 185 | 328 | 0.1337 | 1.579 | 0.911–2.790 |
| NC | 22 | 45 | 67 | | 1.000 | |
| Total | 165 | 230 | 395 | | | |
 | Table IIAssociation of the KRAS-LCS6 and
SLC23A2-05 polymorphisms with location and differentiation degree
in patients with squamous cell carcinoma of the head and neck
identified by the χ2 test. |
Table II
Association of the KRAS-LCS6 and
SLC23A2-05 polymorphisms with location and differentiation degree
in patients with squamous cell carcinoma of the head and neck
identified by the χ2 test.
| A, Association of
the KRAS-LCS6 and SLC23A2-05 polymorphisms with location |
|---|
|
|---|
| KRAS-LCS6 | | | SLC23A2-05 | | |
|---|
|
| | |
| | |
|---|
| Location | GG + TG | TT | Total | P-value | OR | 95% CI | CC | OR (95% CI) | CG | OR (95% CI) | GG | OR (95% CI) | Total | P-value |
|---|
| Oral cavity | 1 | 0 | 1 | 0.140 | - | - | 0 | - | 1 | - | 0 | - | 1 | 0.186 |
| Larynx | 16 | 84 | 100 | | 0.695 | 0.310–1.568 | 16 | 0.463
(0.214–0.992)a | 56 | 1.676
(0.892–3.173) | 28 | 1.015
(0.505–2.067) | 100 | |
| Opharynx | 13 | 51 | 64 | | 1.258 | 0.553–2.821 | 19 | 2.232
(1.042–4.829)a | 27 | 0.565
(0.298–1.065) | 18 | 1.02
(0.501–2.052) | 64 | |
| Total | 30 | 135 | 165 | | | | 35 | | 84 | | 46 | | 165 | |
|
| B, Association of
the KRAS-LCS6 and SLC23A2-05 polymorphisms with degree of
differentiation |
|
| KRAS-LCS6 | | | SLC23A2-05 | | |
|
| | |
| | |
| Degree of
differentiation | GG + TG | TT | Total | P-value | OR | 95% CI | CC | OR (95% CI) | CG | OR (95% CI) | GG | OR (95% CI) | Total | P-value |
|
|
Well-differentiated | 4 | 16 | 20 | 0.640 | 1.282 | 0.340–4.052 | 2 | 0.345
(0.052–1.381) | 10 | 0.985
(0.376–2.581) | 8 | 1.971
(0.712–5.267) | 20 | |
|
Moderately-differentiated | 19 | 105 | 124 | | 0.623 | 0.238–1.751 | 29 | 1.27
(0.488–3.687) | 63 | 1.101
(0.496–2.453) | 32 | 0.732
(0.313–1.781) | 124 | |
|
Non-keratinizing | 0 | 1 | 1 | | - | - | 1 | - | 0 | - | 0 | - | 1 | 0.447 |
|
Poorly-differentiated | 3 | 6 | 9 | | 2.652 | 0.51–11.46 | 3 | 1.774
(0.346–7.537) | 4 | 0.780
(0.180–3.197) | 2 | 0.758
(0.104–3.573) | 9 | |
| Superficially
invasive | 0 | 1 | 1 | | - | - | 0 | - | 1 | - | 0 | - | 1 | |
| Total | 26 | 129 | 155 | | | | 35 | | 78 | | 42 | | 155 | |
 | Table IIIAssociation between the KRAS-LCS6 and
SLC23A2-05 polymorphisms as a risk factor for squamous cell
carcinoma of the head and neck as identified by regression
analysis. |
Table III
Association between the KRAS-LCS6 and
SLC23A2-05 polymorphisms as a risk factor for squamous cell
carcinoma of the head and neck as identified by regression
analysis.
| A, KRAS-LCS6
polymorphism |
|---|
|
|---|
| Patients, n | Frequency | | |
|---|
|
|
| | |
|---|
| Feature | Case group | Control group | Case group (%) | Control group
(%) | P-value | OR (95% CI) |
|---|
| Genotype |
| GG | 1 | 2 | 0.61 | 0.87 | 0.8597 | 0.83
(0.08–8.70) |
| TG | 29 | 41 | 17.57 | 17.83 | 0.9603 | 0.99
(0.58–1.67) |
| TT | 135 | 187 | 81.81 | 81.30 | 0.9080 | 1.03
(0.62–1.73) |
| Allele |
| G | 31 | 45 | 9.39 | 9.78 | 0.9080 | 0.97
(0.58–1.63) |
| T | 299 | 415 | 90.61 | 90.22 | 0.8597 | 1.20
(0.11–12.53) |
|
| B, SLC23A2-05
polymorphism |
|
| Patients, n | Frequency | | |
|
|
| | |
| Feature | Case group | Control group | Case group (%) | Control group
(%) | P-value | OR (95% CI) |
|
| Genotype |
| CC | 35 | 43 | 21.21 | 18.69 | 0.5304 | 1.17
(0.71–1.93) |
| CG | 84 | 132 | 50.90 | 57.39 | 0.2026 | 0.77
(0.52–1.15) |
| GG | 46 | 55 | 27.88 | 23.91 | 0.3711 | 1.23
(0.78–1.94) |
| Allele |
| C | 154 | 218 | 46.67 | 47.39 | 0.3711 | 0.81
(0.52–1.28) |
| G | 176 | 242 | 53.33 | 52.61 | 0.5304 | 0.85
(0.52–1.40) |
 | Table IVTNM staging and correlation with the
KRAS-LCS6 and SLC23A2-05 polymorphisms in patients with squamous
cell carcinoma of the head and neck as identified by regression
analysis. |
Table IV
TNM staging and correlation with the
KRAS-LCS6 and SLC23A2-05 polymorphisms in patients with squamous
cell carcinoma of the head and neck as identified by regression
analysis.
| A, KRAS-LCS6
gene |
|---|
|
|---|
| Stage I | Stage II | Stage III | Stage IV |
|---|
|
|
|
|
|
|---|
| Feature | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) |
|---|
| Genotype |
| GG | 0.7263 | 1.79
(0.04–75.55) | 0.5613 | 2.77
(0.06–118.05) | 0.9512 | 1.10
(0.03–46.06) | 0.6419 | 1.64
(0.16–17.23) |
| TG | 0.2017 | 1.85
(0.74–4.65) | 0.4044 | 1.64
(0.53–5.13) | 0.2259 | 0.55
(0.19–1.56) | 0.9559 | 1.02
(0.53–1.95) |
| TT | 0.2445 | 0.57
(0.23–1.43) | 0.4586 | 0.64
(0.21–2.01) | 0.1810 | 1.93
(0.68–5.48) | 0.9073 | 0.96
(0.51–1.82) |
| Allele |
| G | 0.2445 | 1.75
(0.70–4.37) | 0.4586 | 1.55
(0.50–4.83) | 0.1810 | 0.52
(0.18–1.47) | 0.9073 | 1.04
(0.55–1.96) |
| T | 0.7263 | 0.56
(0.01–23.52) | 0.5613 | 0.36
(0.01–15.39) | 0.9512 | 0.91
(0.02–37.98) | 0.6419 | 0.61
(0.06–6.39) |
|
| B,
SLC23A2-05 gene |
|
| Stage I | Stage II | Stage III | Stage IV |
|
|
|
|
|
| Feature | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) |
|
| Genotype |
| CC | 0.0627 | 0.26
(0.05–1.44) | 0.8598 | 1.12
(0.33–3.83) | 0.3675 | 1.44
(0.66–3.13) | 0.2147 | 1.46
(0.81–2.64) |
| CG | 0.5475 | 1.29
(0.56–3.00) | 0.9055 | 0.94
(0.35–2.55) | 0.6492 | 0.86
(0.44–1.66) | 0.0566 | 0.62
(0.37–1.02) |
| GG | 0.3456 | 1.54
(0.64–3.70) | 0.8220 | 1.14
(0.37–3.51) | 0.8431 | 0.92
(0.42–2.03) | 0.2859 | 1.36
(0.78–2.36) |
| Allele |
| C | 0.3456 | 0.65
(0.27–1.57) | 0.8220 | 0.88
(0.28–2.71) | 0.8431 | 1.08
(0.49–2.38) | 0.2859 | 0.74
(0.42–1.29) |
| G | 0.0627 | 3.79
(0.69–20.73) | 0.8598 | 0.89
(0.26–3.07) | 0.3675 | 0.70
(0.32–1.52) | 0.2147 | 0.69
(0.38–1.24) |
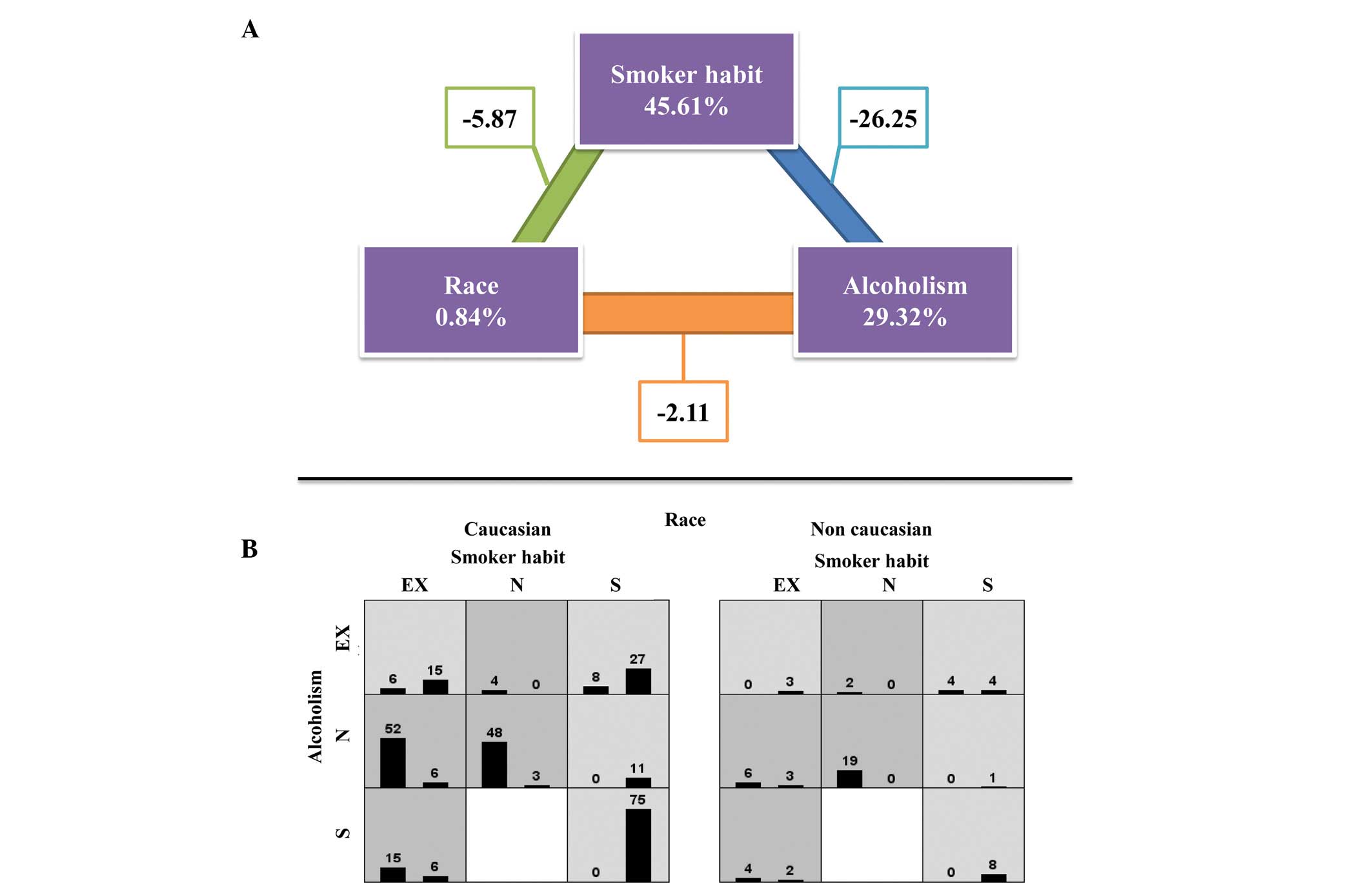
The combination of the KRAS-LCS6 and SLC23A2-05
polymorphisms with cancer presence is shown in Table V (P=0.646) and the sample
distribution is shown in Fig.
1.
 | Table VCombination of the KRAS-LCS6 and
SLC23A2-05 polymorphisms and the associated risk of squamous cell
carcinoma of the head and neck as determined by the χ2
test. |
Table V
Combination of the KRAS-LCS6 and
SLC23A2-05 polymorphisms and the associated risk of squamous cell
carcinoma of the head and neck as determined by the χ2
test.
| KRAS-LCS6 and
SLC23A2-05 polymorphisms | Cancer group | Control group | Total | P-valuea | OR | 95% CI |
|---|
| Combination
code |
| 1 | 6 | 6 | 12 | 0.826 | 1.408 | 0.422–4.693 |
| 2 | 15 | 28 | 43 | | 0.722 | 0.364–1.392 |
| 3 | 9 | 9 | 18 | | 1.415 | 0.534–3.754 |
| 4 | 28 | 37 | 65 | | 1.066 | 0.618–1.826 |
| 5 | 70 | 104 | 174 | | 0.893 | 0.595–1.337 |
| 6 | 37 | 46 | 83 | | 1.156 | 0.796–1.885 |
| Total | 165 | 230 | 395 | | | |
The MDR program analysis (Fig. 2) did not identify any statistically
significant association between cancer and the KRAS-LCS6 and
SLC23A2-05 polymorphisms (ratio, 0.7174; testing balanced accuracy,
0.5157; P=0.4090). For the environmental factors, there was an
association between smoking habit and alcohol consumption with
cancer; however, this was not identified with ethnicity (ratio,
1.0244; testing balanced accuracy, 0.8733; P=0.0000–0.0010).
 | Figure 2Genotype distribution of KRAS-LCS6 and
SLC23A2-05 polymorphisms, and the combination between the
polymorphisms. χ2 test was performed. 1. OR, 1.408 (CI,
0.422–4.693); 2. OR, 0.722 (CI, 0.364–1.392); 3. OR, 1.415 (CI,
0.534–3.754); 4. OR, 1.066 (CI, 0.618–1.826); 5. OR, 0.893 (CI,
0.595–1.337); 6. OR, 1.156 (CI, 0.796–1.885). SLC23A2,
solute carrier family 23 member 2; KRAS, v-Ki-ras2 Kirsten
rat sarcoma viral oncogene homolog; G, guanine; T, thymine; C,
cytosine; OR, odds ratio, CI, confidence interval. |
Discussion
Cancer is a complex disease and is a significant
health problem. Cancer is a genetic disease, which is highly
influenced by environmental factors. Determining the risk factors,
including genes and environmental variables, improves current
knowledge regarding cancer, provides improved tools for its
diagnosis and treatment, as well as a method to determine the risk
within a given population. In the present study, squamous cell
carcinoma of the head and neck was analyzed considering two
polymorphisms, SLC23A2-05 (rs4987219) and KRAS-LCS6 (rs61764370),
and environmental factors (smoking habit and alcohol
consumption).
For the KRAS-LCS6 polymorphism, the case and control
groups were in Hardy-Weinberg equilibrium. In the literature, there
is a hypothesis that the activation of the KRAS pathway may occur
in squamous cell carcinomas by the action of the KRAS-LCS6 normal
variant, which may therefore be associated with the susceptibility
and clinical outcome of cancer (10). The present study did not show an
association with the overall risk of squamous cell carcinoma of the
head and neck, however, cases with the G allele showed a reduction
in survival: Hazard ratio (HR), 1.6 and 95% confidence interval;
CI, 1.0–2.5. This risk was higher in oral cavity carcinoma (HR,
2.7; 95% CI, 1.4–5.3). The comparison of allele frequencies between
the present study and Christensen et al (10) is not possible; in the present study,
the polymorphisms were analyzed in blood samples, whereas
Christensen et al (10)
analyzed tumor tissue.
In the present study the G allele occurred with a
low frequency, in the case and control groups. Therefore, this
allele is not considered to be a risk factor to disease
susceptibility. The same was found when the sample was evaluated by
cancer location. Furthermore, when correlated with the disease
stage, no association was found, which indicates no influence on
disease progression. However, in the present study the polymorphism
was analyzed in the normal tissue of the case patients. Therefore,
the hypothesis that this polymorphism may be significant in tumor
development could not be excluded, and further investigation of
this polymorphism in tumor tissue is required.
In the present study, the SLC23A2-05 polymorphism in
the case and control groups demonstrated the same allele frequency.
The allele frequency of C and G were 0.47 and 0.53, respectively.
In a previous study, C and G allele frequency in an
African-American population was 0.80 and 0.20, and in Caucasians
was 0.42 and 0.58, respectively (28). In a cancer study, the G and C allele
frequency was 0.59 and 0.41 in cancer patients, and 0.57 and 0.43
in control groups. There was a predominance of Caucasian subjects
(92 and 93% in the cancer and case groups, respectively) (17). The data of the present study are
comparable to the frequencies that were identified in the Caucasian
population.
A study by Chen et al (17) evaluated 319 patients with squamous
cell carcinoma of the head and neck, and 495 control subjects in
association with HPV16 status, citrus consumption and the single
nucleotide polymorphism (SNP) panel (SNP500Cancer). The SLC23A2-05
polymorphism plus HPV16 infection were considered to be a risk
factor for cancer. Among G allele patients with the SLC23A2-05
polymorphism, the cancer risk in association with HPV16 was 5.0
(95% CI, 3.2–7.8). Among the homozygous variant (CC) subjects, the
cancer risk associated with HPV16 was attenuated (odds ratio; OR,
2.8 and 95% CI, 1.2–6.2). In determining if the genotype modifies
the interaction between citrus consumption, HPV16 and cancer risk,
there was an increased risk of cancer found in individuals with one
G allele to SLC23A2-5 polymorphism, HPV16 seropositive and high
citrus consumption (OR, 7.4; 95% CI, 3.6–15.1). In the present
study the polymorphism was not associated with cancer presence or
cancer progression.
In TNM staging, there was no statistically
significant association identified with the SLC32A2-05
polymorphism. In this context, these polymorphisms may not be
cancer severity modulators. As for the KRAS-LCS6 polymorphism, the
SLC32A2-05 polymorphism was analyzed in normal tissue.
Concerning smoking habit and alcohol consumption,
there was an association with cancer risk. In this context, the
squamous cell carcinoma of the head and neck was influenced by
environmental factors, however, there was not a modifier gene
associated with the polymorphisms that were investigated in the
present study.
In conclusion, squamous cell carcinoma of the head
and neck is a significant disease worldwide. Therefore, determining
the factors that are associated with the severity and presence of
this type of cancer is considered to be important. Globally,
smoking habit and alcohol consumption are considered to be risk
factors, resulting in individuals exhibiting squamous cell
carcinoma of the head and neck. In the present study, the KRAS-LCS6
and SLC32A2-05 polymorphisms were not considered to be cancer risk
factors, however, they were analyzed in the blood and not in tumor
tissue. Potentially, the observation of these polymorphisms in a
localized tumor may provide a significant tool for the detection of
cancer. In future studies, the investigation of additional genes
and tumor tissue is required to clarify the complex mechanisms
associated with cancer, as well as protein quantification to
determine additional data regarding cancer severity.
Acknowledgements
The authors would like to thank Dr Gustavo Jacob
Lourenço for his contribution regarding the DNA samples.
References
|
1
|
Dedivitis RA, França CM, Mafra ACB,
Guimarães FT and Guimarães AV: Clinic and epidemiologic
characteristics in the with squamous cell carcinoma of the mouth
and oropharynx. Rev Bras Otorrinolaringol. 70:35–40. 2004.
|
|
2
|
Kamangar F, Dores GM and Anderson WF:
Patterns of cancer incidence, mortality, and prevalence across five
continents: defining priorities to reduce cancer disparities in
different geographic regions of the world. J Clin Oncol.
24:2137–2150. 2006. View Article : Google Scholar
|
|
3
|
Chaikhoutdinov I and Goldenberg D: Impact
of genetic targets on therapy in head and neck squamous cell
carcinoma. Adv Exp Med Biol. 779:165–177. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rothenberg SM and Ellisen LW: The
molecular pathogenesis of head and neck squamous cell carcinoma. J
Clin Invest. 122:1951–1957. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Feng Z, Xu Q and Chen W: Epigenetic and
genetic alterations-based molecular classification of head and neck
cancer. Expert Rev Mol Diagn. 12:279–290. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Little JW, Falace DA, Miller CS and Rhodus
NL: Cancer and oral care of the patient. Little and Falace’s Dental
Management of the Medically Compromised Patient. 8th edition. 1.
Elsevier Inc; St. Louis, MO: pp. 459–492. 2013
|
|
7
|
Van der Vorst S, Dekairelle AF, Weynand B,
Hamoir M and Gala JL: Assessment of p53 functional activity in
tumor cells and histologically normal mucosa from patients with
head and neck squamous cell carcinoma. Head Neck. 34:1542–1550.
2012.PubMed/NCBI
|
|
8
|
Turksma AW, Bontkes HJ, van den Heuvel H,
et al: Effector memory T cell frequencies in relation to tumour
stage, location and HPV status in HNSCC patients. Oral Dis.
19:577–584. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
O’Connell P, Lathrop GM, Law M, et al: A
primary genetic linkage map for human chromosome 12. Genomics.
1:93–102. 1987.PubMed/NCBI
|
|
10
|
Christensen BC, Moyer BJ, Avissar M, et
al: A let-7 microRNA-binding site polymorphism in the KRAS 3′ UTR
is associated with reduced survival in oral cancers.
Carcinogenesis. 30:1003–1007. 2009.
|
|
11
|
Online Mendelian Inheritance in Man
(OMIM). http://www.ncbi.nlm.nih.gov/omim.
Accessed March 21st, 2013
|
|
12
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
13
|
Chin LJ, Ratner E, Leng S, Zhai R, Nallur
S, Babar I, Muller RU, Straka E, Su L, Burki EA, et al: A SNP in a
let-7 microRNA complementary site in the KRAS 3′ untranslated
region increases non-small cell lung cancer risk. Cancer Res.
68:8535–8540. 2008.PubMed/NCBI
|
|
14
|
Johnson SM, Grosshans H, Shingara J, Byrom
M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D and Slack
FJ: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Faaland CA, Race JE, Ricken G, Warner FJ,
Williams WJ and Holtzman EJ: Molecular characterization of two
novel transporters from human and mouse kidney and from LLC-PK1
cells reveals a novel conserved family that is homologous to
bacterial and Aspergillus nucleobase transporters. Biochim Biophys
Acta. 1442:353–360. 1998. View Article : Google Scholar
|
|
17
|
Chen AA, Marsit CJ, Christensen BC,
Houseman EA, McClean MD, Smith JF, Bryan JT, Posner MR, Nelson HH
and Kelsey KT: Genetic variation in the vitamin C transporter,
SLC23A2, modifies the risk of HPV16-associated head and neck
cancer. Carcinogenesis. 30:977–981. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Meyer MS, Applebaum KM, Furniss CS, Peters
ES, Luckett BG, Smith JF, Bryan J, McClean MD, Marsit C and Kelsey
KT: Human papillomavirus-16 modifies the association between fruit
consumption and head and neck squamous cell carcinoma. Cancer
Epidemiol Biomarkers Prev. 17:3419–3426. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Woodhead NE, Long WF and Williamson FB:
Binding of zinc ions to heparin. Analysis by equilibrium dialysis
suggests the occurrence of two, entropy-driven, processes. Biochem
J. 237:281–284. 1986.PubMed/NCBI
|
|
20
|
Morson BC and Sobin LH: Histological
typing of intestinal tumours. International Histological
Classification of Tumours. (15)World Health Organization; Geneva:
1976
|
|
21
|
R Development Core Team. R: A Language and
Environment for Statistical Computing. 2.12.2 edition. R Foundation
for Statistical Computing; Vienna, Austria: 2011
|
|
22
|
Barrett JC, Fry B, Maller J and Daly MJ:
Haploview: analysis and visualization of LD and haplotype maps.
Bioinformatics. 21:263–265. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
SPSS 17.0 for Windows (computer program).
Statistical Package for Social Science (SPSS). Release Version
17.0.1. SPSS, Incorporation; Chicago, IL: 2011, http://www.spss.com.
Accessed July 19, 2013
|
|
24
|
Hahn LW, Ritchie MD and Moore JH:
Multifactor dimensionality reduction software for detecting
gene-gene and gene-environment interactions. Bioinformatics.
19:376–382. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ritchie MD, Hahn LW and Moore JH: Power of
multifactor dimensionality reduction for detecting gene-gene
interactions in the presence of genotyping error, missing data,
phenocopy, and genetic heterogeneity. Genet Epidemiol. 24:150–157.
2003. View Article : Google Scholar
|
|
26
|
Moore JH, Gilbert JC, Tsai CT, Chiang FT,
Holden T, Barney N and White BC: A flexible computational framework
for detecting, characterizing, and interpreting statistical
patterns of epistasis in genetic studies of human disease
susceptibility. J Theor Biol. 241:252–261. 2006. View Article : Google Scholar
|
|
27
|
Faul F, Erdfelder E, Lang AG and Buchner
A: G*Power 3: a flexible statistical power analysis
program for the social, behavioral, and biomedical sciences. Behav
Res Methods. 39:175–191. 2007.
|
|
28
|
Erichsen HC, Engel SA, Eck PK, Welch R,
Yeager M, Levine M, Siega-Riz AM, Olshan AF and Chanock SJ: Genetic
variation in the sodium-dependent vitamin C transporters, SLC23A1,
and SLC23A2 and risk for preterm delivery. Am J Epidemiol.
163:245–254. 2006. View Article : Google Scholar : PubMed/NCBI
|