Introduction
Lung cancer is the most frequently diagnosed cancer
and the leading cause of cancer-related mortality in economically
developed and developing countries (1,2). In
addition, lung cancer was predicted to represent 26% of all female
and 29% of all male cancer-related mortalities in 2012 (3). Non-small cell lung cancer (NSCLC) is
the most common histological type, accounting for ~85% of lung
cancer diagnoses in the USA (4).
The most common forms of NSCLC include adenocarcinoma (AC),
squamous cell carcinoma (SCC) and large cell carcinoma (5). Despite recent advances in the
treatment of lung cancer, the overall five-year survival rate for
such tumors remains poor at <15% (3). Thus, in order to develop rational and
targeted therapies for NSCLC, an improved understanding of the
molecular etiology of these tumors is required.
The human leucine-rich repeats and
immunoglobulin-like domains (LRIG) gene family is comprised of
three members, which are located at chromosome bands 3p14.3
(LRIG1), 1p13 (LRIG2) and 12q13 (LRIG3) (6–8). The
LRIG genes encode integral membrane proteins consisting of a signal
peptide, a leucine-rich repeat domain, a transmembrane domain,
three LRIGs and a cytoplasmic tail (7). Increasing evidence indicates that, in
certain cancer types, LRIG1 is a tumor suppressor (9). For example, LRIG1 expression is
decreased in renal cell carcinoma (10), while high LRIG1 expression is
associated with an improved prognosis in breast cancer (11), early stage invasive squamous
cervical cancer (12) and cutaneous
SCC (13). By contrast, in
oligodendroglioma and uterine cervical carcinoma patients, LRIG2
expression in the cytoplasm has been demonstrated to correlate with
poor survival (14,15). LRIG3 may also have a similar tumor
suppressive function compared with LRIG1 (16). Furthermore, the LRIG proteins may
have different roles depending on their subcellular localization.
LRIG1 has not been associated with improved survival when expressed
in the perinulear region, while the protein expression of LRIG2 and
LRIG3 in the perinuclear area of astrocytoma cells has been found
to correlate with improved patient survival (17). However, the expression profiles of
LRIG2 have not been described in NSCLC. In the present study,
quantitative polymerase chain reaction (qPCR) method and
immunohistochemistry (IHC) were used to detect the mRNA and protein
expression status of LRIG2. In addition, the potential associations
between LRIG2 protein expression in NSCLC and the histological
subtypes, clinical stage, differentiation status and survival were
analyzed.
Materials and methods
Sample collection
The study population included two groups. Firstly,
for the qPCR detection of LRIG2 mRNA expression, 39 NSCLC tissues
and matched paracancerous and normal tissues were collected from
patients with NSCLC at the Department of Thoracic Surgery at the
First Affiliated Hospital of Liaoning Medical University (Jinzhou,
China) between May 2010 and August 2012. Patients provided written
informed consent prior to the specimen collection. None of the
patients had received chemotherapy, radiotherapy or immunotherapy
prior to the surgery. All samples were randomly selected regardless
of age, gender or duration of the disease, and all cases were
diagnosed pathologically. The clinicopathological data were
retrospectively collected by reviewing the patients’ medical
charts. Secondly, 125 formalin-fixed paraffin-embedded tissues of
NSCLC samples were obtained for the immunohistochemical analysis of
LRIG2. Patients enrolled in the study were followed to obtain
five-year survival data. Survival was defined as the time between
the surgery of the primary tumor and mortality or final follow-up
of the patient. Due to loss to follow-up, nine patients were
excluded from the study. All cases were classified according to the
World Health Organization (WHO) revised proposal for histological
types of lung and pleural tumors (18) and the tumor-node-metastasis staging
was performed according to the UICC 1997 criteria. The study was
performed with respect to the ethical standards of the 1975
Declaration of Helsinki, as revised in 2000, and was approved by
the Ethics Committee of Liaoning Medical University (China). The
clinicopathological data are summarized in Table I.
 | Table ICharacterization of the NSCLC patients
included in the study. |
Table I
Characterization of the NSCLC patients
included in the study.
| Patients, n |
|---|
|
|
|---|
| Clinicopathological
features | qPCR analysis
(n=39) | IHC analysis
(n=116) |
|---|
| Gender,
female/male | 14/25 | 45/71 |
| Median age at
diagnosis, years (range) |
| Female | 60.5 (44–71) | 59.5 (42–70) |
| Male | 60.0 (39–79) | 57.5 (38–79) |
| Histological
subtypes |
| Adenocarcinoma | 21 | 86 |
| Squamous cell
carcinoma | 18 | 30 |
| Differentiation
status |
| Well | 14 | 45 |
| Moderate | 12 | 47 |
| Poor | 13 | 24 |
| Tumor staging |
| IA-IB | 15 | 48 |
| IIA-IIB | 16 | 44 |
| IIIA | 8 | 24 |
qPCR
Total RNA was extracted using the TRIzol RNA kit
(Invitrogen Life Technologies, Carlsbad, CA, USA) according to the
manufacturer’s instructions. The first-strand complementary DNA
(cDNA) was prepared from total RNA using a first-strand
PrimeScript™ RT reagent kit with gDNA Eraser (Takara Bio, Inc.,
Shiga, Japan). Next, 1 μg total RNA was used as a template for
reverse transcription (RT). The RT reaction was performed under the
conditions of 37°C for 15 min and 85°C for 5 sec, followed by 42°C
for 2 min with the gDNA Eraser. The PCR was performed using a
primer specific for LRIG2 and the housekeeping gene,
glyceraldehyde-3-phosphate dehydrogenase (GAPDH), sequence. Primers
spanning at least one intron were selected to minimize inaccuracies
due to genomic DNA contamination. The following primer sequences
were used: LRIG2 sense, 5′-TGTGCACACCCTGAATGGCTA-3′ and antisense,
5′-TGT GTCCTTATCTGTGGCTTGAGAA-3′ (PCR product length, 98 bp); and
GAPDH sense, 5′-GCACCGTCAAGGCTGAGAAC-3′ and antisense
5′-TGGTGAAGACGCCAGTGGA-3′ (PCR product length, 138 bp).
The qPCR was run on a Mastercycler® ep
realplex (Eppendorf, Hamburg, Germany) using the SYBR®
Premix Ex Taq™ kit (Takara Bio, Inc.). Each reaction consisted of a
20-μl sample containing 2 μl cDNA, 0.2 μM of each primer and 10 μl
2X SYBR Premix Ex Taq. Each PCR also included a non-template
negative control to check for primer-dimer. The cycling conditions
were one cycle of denaturation at 95°C for 30 sec, followed by 40
three-segment cycles of amplification (95°C for 5 sec, 55°C for 30
sec and 72°C for 30 sec), where the fluorescence was automatically
measured during PCR, and one three-segment cycle of product melting
(95°C for 15 sec, 60°C for 15 sec and 95°C for 15 sec). The
baseline adjustment method of the Mastercycler ep realplex
(Eppendorf) software was used to determine the cycle threshold
(CT) in each reaction. A melting curve was constructed
for each primer pair to verify the presence of one gene-specific
peak and the absence of primer dimer. All samples were amplified in
triplicate and the mean was used for further analysis. The PCR
products were electrophoresed on a 3% agarose gel stained with
ethidium bromide, and calculations were made using the
ΔΔCT method, as previously described (19). GAPDH was used as an internal control
gene in order to normalize the PCR for the amount of RNA added to
the RT reactions.
IHC
IHC was performed on the formalin-fixed paraffin
sections. In brief, 5-μm sections were dewaxed, rehydrated and
incubated in 0.3% (V/V) hydrogen peroxide in 0.01 M
phosphate-buffered saline (pH 7.6) for 20 min to inactivate the
endogenous peroxidase. Antigen retrieval was performed using 0.01 M
sodium citrate buffer (pH 6.0) under high pressure for 2 min. Next,
the sections were immunostained with 2 μg/ml anti-LRIG2 primary
antibody (Abcam, Cambridge, UK) at 4°C overnight and then stained
with a horseradish peroxidase/Fab polymer-conjugated secondary
antibody (Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.,
Beijing, China) for 30 min at room temperature. Finally, the
antibody was revealed by diaminobenzidine at room temperature for 1
min and counterstained with hematoxylin for 15 min. All sections
were examined and scored independently by two investigators who
were blinded to the clinical details, and at least five fields were
randomly selected. The immunostaining was scored as previously
described (15). The expression was
scored as high when ≥50% of the cancer cells were immunopositive
and as low when <50% of the cancer cells were immunopositive or
negative. This cut-off was selected as it showed the best
explanatory power of the various cut-offs tested (0, 20, 50 and
100%). The subcellular localization of the staining was also
evaluated.
Statistical analysis
All statistical analyses were performed using the
SPSS 16.0 statistical software package (SPSS, Inc., Chicago, IL,
USA). A two-sample t-test for independent samples and a
χ2 test were used for continuous and categorical
variables, respectively. The one-way analysis of variance was used
to compare the means of two or more independent groups. The
Kaplan-Meier estimator was used to calculate the survival rate
probability as a function of time and the log-rank test was used to
compare survival time between the groups. All statistical tests
were two-sided and P<0.05 was considered to indicate a
statistically significant difference.
Results
Correlation between LRIG2 mRNA expression
status and clinicopathological variables
Melting curve analysis confirmed the specific
amplification of the target and reference genes. Furthermore, the
gel electrophoresis analysis of the amplification products revealed
a single band with the predicted size for LRIG2 (98 bp) and GAPDH
(138 bp) (Fig. 1). The slopes of
the standard curves were −3.242 and −3.238 for the GAPDH and LRIG2
genes, respectively. The reliability of the PCR reaction
efficiencies was also assessed by plotting ΔCT values
(CT LRIG2 - CT GAPDH), and the absolute value
of the trend line slopes was ≤0.1, which indicated the validity of
the relative quantitative assay by ΔΔCT method.
 | Figure 1qPCR assay. (A) Gel electrophoresis
analysis of the target and reference gene PCR products. Lanes 1 and
6, 500-bp molecular marker; 2, LRIG2; 3, NTC for LRIG2; 4, GAPDH;
and 5, NTC for GAPDH. (B) Melting curve analysis of the target and
reference gene PCR products. The SYBR Green qPCR reactions of the
(a) LRIG2 and (b) GAPDH genes were performed using a normal sample
of the complementary DNA (cDNA). Each peak corresponds to a unique
PCR product. (c) NTC reactions showed no PCR product. LRIG2,
leucine-rich repeats and immunoglobulin-like domains 2; NTC,
non-template negative control; qPCR, quantitative polymerase chain
reaction; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; −dI/dT,
negative first derivative of the melting curves as a function of
temperature. |
The LRIG2 mRNA expression was examined in 39 pairs
of NSCLC and adjacent cancerous tissues using the ΔΔCT
method. The results showed that the mRNA expression of LRIG2 was
decreased in the cancer and adjacent cancerous tissues. The mean
mRNA expression level of LRIG2 in the 39 NSCLC cancer and adjacent
cancerous tissues was 0.2288±0.0230 and 0.6185±0.0321,
respectively. The correlation between LRIG2 mRNA expression in the
cancer tissues and the various clinicopathological parameters were
further analyzed and are shown in Table II. The expression of LRIG2 mRNA was
significantly higher in AC compared with that in SCC (P=0.005).
According to the tumor differentiation status, a significant
downregulation of LRIG2 (P=0.013) was also observed, while no
significant correlation was observed between LRIG2 expression and
tumor staging (P=0.822). In addition, no correlation was observed
between LRIG2 mRNA expression and age, gender and smoking habits
(data not shown). The correlation between LRIG2 mRNA expression in
adjacent cancerous tissues and the various clinicopathological
parameters were also analyzed. However, no correlations were
identified between LRIG2 mRNA expression and the
clinicopathological parameters.
 | Table IICorrelation between LRIG2 mRNA
expression status and the clinicopathological features of
patients. |
Table II
Correlation between LRIG2 mRNA
expression status and the clinicopathological features of
patients.
| | LRIG2 expression,
mean ± SEM |
|---|
| |
|
|---|
| Variables | Patients, n | Cancer tissues | P-value | Adjacent to cancer
tissues | P-value |
|---|
| Histological
subtypes |
| Adenocarcinoma | 21 | 0.2869±0.0318 |
0.005a | 0.6744±0.0396 |
0.059a |
| Squamous cell
carcinoma | 18 | 0.1609±0.0258 | | 0.5533±0.0489 | |
| Differentiation
status |
| Well | 14 | 0.3140±0.0424 |
0.013b | 0.6695±0.0478 |
0.488b |
| Moderate | 12 | 0.2012±0.0302 | | 0.6011±0.0534 | |
| Poor | 13 | 0.1625±0.0327 | | 0.5795±0.0659 | |
| Tumor staging |
| IA-IB | 15 | 0.2430±0.0398 |
0.822b | 0.5808±0.0595 |
0.228b |
| IIA-IIB | 16 | 0.2113±0.0351 | | 0.6845±0.0424 | |
| IIIA | 8 | 0.2370±0.0507 | | 0.5573±0.0651 | |
Correlation between LRIG2 protein
expression and the clinicopathological features of NSCLC
By IHC analysis, specific LRIG2 immunoreactivity was
generally only observed in the cytoplasm (Fig. 2). According to the LRIG2
immunoreactive intensity, in the total 116 cases of NSCLC, 80
patients (68.97%) were classified into the low-LRIG2 group, and 36
(31.03%) were classified into the high-LRIG2 group. A statistically
significant correlation was found between LRIG2 expression and the
two major histological subtypes (AD and SCC; P=0.048), which also
conformed to the results of the qPCR. The correlation between LRIG2
expression and the various clinicopathological parameters of 116
cases of NSCLC was also analyzed. Briefly, the LRIG2 staining level
was found to significantly correlate with differentiation status
(P=0.034), as shown in Table III.
No significant correlation was identified between LRIG2 expression
and patient gender, age, smoking history and tumor staging (all
P>0.05).
 | Table IIICorrelation between LRIG2 protein
expression status and the clinicopathological features of
patients. |
Table III
Correlation between LRIG2 protein
expression status and the clinicopathological features of
patients.
| | LRIG2 expression,
n | |
|---|
| |
| |
|---|
| Clinicopathological
features | Patients, n | High score | Low score | P-value |
|---|
| Histological
subtypes |
| Adenocarcinoma | 86 | 31 | 55 | 0.048 |
| Squamous cell
carcinoma | 30 | 5 | 25 | |
| Differentiation
status |
| Well | 45 | 20 | 25 | 0.034 |
| Moderate | 47 | 12 | 35 | |
| Poor | 24 | 4 | 20 | |
| Tumor staging |
| IA-IB | 48 | 15 | 33 | 0.948 |
| IIA-IIB | 44 | 13 | 31 | |
| IIIA | 24 | 8 | 16 | |
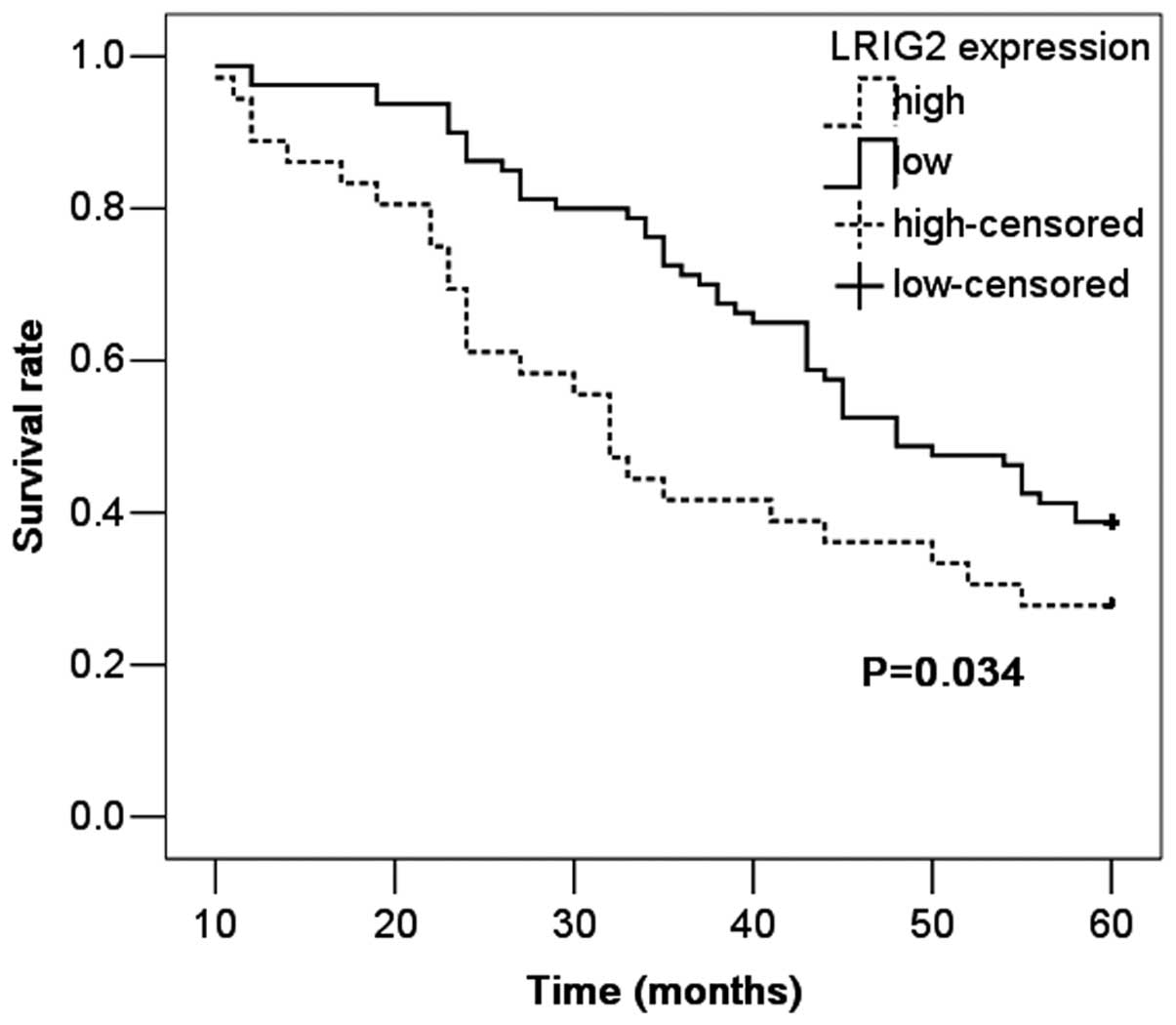
Correlation between LRIG2 protein
expression and overall survival
The prognostic value of LRIG2 protein expression for
overall survival in NSCLC patients was evaluated by comparing the
patients with high and low LRIG2 expression. According to the
Kaplan-Meier survival analysis, the patients with high LRIG2
expression exhibited evidently lower overall survival rates than
those with low LRIG2 expression (P=0.034; Fig. 3). The five-year survival rate for
patients with high LRIG2 expression was 27.8%, compared with 38.8%
for patients with low expression. Multivariate analysis was
conducted using the Cox proportional hazards model to examine the
impact of LRIG2 expression and other clinicopathological
parameters, including tumor differentiation status and tumor stage.
The expression of LRIG2 emerged as an independent and significant
factor associated with poor five-year survival rates (P=0.019).
Thus, LRIG2 expression levels may have a prognostic value in NSCLC
patients.
Discussion
LRIG2 is an integral membrane protein that is widely
expressed in human tissues (7,8).
Although LRIG2 has been proven to be of prognostic value in several
types of human cancers (16), the
expression status in NSCLC remains unknown. The current study
provides the first characterization of the LRIG2 expression status
in human NSCLC. It was found that the expression of LRIG2 was
decreased in the cancer tissues, indicating a potential role of the
LRIG2 protein in the pathogenesis of NSCLC. In addition, the
cytoplasmic expression level of LRIG2 was associated with poor
prognosis, suggesting that LRIG2 may have prognostic value in NSCLC
patients. These conclusions were consistent with the results
obtained in other types of human cancer (14,15).
LRIG2 expression has been investigated in several
types of cancer. Guo et al (17) found that perinuclear staining of
LRIG2 was associated with a low WHO grade in astrocytic tumors, and
compared with the normal pituitary samples, the expression of LRIG2
was lower in the human pituitary adenoma HP75 cell line. Wang et
al (20) also described the
consequences of selectively knocking down LRIG2 expression in the
glioma GL-15 cell line. The study found that the downregulation of
LRIG2 expression decreased the proliferation rate, which resulted
in G0/G1 arrest and the increased spontaneous
apoptosis, cell adhesion and invasion capability of the glioma cell
line. In addition, in cells lacking LRIG2, the activation of ErbB1
(epidermal growth factor receptor) was reduced through increased
ErbB1 degradation and decreased ErbB phosphorylation (20). LRIG2 expression was also found in
the precancerous cervical epithelium and shown to increase with
increasing cervical intraepithelial neoplasia grade (21). An association was also found between
the expression of LRIG2 and specific tumor markers. Similarly,
LRIG2 expression was found to correlate with increased FHIT and
p164INKa, as well as IL-10 expression, while a negative
correlation was observed with Rb and Ki-67 expression (21). In meningiomas, the LRIG2 expression
in the cytoplasm of has been found to correlate with estrogen
receptor (ER) status and histological subtype, with the benign
subtypes most frequently expressing LRIG2 (22). Recently, emerging evidence has
indicated that the hormones, estrogen and progesterone, are key in
the progression of NSCLC (23).
LRIG2 is a glycoprotein with N-linked oligosaccharides, and a
recent study found that LRIG2 has a physical association with FBXO6
(also known as FBX6) (24), which
is involved in the endoplasmic reticulum-associated degradation
pathway by mediating the ubiquitination of glycoproteins. The F box
protein, Fbx6, also regulates Chk1 (a key protein kinase in the
replication checkpoint) stability and cellular sensitivity to
replication stress (25). In the
present study, the mRNA expression of LRIG2 was decreased in NSCLC
cancer tissues and found to correlate with histological subtypes
and tumor differentiation status. The protein expression of LRIG2
also conformed to the mRNA expression results. This indicates a
potential role for LRIG2, which may interact with ER and FBX6 in
the pathogenesis of NSCLC. However, further study is required to
confirm this hypothesis.
The LRIG2 protein localizes to different subcellular
compartments, including the nucleus, perinuclear area, cytoplasm
and cell surface (9). The
subcellular localization of LRIG2 also appears clinically
important. Hedman et al (15) found that high LRIG2 expression
correlates with poor survival in invasive early-stage squamous
cervical cancer. In addition, Holmlund et al (14) reported that the expression of
cytoplasmic LRIG2 is a negative prognostic factor for
oligodendroglioma. As for pituitary adenoma, LRIG2 expression has
been found to predict the invasiveness of pituitary tumors and a
poor prognosis (26). In esophageal
carcinoma, a trend towards decreased survival was found for the
high expression of LRIG2, however, this trend was not statistically
significant (27). By contrast, the
protein expression of LRIG2 in the perinuclear area of astrocytoma
cells has been found to correlate with improved patient survival
(17). In the present study, only
the cytoplasmic expression of LRIG2 was observed and the patients
with high LRIG2 expression were associated with poor survival,
which is consistent with the results of several other studies
(14,15). Certain studies (28,29)
have indicated that the LRIG2 protein exhibits different roles in
human tumors depending on their subcellular localization. LRIG2
localization in the cytoplasm may augment its action as a tumor
promoter, whereas the perinuclear localization of LRIG2 may act as
a tumor suppressor. The significance of the specific subcellular
localization of LRIG2 protein requires further investigation, as
well as the mechanism of its different functions.
In conclusion, the present study showed that LRIG2
expression is decreased in NSCLC tissues, which indicates a
potential role in the pathogenesis of NSCLC. In addition, the
cytoplasmic expression of LRIG2 was found to be an independent
prognostic factor associated with poor survival in NSCLC. This may
have prognostic value in NSCLC patients. However, to fully
elucidate the exact function of LRIG2 in NSCLC, further and larger
studies are required.
Acknowledgements
This study was financially supported by the Youth
Science and Technology Foundation of Liaoning Medical University
(grant no. Y2012Z012).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011.
|
|
2
|
Saintigny P and Burger JA: Recent advances
in non-small cell lung cancer biology and clinical management.
Discov Med. 13:287–297. 2012.
|
|
3
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012.
|
|
4
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594.
2008.
|
|
5
|
Collins LG, Haines C, Perkel R and Enck
RE: Lung cancer: diagnosis and management. Am Fam Physician.
75:56–63. 2007.
|
|
6
|
Nilsson J, Vallbo C, Guo D, et al:
Cloning, characterization, and expression of human LIG1. Biochem
Biophys Res Commun. 284:1155–1161. 2001.
|
|
7
|
Holmlund C, Nilsson J, Guo D, et al:
Characterization and tissue-specific expression of human LRIG2.
Gene. 332:35–43. 2004.
|
|
8
|
Guo D, Holmlund C, Henriksson R and Hedman
H: The LRIG gene family has three vertebrate paralogs widely
expressed in human and mouse tissues and a homolog in Ascidiacea.
Genomics. 84:157–165. 2004.
|
|
9
|
Hedman H and Henriksson R: LRIG inhibitors
of growth factor signalling - double-edged swords in human cancer?
Eur J Cancer. 43:676–682. 2007.
|
|
10
|
Thomasson M, Hedman H, Guo D, Ljungberg B
and Henriksson R: LRIG1 and epidermal growth factor receptor in
renal cell carcinoma: A quantitative RT-PCR and immunohistochemical
analysis. Br J Cancer. 89:1285–1289. 2003.
|
|
11
|
Krig SR, Frietze S, Simion C, et al: Lrig1
is an estrogen-regulated growth suppressor and correlates with
longer relapse-free survival in ERalpha-positive breast cancer. Mol
Cancer Res. 9:1406–1417. 2011.
|
|
12
|
Lindstrom AK, Ekman K, Stendahl U, et al:
LRIG1 and squamous epithelial uterine cervical cancer: correlation
to prognosis, other tumor markers, sex steroid hormones, and
smoking. Int J Gynecol Cancer. 18:312–317. 2008.
|
|
13
|
Tanemura A, Nagasawa T, Inui S and Itami
S: LRIG-1 provides a novel prognostic predictor in squamous cell
carcinoma of the skin: immunohistochemical analysis for 38 cases.
Dermatol Surg. 31:423–430. 2005.
|
|
14
|
Holmlund C, Haapasalo H, Yi W, et al:
Cytoplasmic LRIG2 expression is associated with poor
oligodendroglioma patient survival. Neuropathology. 29:242–247.
2009.
|
|
15
|
Hedman H, Lindstrom AK, Tot T, Stendahl U,
Henriksson R and Hellberg D: LRIG2 in contrast to LRIG1 predicts
poor survival in early-stage squamous cell carcinoma of the uterine
cervix. Acta Oncol. 49:812–815. 2010.
|
|
16
|
Muller S, Lindquist D, Kanter L, et al:
Expression of LRIG1 and LRIG3 correlates with human papillomavirus
status and patient survival in cervical adenocarcinoma. Int J
Oncol. 42:247–252. 2013.
|
|
17
|
Guo D, Nilsson J, Haapasalo H, et al:
Perinuclear leucine-rich repeats and immunoglobulin-like domain
proteins (LRIG1–3) as prognostic indicators in astrocytic tumors.
Acta Neuropathol. 111:238–246. 2006.
|
|
18
|
Shimosato Y: Histological typing of lung
and pleural tumors (3rd edition, 1999): Malignant epithelial
tumors. Nihon Rinsho. 60:123–131. 2002.(In Japanese).
|
|
19
|
Kontos CK, Papadopoulos IN, Fragoulis EG
and Scorilas A: Quantitative expression analysis and prognostic
significance of L-DOPA decarboxylase in colorectal adenocarcinoma.
Br J Cancer. 102:1384–1390. 2010.
|
|
20
|
Wang B, Han L, Chen R, et al:
Downregulation of LRIG2 expression by RNA interference inhibits
glioblastoma cell (GL15) growth, causes cell cycle redistribution,
increases cell apoptosis and enhances cell adhesion and invasion
in vitro. Cancer Biol Ther. 8:1018–1023. 2009.
|
|
21
|
Lindstrom AK, Asplund A and Hellberg D:
Correlation between LRIG1 and LRIG2 expressions and expression of
11 tumor markers, with special reference to tumor suppressors, in
CIN and normal cervical epithelium. Gynecol Oncol. 122:372–376.
2011.
|
|
22
|
Ghasimi S, Haapasalo H, Eray M, et al:
Immunohistochemical analysis of LRIG proteins in meningiomas:
correlation between estrogen receptor status and LRIG expression. J
Neurooncol. 108:435–441. 2012.
|
|
23
|
Kazmi N, Marquez-Garban DC, Aivazyan L, et
al: The role of estrogen, progesterone and aromatase in human
non-small-cell lung cancer. Lung Cancer Manag. 1:259–272. 2012.
|
|
24
|
Liu B, Zheng Y, Wang TD, et al: Proteomic
identification of common SCF ubiquitin ligase FBXO6-interacting
glycoproteins in three kinds of cells. J Proteome Res.
11:1773–1781. 2012.
|
|
25
|
Zhang YW, Brognard J, Coughlin C, et al:
The F box protein Fbx6 regulates Chk1 stability and cellular
sensitivity to replication stress. Mol Cell. 35:442–453. 2009.
|
|
26
|
Zhang H, Yan Q, Xu S, et al: Association
of expression of Leucine-rich repeats and immunoglobulin-like
domains 2 gene with invasiveness of pituitary adenoma. J Huazhong
Univ Sci Technolog Med Sci. 31:520–523. 2011.
|
|
27
|
Wu X, Hedman H, Bergqvist M, et al:
Expression of EGFR and LRIG proteins in oesophageal carcinoma with
emphasis on patient survival and cellular chemosensitivity. Acta
Oncol. 51:69–76. 2012.
|
|
28
|
Musacchio M and Perrimon N: The Drosophila
kekkon genes: novel members of both the leucine-rich repeat and
immunoglobulin superfamilies expressed in the CNS. Dev Biol.
178:63–76. 1996.
|
|
29
|
Eisenstat DD and Gibson SB: RIGging
functional outcomes in glioma cells: new insights into LRIG
proteins in malignant gliomas. Cancer Biol Ther. 8:1024–1026.
2009.
|