Introduction
Thymomas are primary tumors that arise from thymic
epithelial cells (TEC) (1). The
thymus is a primary lymphoid organ that plays a role in regulating
the proliferation and differentiation of T cells. Although the
thymus typically starts to coalesce and becomes completely atrophic
with remnant adipose tissue by the late teens, lymphopoiesis of the
T cells continues during adult life (2). Thymomas retain thymic cortical
epithelial function to induce T-cell differentiation (3); however, they may lack normal
mechanisms for selection of the T cell repertoire. Autoreactive T
cells possibly emerging in a thymoma may trigger autoimmune
disorders (4). Thymomas are
well-known for their significant association with multiple
autoimmune diseases, particularly myasthenia gravis (MG). It has
been reported that up to 50% of thymoma patients develop MG
(5).
MG is a prototypical antibody-mediated autoimmune
disease characterized by the production of autoantibodies against
the skeletal muscle acetylcholine receptor (AChR) at the
neuromuscular junction (6). An
increasing number of muscle autoantibodies, such as muscle-specific
tyrosine kinase, titin and ryanodine receptor (RyR) antibodies,
have been found in patients with MG (7). MG is paraneoplastic in association
with thymoma, which is detected in 10–15% of MG patients (8). Histologically, thymomas are epithelial
neoplastic cells surrounded by maturing T cells. The epithelial
cells are capable of expressing epitopes cross-reactive with
skeletal muscle proteins, such as AChR, titin and RyR (9). The muscle-like epitopes are presented
to T cells together with costimulatory molecules (9). Autoreactive T cells that are specific
for AChR and titin are found in the sera of thymoma patients and
thymoma patients with MG (10).
Thus, autoreactive T cells play a vital role in the incidence of
thymoma and MG.
The T-cell immunoglobulin domain and mucin domain
(TIM) family of genes, positionally cloned in 2001 from within the
T cell and airway phenotype regulator (Tapr) locus (11), consists of three members (Tim-1, -3
and -4) on the human chromosome 5q33.2 (12). TIM proteins are involved in the
regulation of T helper (Th) cell immune responses and thus are key
regulators of immune responses (13,14).
The Th cells are subdivided into Th1 or Th2 cells based on the
cytokines produced and distinct functions performed (15). The Th1 and Th2 cells play critical
roles in the regulation of cellular and humoral immune responses.
The balance of Th1 and Th2 cells is crucial in the immune response
to several organ-specific autoimmune diseases. Tim-1, the first
member of the TIM gene family, which is tightly linked to the
immune system, plays an important role in the generation and/or
maintenance of the balance between Th1 and Th2 cells, and is
upregulated in Th2 cells following activation and interacts with
its ligand expressed on antigen-presenting cells (16). It has been reported that Tim-1
polymorphisms are associated with various immune-related diseases,
including rheumatoid arthritis (17), systemic lupus erythematosus
(18), multiple sclerosis (19), diabetes (20), tumors (21,22)
and asthma (23). However, the
association of Tim-1 gene polymorphisms with thymoma and MG has not
yet been studied, although it has been reported that thymoma is a
tumor of the thymus, the primate lymphoid organ of T cells, and MG
is an autoimmune disorder closely associated with an imbalance of
Th1 and Th2 (24). The present
study aimed to investigate the expression of Tim-1 in thymoma
patients with and without MG and to examine whether the
single-nucleotide polymorphism (SNP) -1637A/G in the promoter
region of the Tim-1 gene contributes to the susceptibility of
thymoma with MG. The study was approved by the ethics committee of
Tianjin Medical University General Hospital (Tianjin, China).
Materials and methods
Reagents
Mouse anti-human Tim-1 monoclonal antibodies (mAbs),
manufactured by Abcam Corporation, were purchased from Indole
Biological Technology Co., Ltd (Shanghai, China). A streptavidin
peroxidase (SP) test agent box was purchased from Gene Technology
Co., Ltd. (Shanghai, China).
Patients
The thymoma tissues were obtained from patients at
the Tianjin Medical University General Hospital (Heping, China)
from January, 2007 to April, 2013. All the samples were obtained
from individuals from Northern China who were diagnosed with
thymoma by clinical pathological examination. There were 58 cases
of thymoma with MG, including 28 males and 30 females (mean age,
47.3 years), and 62 cases of thymoma without MG, including 38 males
and 24 females (mean age, 52.7 years). Blood samples were collected
from the patients in the two groups. Informed consent was obtained
from all participants.
Immunohistochemical staining
The SP immunohistochemical staining method was used
according to the manufacturer’s instructions. Serial paraffin
sections (five slices, 4 μm thick) were prepared. One slice was
stained with hematoxylin and eosin (H&E) and the others were
used for immunohistochemical analyses. The paraffin pretreatment
involved xylene and alcohol graded hydration. The preparations were
incubated in a 3% H2O2 for 10 min to allow
endogenous peroxidase activity and microwave repair of the antigen
was performed. Following three washes with phosphate-buffered
saline (PBS), the samples were blocked with 5% goat serum (Shanghai
Vita Chemical Reagent Co., Ltd., Shanghai, China) in PBS for 2 h at
room temperature, and then incubated overnight at 4°C after adding
the primary antibodies. The following day, the samples were warmed
for 30 min and washed with PBS thrice for 5 min each. The samples
were then incubated with the secondary goat anti-mouse polyclonal
antibody (Abcam, Shanghai, China) for 30 min at room temperature,
washed with PBS thrice for 5 min each, stained with
diaminobenzidine for 5–10 min, rinsed with distilled water, stained
with H&E, dehydrated and rinsed with xylene for 5 min. The
mounting process was divided into the following two steps: i) a few
drops of acacia were added to orient the samples and ii) the
samples were then fixed and drops of acacia were added before
covering the samples with a slide. The slides were visualized under
a light microscope (XSP148AT; Shanghai Taiyi Medical Apparatus
Equipment Co., Ltd., Shanghai, China). Three randomly selected
fields were obtained from each slide to obtain a mean value
(optical density − mean value of immunostaining intensity).
Determining positive results
In a clear background, cells whose cytoplasm was a
clear brown-yellow color or had brown granules were considered as
positive cells. Cell counting was performed under a light
microscope (magnification, ×400) and expressed in powers of 10. The
ratio of positive to negative cells accounted for the total number
of cells in a slice. The results were divided into three grades as
follows: i) negative (−), no clear positive cells or <10%
positive cells; ii) positive (+), 10–50% positive cells; and iii)
strongly positive (++), >50% positive cells.
SNP analysis
The genomic DNA of leukocytes from peripheral blood
was extracted using sodium dodecyl sulfate lysis and proteinase K
digestion (both Shanghai Vita Chemical Reagent Co., Ltd.), followed
by a standard phenol-chloroform (Shanghai Vita Chemical Reagent
Co., Ltd.) extraction method (25).
Single allele-specific primer polymerase chain reaction was
performed on 1637A/G in the promoter region of the TIM-1 gene.
Primers used for polymerase chain reaction (PCR) to amplify the
-1637A/G SNP in the promoter region of the TIM-1 gene are shown in
Table I (OMIM: 606518) (http://omim.org/entry/606518). Wherein, allele A
fragment was amplified with forward (F) 1 and reverse (R), and
allele G fragment was amplified with F2 and R. The length of
amplified products was 452 bp. PCR was performed with F1 and R and
with F2 and R, respectively, in each sample. Touchdown PCR (Beijing
AuGCT DNA-Syn Biotechnology Co. Ltd., Beijing, China) was performed
in a 25-μl total reaction volume, including 1.0 μl DNA template,
2.5 μl 10× Advantage PCR buffer, 1.5 μl dNTP (2.5 mmol/l), 1.0 μl
F1/F2 (5 μmol/l), 1.0 μl R (5 μmol/L), 0.5 μl DNA polymerase (2.5
U/μl) and 17.5 μl ddH2O. The PCR conditions were as
follows: Denaturing step (95°C for 5 min), 27 cycles of chain
reaction (94°C for 30 sec, annealing temperature was decreased by
1°C from 65°C to 57°C every three cycles, 72°C for 30 sec) and a
final extension (72°C for 10 min). The PCR products were then
detected with 1.2% agarose gel (Shanghai Vita Chemical Reagent Co.,
Ltd.,) electrophoresis. The PCR-amplified products of alleles G and
A were directly sequenced by Beijing AuGCT DNA-Syn Biotechnology
Co. Ltd.
 | Table IPrimers used for amplifying the
-1637A/G SNP in the promoter region of the Tim-1 gene. |
Table I
Primers used for amplifying the
-1637A/G SNP in the promoter region of the Tim-1 gene.
| SNP | Primer sequences |
|---|
| -1637A/G | F1:
5′-CTTCCAGGTTCAAGCAATTCTTCTA-3′
F2: 5′-CTTCCAGGTTCAAGCAATTCTTCTG-3′
R: 5′-AATCGGGCTGTTGACTTCTGCT-3′ |
Statistical analysis
The χ2 test performed using SPSS
software, version 17.0 (SPSS, Inc., Chicago, IL, USA) tested for
deviation from the Hardy-Weinberg equilibrium and compared the
frequency of discrete variables among thymoma patients with and
without MG. P<0.05 was considered to indicate a statistically
significant difference.
Results
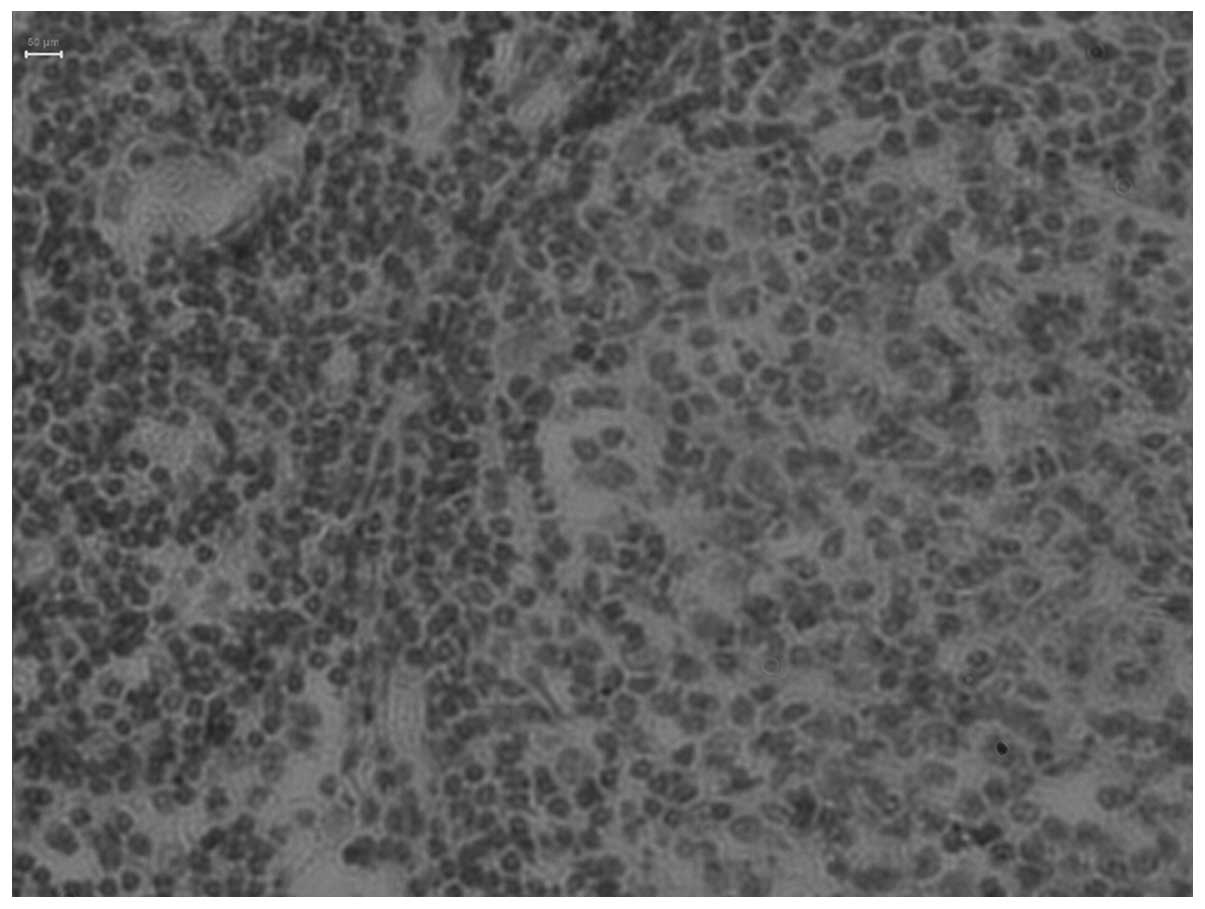
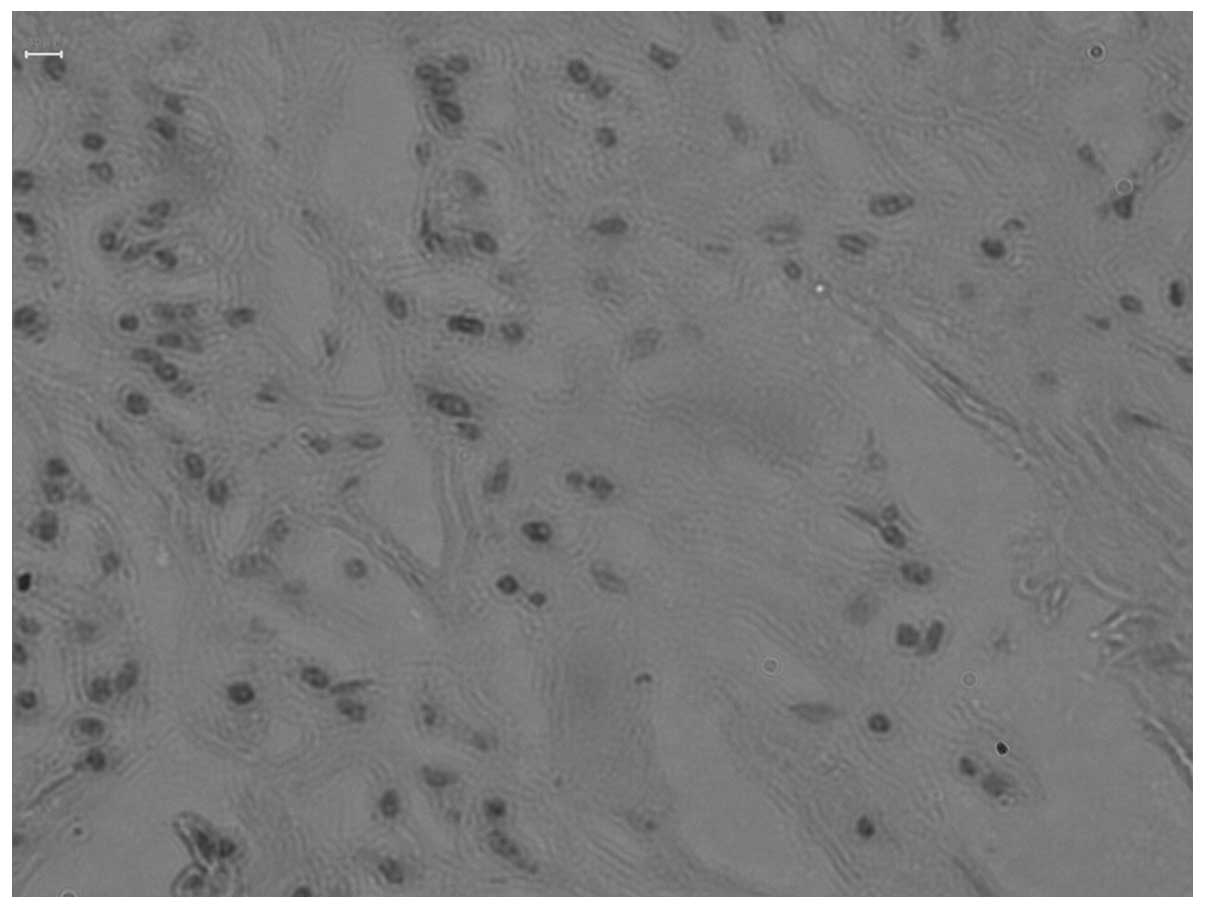
Positive expression and -1637A/G
polymorphism of Tim-1 in thymoma patients with MG
The expression of Tim-1 in thymoma patients with and
without MG is shown in Figs. 1 and
2. Positive Tim-1 expression was
determined by the presence of brown particles in the cytoplasm. The
detailed data of the positive rate of Tim-1 expression are
presented in Table II. The
positive rate of Tim-1 expression in the thymoma with MG was
significantly higher compared with thymoma patients without MG
(P=0.002). The difference between the two groups was statistically
significant (P<0.05).
 | Table IITim-1 expression levels in thymoma
patients with MG and thymoma patients without MG. |
Table II
Tim-1 expression levels in thymoma
patients with MG and thymoma patients without MG.
| Tim-1 expression
level (%) | | |
|---|
|
| | |
|---|
| Thymoma patients | +/++ | − | χ2 | P-value |
|---|
| With MG | 36 | 22 | 9.555 | 0.002 |
| Without MG | 21 | 42 | | |
GG and GA genotypes and the PCR products
of alleles G and A
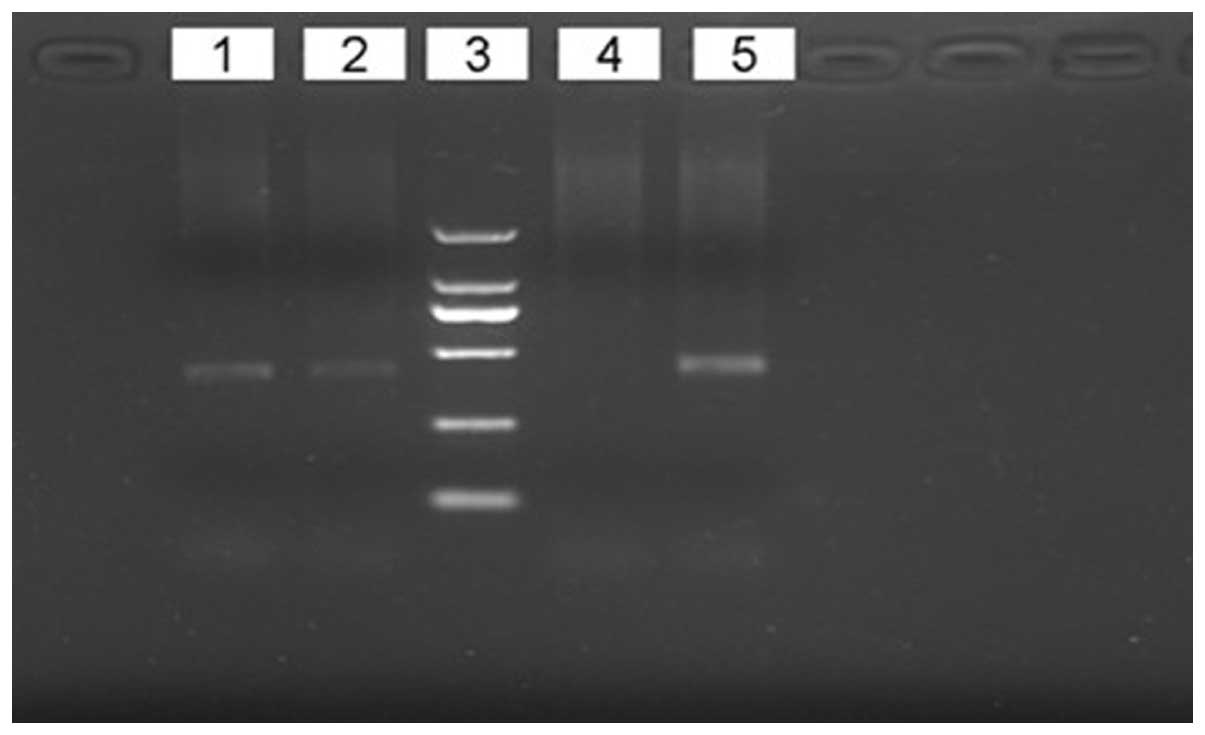
The GG and GA genotypes, without AA, were detected
at the site of -1637A/G of the Tim-1 gene in all the cases. The
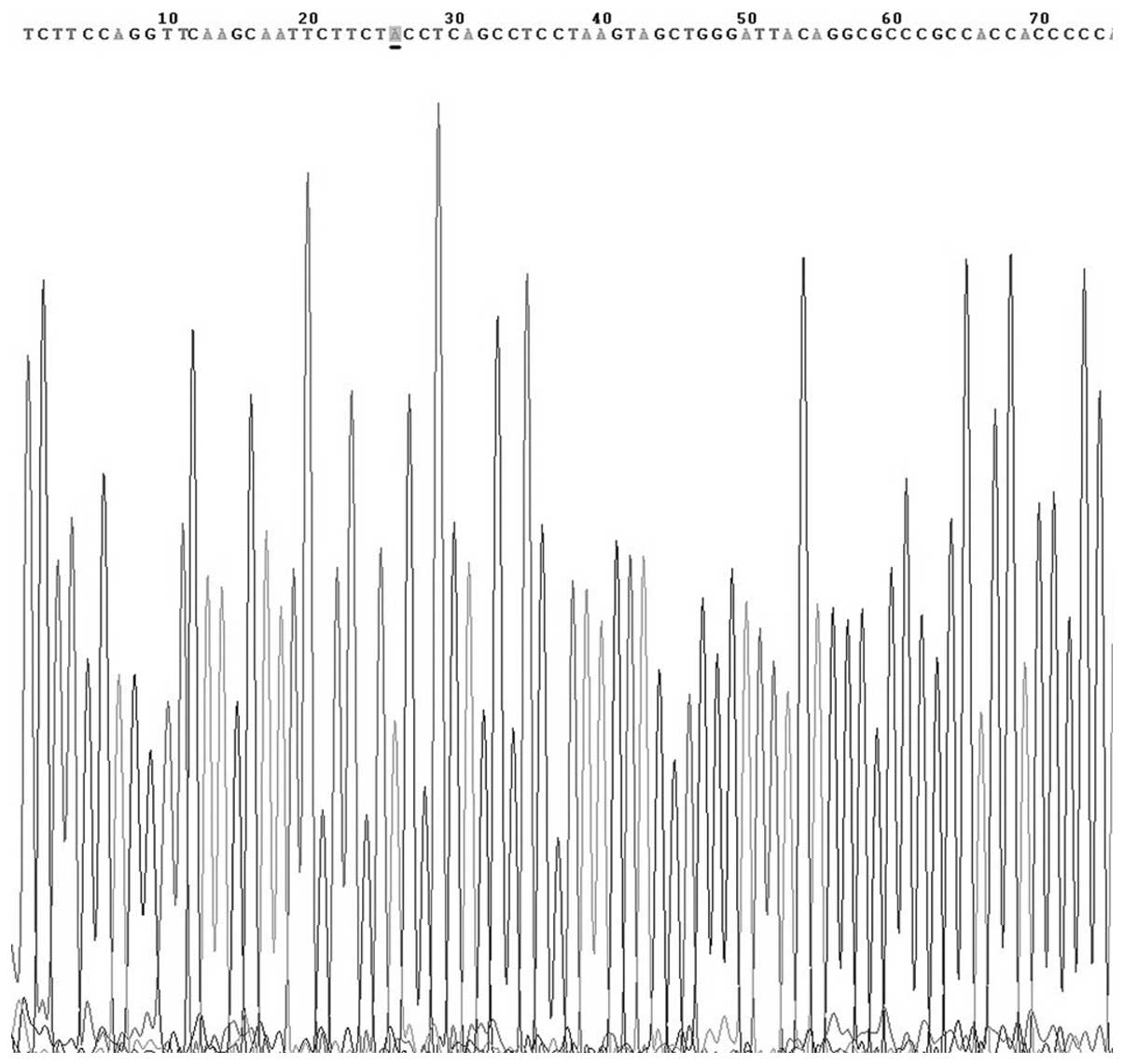
results of genotype analysis are shown in Fig. 3. The PCR-amplified products of
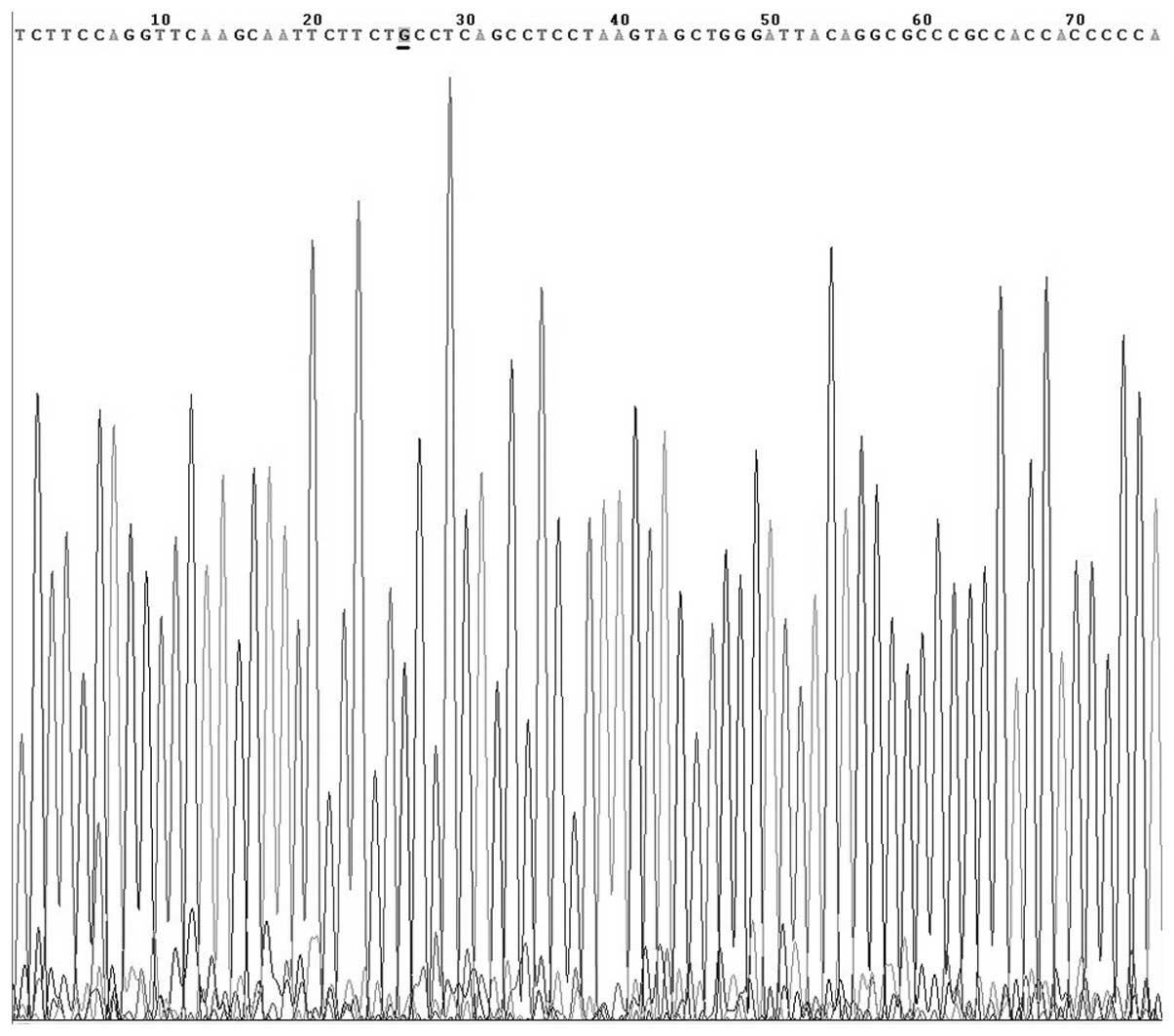
alleles G and A were sequenced and confirmed by Beijing AUGCT
DNA-SYN Biotechnology Co., Ltd. (Beijing, China) (Figs. 4 and 5). In the thymoma with MG group, the G and
A allele frequencies were 84.50 and 15.50%, respectively and 93.55
and 6.45%, respectively, in the thymoma without MG group.
Positive polymorphisms of -1637A/G loci
in Tim-1 with thymoma with MG
The -1637A/G polymorphism in the promoter region of
Tim-1 was analyzed in thymoma patients with and without MG, and in
the normal thymus group. The genotype and allele frequency
distribution of the -1637A/G loci of the Tim-1 promoter region was
confirmed according to the Hardy-Weinberg equilibrium principal,
which suggested that the gene frequency of Tim-1 met the genetic
equilibrium and was fully representative. At the -1637A/G loci,
genotypes GG>GA, but not AA, were observed in all the detected
samples. Additionally, the genotype frequencies at the -1637A/G
polymorphic site were significantly different between thymoma
patients with and without MG (P=0.031). The allele frequencies at
the -1637A/G polymorphic site were significantly different between
thymoma patients with and without MG (P=0.024) (Table III).
 | Table IIIGenotype and allele analyses of the
-1637A/G loci polymorphism of the Tim-1 gene promoter region. |
Table III
Genotype and allele analyses of the
-1637A/G loci polymorphism of the Tim-1 gene promoter region.
| Genotype/allele | Thymoma with MG, n
(%) | Thymoma without MG, n
(%) | P-value |
|---|
| GG | 46 (79.31) | 38 (61.29) | 0.031 |
| GA | 12 (20.69) | 24 (38.71) | - |
| AA | 0 | 0 | - |
| G | 98 (84.50) | 116 (93.55) | 0.024 |
| A | 18 (15.50) | 8 (6.45) | - |
Discussion
The TIM gene family has received significant
attention since it was positionally cloned in 2001 from within the
Tapr locus as a novel allergy and asthma susceptibility gene
(11). Tim-1, the first family
member, was initially identified in 1996 as the receptor for the
hepatitis A virus (HAVCR1) in monkeys (26) and then in humans in 1998 (27). Subsequently, Tim-1 was identified as
a kidney injury molecule (KIM-1) in 1998 (28). Tim-1, akin to all of the TIMs,
posses a similar structure to Type 1 membrane proteins consisting
of an N-terminal Cys-rich immunoglobulin variable-like domain, a
mucin-like domain, a transmembrane domain and an intracellular
tail. The intracellular structure of Tim-1 contains tyrosine
phosphorylation motifs that are involved in transmembrane signaling
(29). The signaling pathways
triggered downstream of Tim-1 cross-linking have been investigated
using either Tim-1 antibodies or Tim-4 as ligands. Reporter assays
have shown that the overexpression of Tim-1 resulted in increased
transcription from the interleukin (IL)-4 promoter and nuclear
factor of activated T-cells/activator protein-1 transcriptional
activation, dependent on Y276 in the Tim-1 cytoplasmic tail
(30). Capping experiments using
human Jurkat T cells that expressed Tim-1 suggest that Tim-1 is
associated with cluster of differentiation (CD) 3 and is recruited
to the T cell receptor (TCR) signaling complex in human T cells
(31). In addition, this study
showed that engagement of Tim-1 with agonistic Tim-1 mAbs resulted
in rapid tyrosine phosphorylation of Tim-1, phosphorylation of
ζ-chain-associated protein kinase 70 and IL-2-inducible T-cell
kinase (ITK), as well as the recruitment of an ITK and
phosphoinositide-3 kinase complex to the TCR signaling complex
(31). Tim-1 is primarily expressed
in activated CD4+ T cells (11), Th2 cells (19), at a low level on mast cells
(32) and a subpopulation of B
cells (33), whereas Tim-3, but not
Tim-1, is expressed in Th1 cells (19). The selectively positive expression
of Tim-1 between Th1/Th2 suggests that Tim-1 may be involved in
diseases of immune deviation or Th1/Th2 imbalance, such as MG. The
expression of Tim-1 in tumors (21,22)
and the potential association between Tim-1 and thymoma, which was
previously confirmed using a Tim-4-Ig fusion protein that showed
the activation of T cells with Tim-4-Ig contributed to the
phosphorylation of Tim-1 and thymoma viral proto-oncogene 1,
indicated that Tim-1 may be involved in thymoma. These data suggest
that Tim-1 may play a vital role in thymoma and MG, and formed the
basis for the present study.
Previous studies have aimed to determine whether
Tim-1 gene polymorphisms were associated with the incidence of
asthma (11), rheumatoid arthritis
(13) and hepatitis A virus
infection (34). However, the
association of Tim-1 with thymoma and MG has not been studied in
the literature to date. To the best of our knowledge, this study is
the first to investigate the expression of Tim-1 in thymoma
patients with and without MG, and to examine whether the -1637A/G
SNP in the promoter region of Tim-1 contributes to the
susceptibility of thymoma with MG. The positive rate of Tim-1
expression in thymoma patients with MG was significantly higher
compared with that of thymoma patients without MG. The genotype
frequencies at the -1637A/G polymorphic site were significantly
different between thymoma patients with and without MG (P=0.031).
In addition, the allele frequencies at the -1637A/G polymorphic
site were significantly different between thymoma patients with and
without MG (P=0.024). These data suggest that Tim-1 may play a role
in the development of thymoma and MG, particularly the development
of thymoma with MG. However, the exact pathogenesis remains
unclear. The effects of Tim-1 polymorphism on transcription and
translation, and whether Tim-1 is involved in thymoma with MG via
the TCR signaling pathway, requires further investigation.
In conclusion, this study demonstrated that the
expression of Tim-1 in thymoma patients with MG is positive and the
-1637A/G polymorphism in the promoter region of the Tim-1 gene is a
potential genetic variant for the susceptibility of thymoma with
MG. Further genetic studies are required to clarify the specific
mechanisms involved.
Acknowledgements
This study was supported by the Department of
Cardiothoracic Surgery of Tianjin Medical University General
Hospital (Heping, China).
References
|
1
|
Falkson CB, Bezjak A, Darling G, et al;
Lung Cancer Disease Site Group of Cancer Care Ontario’s Program in
Evidence-Based Care. The management of thymoma: a systematic review
and practice guideline. J Thorac Oncol. 4:911–919. 2009.
|
|
2
|
Harris K, Elsayegh D, Azab B, Alkaied H
and Chalhoub M: Thymoma calcification: Is it clinically meaningful?
World J Surg Oncol. 9:952011.
|
|
3
|
Okumura M, Fujii Y, Shiono H, Inoue M,
Minami M, Utsumi T, Kadota Y and Sawa Y: Immunological function of
thymoma and pathogenesis of paraneoplastic myasthenia gravis. Gen
Thorac Cardiovasc Surg. 56:143–150. 2008.
|
|
4
|
Okumura M, Inoue M, Kadota Y, Hayashi A,
Tokunaga T, Kusu T, Sawabata N and Shiono H: Biological
implications of thymectomy for myasthenia gravis. Surg Today.
40:102–107. 2010.
|
|
5
|
Müller-Hermelink HK and Marx A:
Pathological aspects of malignant and benign thymic disorders. Ann
Med. 31(Suppl 2): 5–14. 1999.
|
|
6
|
Hong YH, Kwon SB, Kim BJ, et al; Korean
Research Group for Neuromuscular Diseases. Prognosis of ocular
myasthenia in Korea: a retrospective multicenter analysis of 202
patients. J Neurol Sci. 273:10–14. 2008.
|
|
7
|
Romi F: Thymoma in myasthenia gravis: from
diagnosis to treatment. Autoimmune Dis. 2011:4745122011.
|
|
8
|
Vincent A, Palace J and Hilton-Jones D:
Myasthenia gravis. Lancet. 357:2122–2128. 2001.
|
|
9
|
Romi F, Bø L, Skeie GO, Myking A, et al:
Titin and ryanodine receptor epitopes are expressed in cortical
thymoma along with costimulatory molecules. J Neuroimmunol.
128:82–89. 2002.
|
|
10
|
Skeie GO, Bentsen PT, Freiburg A, Aarli JA
and Gilhus NE: Cell-mediated immune response against titin in
myasthenia gravis: evidence for the involvement of Th1 and Th2
cells. Scand J Immunol. 47:76–81. 1998.
|
|
11
|
McIntire JJ, Umetsu SE, Akbari O, Potter
M, Kuchroo VK, Barsh GS, Freeman GJ, Umetsu DT and DeKruyff RH:
Identification of Tapr (an airway hyperreactivity regulatory locus)
and the linked Tim gene family. Nat Immunol. 2:1109–1116. 2001.
|
|
12
|
McIntire JJ, Umetsu DT and DeKruyff RH:
TIM-1, a novel allergy and asthma susceptibility gene. Springer
Semin Immunopathol. 25:335–348. 2004.
|
|
13
|
Xu JR, Yang Y, Liu XM, Sun JY and Wang YJ:
Polymorphisms of the TIM-1 gene are associated with rheumatoid
arthritis in the Chinese Hui minority ethnic population. Genet Mol
Res. 11:61–69. 2012.
|
|
14
|
Vega-Carrascal I, Reeves EP and McElvaney
NG: The role of TIM-containing molecules in airway disease and
their potential as therapeutic targets. J Inflamm Res. 5:77–87.
2012.
|
|
15
|
Abbas AK, Murphy KM and Sher A: Functional
diversity of helper T lymphocytes. Nature. 383:787–793. 1996.
|
|
16
|
Ohtani H, Naruse TK, Iwasaki Y, Akari H,
Ishida T, Matano T and Kimura A: Lineage-specific evolution of
T-cell immunoglobulin and mucin domain 1 gene in the primates.
Immunogenetics. 64:669–678. 2012.
|
|
17
|
Seki M, Oomizu S, Sakata KM, et al:
Galectin-9 suppresses the generation of Th17, promotes the
induction of regulatory T cells, and regulates experimental
autoimmune arthritis. Clin Immunol. 127:78–88. 2008.
|
|
18
|
Wang Y, Meng J, Wang X, Liu S, Shu Q, Gao
L, Ju Y, Zhang L, Sun W and Ma C: Expression of human TIM-1 and
TIM-3 on lymphocytes from systemic lupus erythematosus patients.
Scand J Immunol. 67:63–70. 2008.
|
|
19
|
Khademi M, Illés Z, Gielen AW, et al: T
Cell Ig- and mucin-domain-containing molecule-3 (TIM-3) and TIM-1
molecules are differentially expressed on human Th1 and Th2 cells
and in cerebrospinal fluid-derived mononuclear cells in multiple
sclerosis. J Immunol. 172:7169–7176. 2004.
|
|
20
|
Sánchez-Fueyo A, Tian J, Picarella D, et
al: Tim-3 inhibits T helper type 1-mediated auto- and alloimmune
responses and promotes immunological tolerance. Nat Immunol.
4:1093–1101. 2003.
|
|
21
|
Ngiow SF, von Scheidt B, Akiba H, Yagita
H, Teng MW and Smyth MJ: Anti-TIM3 antibody promotes T cell
IFN-γ-mediated antitumor immunity and suppresses established
tumors. Cancer Res. 71:3540–3551. 2011.
|
|
22
|
Sakuishi K, Jayaraman P, Behar SM,
Anderson AC and Kuchroo VK: Emerging Tim-3 functions in
antimicrobial and tumor immunity. Trends Immunol. 32:345–349.
2011.
|
|
23
|
Rennert PD, Ichimura T, Sizing ID, Bailly
V, Li Z, Rennard R, McCoon P, Pablo L, Miklasz S, Tarilonte L and
Bonventre JV: T cell, Ig domain, mucin domain-2 gene-deficient mice
reveal a novel mechanism for the regulation of Th2 immune responses
and airway inflammation. J Immunol. 177:4311–4321. 2006.
|
|
24
|
Wang Z, Wang W, Chen Y and Wei D: T helper
type 17 cells expand in patients with myasthenia-associated
thymoma. Scand J Immunol. 76:54–61. 2012.
|
|
25
|
Chae SC, Park YR, Song JH, Shim SC, Yoon
KS and Chung HT: The polymorphisms of Tim-1 promoter region are
associated with rheumatoid arthritis in a Korean population.
Immunogenetics. 56:696–701. 2005.
|
|
26
|
Kaplan G, Totsuka A, Thompson P, Akatsuka
T, Moritsugu Y and Feinstone SM: Identification of a surface
glycoprotein on African green monkey kidney cells as a receptor for
hepatitis A virus. EMBO J. 15:4282–4296. 1996.
|
|
27
|
Feigelstock D, Thompson P, Mattoo P, Zhang
Y and Kaplan GG: The human homolog of HAVcr-1 codes for a hepatitis
A virus cellular receptor. J Virol. 72:6621–6628. 1998.
|
|
28
|
Ichimura T, Bonventre JV, Bailly V, Wei H,
Hession CA, Cate RL and Sanicola M: Kidney injury molecule-1
(KIM-1), a putative epithelial cell adhesion molecule containing a
novel immunoglobulin domain, is up-regulated in renal cells after
injury. J Biol Chem. 273:4135–4142. 1998.
|
|
29
|
Rodriguez-Manzanet R, DeKruyff R, Kuchroo
VK and Umetsu DT: The costimulatory role of TIM molecules. Immunol
Rev. 229:259–270. 2009.
|
|
30
|
de Souza AJ, Oriss TB, O’malley KJ, Ray A
and Kane LP: TIM-1 is expressed on in vivo-activated T cells and
provides a co-stimulatory signal for T cell activation. Proc Natl
Acad Sci USA. 102:17113–17118. 2005.
|
|
31
|
Binné LL, Scott ML and Rennert PD: Human
TIM-1 associates with the TCR complex and up-regulates T cell
activation signals. J Immunol. 178:4342–4350. 2007.
|
|
32
|
Nakae S, Iikura M, Suto H, Akiba H, Umetsu
DT, Dekruyff RH, Saito H and Galli SJ: Tim-1 and Tim-3 enhancement
of Th2 cytokine production by mast cells. Blood. 110:2565–2568.
2007.
|
|
33
|
Sizing ID, Bailly V, McCoon P, et al:
Epitope-dependent effect of anti-murine Tim-1 monoclonal antibodies
on T cell activity and lung immune responses. J Immunol.
178:2249–2261. 2007.
|
|
34
|
Kim HY, Eyheramonho MB, Pichavant M, et
al: A polymorphism in TIM1 is associated with susceptibility to
severe hepatitis A virus infection in humans. J Clin Invest.
121:1111–1118. 2011.
|