Introduction
Breast cancer is the leading cause of
cancer-associated mortality among females worldwide. Due to the
progress achieved in the treatment of this type of cancer, patient
mortality is now increasingly associated with the occurrence of
distant metastases (1). Certain
organs are favored sites for circulating cancer cells to develop
metastases, which occur as a result of a permissive
microenvironment in the target tissue that facilitates tumor growth
(2). Breast cancer is characterized
by a distinct metastatic pattern involving the regional lymph
nodes, bone marrow, lung and liver.
Tumor cell migration and metastasis is a complex
process, which is regulated by chemokines and their receptors
(3). Chemokines are a superfamily
of small, low molecular weight proteins that induce cytoskeletal
rearrangement, firm adhesion to endothelial cells and directional
migration, via their interaction with G-protein-coupled serpentine
receptors (4). The CXC chemokine
receptor 4 (CXCR4), a member of the G-protein-coupled receptor
superfamily, and is the only physiological receptor of high
specificity for stromal cell derived factor-1 (SDF-1). It is
closely involved and important in various physiological and
pathological processes in vivo, including immune defense and
anti-inflammation. Previous studies (5–8) have
indicated that CXCR4 is expressed in a number of tumor cells, and
its specific binding with SDF-1 in certain tissues is key for tumor
genesis, progression and metastasis.
In the present study, a short-hairpin RNA (shRNA)
eukaryotic expression vector targeting CXCR4 was constructed and
transfected into 293T cells in vitro. The most efficacious
interfering vector was selected and transfected into the highly
invasive breast cancer MDA-MB-231 cell line, and the effect of
silencing the CXCR4 gene on the proliferation, adhesion and
migration of 231 cells was observed in vitro. The results
may provide a basis for cancer therapy which targets CXCR4.
Materials and methods
Reagents
Human renal embryonic 293T cells were obtained from
Shanghai Tongji University (Shanghai, China). The breast cancer
MDA-MB-231 cell line was purchased from the Chinese Academy of
Sciences (Shanghai, China). PGCsi-U6-Neo-green fluorescent protein
(GFP) vacant plasmids were obtained from Shanghai Jiaotong
University (Shanghai, China) and DH5α competent cells were
purchased from Shanghai Hi-tech Bioengineering Co. Ltd. (Shanghai,
China). Fetal bovine serum (FBS) and high-glucose Dulbecco’s
modified Eagle’s medium were purchased from Gibco-BRL (Carlsbad,
CA, USA). RPMI-1640 medium was purchased from Nanjing KeyGen
BioTech Co. Ltd. (Nanjing, China). The incision enzymes,
BamHI, HindIII, NheI and T4DNA ligase were all
purchased from Fementas Life Sciences (Rockford, IL, USA).
Lipofectamine 2000 was purchased from Invitrogen Life Technologies
(Carlsbad, CA, USA). RNA TRIzol reagent and rabbit polyclonal
anti-human CXCR4 multi-clone antibody (1:250) were purchased from
Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). A reverse
transcription reagent kit was purchased from Takara Bio, Inc.
(Shiga, Japan). Cell Counting Kit-8 (CCK-8) was purchased from
Beyotime Biotech. (Jiangsu, Japan) and Matrigel was purchased from
BD Biosciences (Franklin Lakes, NJ, USA).
Construction of plasmid vectors
According to the restriction endonuclease digestion
site of the pGCsi-U6-Neo-GFP vacant plasmid, two interfering
sequences of CXCR4 shRNA and a negative control sequence (Table I) were selected for the construction
of DNA and primers, which were produced by Shanghai Hi-tech
Bioengineering Co. Ltd. Reverse transcription-polymerase chain
reaction (RT-PCR) and western blot analysis were used to screen the
pairs to identify those exhibiting the most efficacious
interference.
 | Table ISequences of interference RNA used in
the study. |
Table I
Sequences of interference RNA used in
the study.
| Gene | Target sequence | ShRNA sequences |
|---|
| CXCR4-1 | TCCTGGCCTTCATCAGTCT
(6) | 5′-GAT CCT CCT GGC
CTT CAT CAG TCT TTC AAG AGA AGA CTG ATG AAG GCC AGG ATT TTT GGA AGC
TAG GA-3′ |
| CXCR4-2 | TGCCCACCATCTACTCCAT
(7) | 5′-GAT CCT GCC CAC
CAT CTA CTC CAT TTC AAG AGA ATG GAG TAG ATG GTG GGC ATT TTT GGA AGC
TAG CA-3′ |
| Negative control | AATCGCATAGCGTATGCCGTT
(8) | 5′-GAT CCA ATC GCA
TAG CGT ATG CCG TTT TCA AGA GAA ACG GCA TAC GCT ATG CGA TTT TTT TGG
AAG CTA GCA-3′ |
Recombinant plasmids
Sense and anti-sense primers (2 μl, respectively)
were mixed with 2 μl annealing buffer and 4 μl double-distilled
water to produce a combined double strand, which was annealed from
95 to 25°C at a velocity of 0.5°C/sec for 2 min. The restriction
endonucleases, BamHI and HindIII, were used to digest
PGCsi-U6-Neo-GFP vacant plasmids at 37°C in a water bath for 3 h.
Agarose gel electrophoresis was used to analyze and retrieve the
linear products. A total of 1 μl diluted annealing primer was then
conjugated with the linear vector in the ratio of 4:1 at 22°C for 1
h, then transfected to the competent bacteria DH5α. Next, the
bacteria solution was added to the LB agar plate with ampicillin
(Hubei Shengtian Hengchuang Biotechnology Co., Ltd., Shanghai,
China) at 37°C overnight. The selected monoclonal bacterial colony
was then amplified for plasmid extraction and NheI
endonuclease digestion. The products were analyzed by agarose gel
electrophoresis and DNA sequencing was performed by Shanghai
Hi-tech Bioengineering Co. Ltd to identify successful recombinants.
The amplified recombinant vectors were maintained at −20°C.
Selection of shRNA eukaryotic expression
vectors targeting CXCR4
293T cells at the logarithmic phase were seeded in
six-well culture plates for 24 h and transfected with recombinant
plasmids by Lipofectamine 2000, when the cells reached ~70%
confluence. GFP expression was observed under a fluorescence
microscope (DM3000; Leica, Mannheim, Germany) at 24, 48 and 72 h
following transfection, to calculate the transfection efficiency of
the plasmids.
Expression of CXCR4 mRNA in 293T cells by
RT-PCR
A total of 2 μl mRNA was extracted from 293T cells
using TRIzol reagent 48 h following transfection. The mRNA was then
reversely transcribed to cDNA using the reverse transcription kit
(Takara bio, Inc.) at 37°C for 15 min, and amplified by PCR for 5
min. The primer sequences were as follows: Forward, 5′-GAC AGG ATG
CAG AAG GAG ATT ACT-3′ and reverse, 5′-TGA TCC ACA TCT GCT GGA AGG
T-3′ for β-actin, yielding a 318-bp amplified fragment; forward,
5′-GGA GGC TGG CAA CAT AAC-3′ and reverse, 5′-TGG CAG GGA ACG TCT
AAT-3′ for CXCR4, yielding a 227-bp amplified fragment. The
reaction procedure was as follows: 94°C for 3 min, 94°C for 30 sec,
59°C for 30 sec, 72°C for 1 min and 72°C for 5 min, for 30 cycles.
Following electrophoresis, the images were scanned and analyzed by
Gel-Pro Analyzer (Media Cybernetics, Silver Spring, MD, USA) to
determine the integrated optical density (OD) and analyzed using
gray values to calculate interference efficiency following 1%
agarose gel electrophoresis.
Western blot analysis of CXCR4 protein
expression in 293T cells
Total protein of 293T cells was extracted by
radioimmunoprecipitation assay buffer and phenylmethanesulfonyl
fluoride 72 h following transfection, and the concentration of
CXCR4 protein was detected using the bicinchoninic acid assay
method. A total of 50 μg extracted protein solution was mixed with
sample buffer for denaturation at 99°C for 10 min to perform sodium
dodecyl sulfate-polyacrylamide gel electrophoresis, whereby the
protein was transferred from gelatin to a polyvinylidene fluoride
membrane, and blocked with non-fat milk for 2 h. Primary antibody
(CXCR4, 1:200; β-actin, 1:500) was added at 4°C and left overnight.
The secondary antibody (CXCR4, 1:10,000, β-actin, 1:10,000) was
added the next day at room temperature for 2 h incubation and
visualized using the EZ-ECL chemiluminescence detection kit for HRP
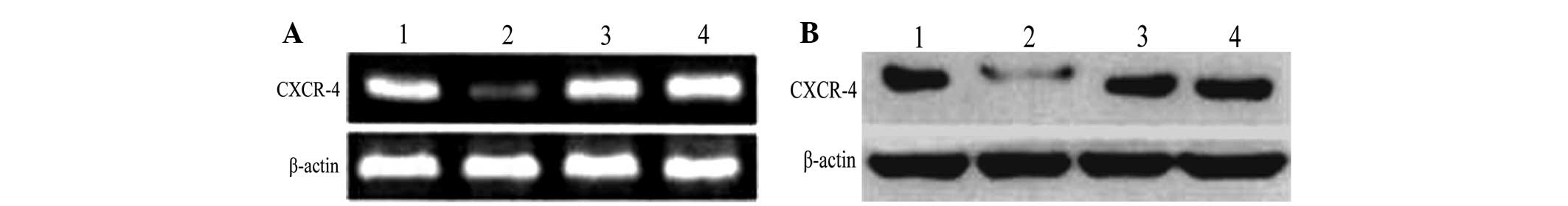
(Bioind, Kibbutz Beit Haemek, Israel). The following groups were
used: 1, pGCsi-CXCR4-1/shRNA; 2, pGCsi-CXCR4-2/shRNA; 3, negative
control (blank plasmid); and 4, untreated group (untransfected
plasmid).
Transfection of MDA-MB-231 cells with
shRNA interfering vector
The most efficacious interfering vector was selected
to transfect MDA-MB-231 cells for two days according to the results
of RT-PCR and western blot analysis. Medium supplemented with G418
(500 mg/l; Gibco-BRL) was added for cell screening. After 10–14
days, monoclonal resistant cells were obtained using limiting
dilution assay and maintained in medium supplemented with G418 (200
mg/l) for amplification.
Assessment of proliferation of MDA-MB-231
cells with silenced CXCR4 by CCK-8 assay
MDA-MB-231 cells were seeded in 96-well culture
plates at a concentration of 1×105 cells/ml. Following
24 h incubation, 100 μl medium was removed from each well and 10 μl
CCK-8 was added and incubated for 1 h at 37°C. Absorbance was
measured using an ultraviolet spectrophotometer (UV-5100; Shanghai
Yuanxi Instrument Co., Ltd., Shanghai, China) at a wavelength of
450 nm for five days. The results were calculated as the mean
values of five wells for each group, and the assay was performed in
triplicate.
Measurement of MDA-MB-231 cell adhesion
by cell-matrix adhesion assays
Fibronectin (FN; Phoenix Pharmaceuticals Inc.,
Burlingame, CA, USA) was used to simulate an extracellular matrix
environment and bovine serum albumin (BSA), simulating basement
membrane, was used as the control. A total of 20 mg/l FN and 10 g/l
BSA (Shenzhen Niubang Bio-technology Company, Shenzhen, China)
coated the 96-well culture plates (50 μl/well), which were air
dried on a sterilized bench and stored at 4°C until use. The cells
were hydrated using phosphate-buffered saline, and 50 μl serum-free
BSA medium (10 g/l) was added to each well at 37°C for 30 min. The
cells were then collected at the logarithmic phase and RPMI-1640
medium supplemented with 0.1% BSA was added, adjusting the
concentration to 1×105 cells/ml. Next, 100 μl of the
previous cell suspension was added to each well and incubated at
37°C. A total of 90 μl serum-free medium and 10 μl CCK-8 reagent
was then added and cells were incubated at 37°C for 1 h, following
discarding former medium, at 30, 60 and 90 min, respectively. The
cells were then washed with serum-free medium three times to remove
floating cells. OD was measured at a wavelength of 450 nm. The cell
adhesion of each group was calculated according to the OD value of
the BSA group, using the following formula: Cell adhesion (%) = OD
value of cells in experimental group/OD value of BSA group × 100.
The results were calculated as the mean values of six wells per
group and the experiment was performed in triplicate.
Measurement of MDA-MB-231 cell migration
by wound-healing assays
MDA-MB-231 cells from each group were seeded in
six-well culture plates at a concentration of 2×106
cells/ml. A scratch in the middle of the wells was established by a
cell knife when cells reached 80% confluence, and the corresponding
positions relative to the wound zone were observed under a
microscope (Olympus BH2-MJLT; Olympus Corporation, Shanghai,
China). Cells were washed three times with serum-free medium and
antibiotic-free RPMI-1640 medium supplemented with 10% FBS, and
then cultured for 24 h. Images were captured using an inverted
microscope [XploRA INV; HORIBA (China) Trading Co., Ltd., Shanghai,
China]. Four marks of equidistance along the scratch were
established as assay points, and the actual migration of cells was
calculated as the mean values according to the distance between the
original wound zone and marks. Experiments were repeated three
times.
Statistical analysis
Statistical significance was assessed by comparing
the mean ± standard deviation values using the Student’s t-test for
independent groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
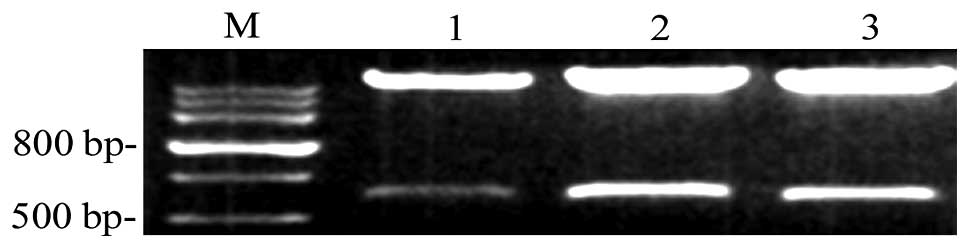
Identification of recombinant CXCR4
Interfering RNA vectors were digested using
restriction endonucleases and agarose gel electrophoresis was
performed. It was found that recombinant plasmids exhibited two DNA
fragments of 5,600 and 650 bp following digestion. The fragments of
CXCR4 shRNA were as expected and were identical to the designed
sequences (Fig. 1).
Transfection efficiency of recombinant
plasmids in 293 T cells
A total of 293T cells were transfected with each
group of plasmids and observed under an inverted microscope 24, 48
and 72 h following transfection. The light of highest intensity was
green fluorescence following 72 h transfection with a transfection
efficiency of ~90% (Fig. 2).
mRNA expression of CXCR4 in 293T
cells
The relative expression levels of CXCR4 mRNA 48 h
following transfection were 1.13±0.19, 0.30±0.09, 1.28±0.11 and
1.60±0.61 for the pGCsi-CXCR4-1/ShRNA, pGCsi-CXCR4-2/ShRNA,
negative control group and blank control groups, respectively (data
not shown). No significant difference was identified between the
negative and control groups, indicating that no RNA interference of
CXCR4 mRNA occurred in cells of the negative control group
(P>0.05). However, a statistically significant difference in
inhibitory rate was identified between pGCsi-CXCR4-2/ShRNA (81.3%)
when compared with the negative and blank control groups
(P<0.05).
CXCR4 protein expression in 293T
cells
RT-PCR analysis of the protein expression of CXCR4
in each group 72 h following transfection demonstrated that CXCR4
protein expression in the pGCsi-CXCR4-2/ShRNA group was the lowest
and no significant differences were identified between the negative
and blank control groups (Fig.
3).
Inhibition of proliferation of MDA-MB-231
cells by CXCR4 silencing
A statistically significant difference was
identified between the CXCR4-shRNA group and blank control group,
as well as the negative control group (P<0.05); however, no
significant difference was identified between the negative and
blank control groups. Therefore, these results suggest that the
proliferation of MDA-MB-231 cells may be significantly inhibited by
RNA interference, which targets CXCR4 gene expression (Fig. 4).
Inhibition of MDA-MB-231 cell adhesion by
CXCR4 silencing
A significant decrease in the number of adhesive
cells in the CXCR4-shRNA transfection group was observed when
compared with the negative and blank control groups (P<0.05);
however, no significant difference was identified between the
negative and blank control groups (P>0.05), which indicated that
breast cancer cells may be inhibited by the downregulation of the
CXCR4 gene in vitro (Table
II).
 | Table IISilencing of CXCR4 expression
inhibited the adhesion of MDA-MB-231 cells. |
Table II
Silencing of CXCR4 expression
inhibited the adhesion of MDA-MB-231 cells.
| Group | 30 min | 60 min | 90 min |
|---|
| PGCsiCXCR4
1–2/ShRNA | 5.97±2.41a | 6.64±2.80a | 7.81±0.77a |
| Negative control | 12.04±3.31 | 13.43±2.50 | 14.51±5.44 |
| Blank control | 12.42±3.49 | 14.37±1.73 | 15.49±1.38 |
Inhibition of MDA-MB-231 cell migration
by CXCR4 silencing
Wound-healing assays revealed that the migration
distance of MDA-MB-231 cells in the CXCR4-shRNA transfection group
(0.42±0.09 mm) was significantly smaller than that in the control
plasmid (2.16±0.44 mm) and the blank control (2.38±0.56 mm) groups
(P<0.01), which indicated that the downregulation of the CXCR4
gene may significantly inhibit the migration of breast cancer
cells.
Discussion
Breast cancer is one of the most common types of
malignant tumors among females, and the incidence is increasing
rapidly in China. Disease recurrence and metastasis are key points
in the prognosis of breast cancer. Cancer metastasis is a complex
process in which malignant cells break away from the primary tumor,
attach to the degraded proteins of the surrounding extracellular
matrix and migrate to other locations via the bloodstream or the
lymphatic system. Tumor cell proliferation, adhesion and migration
are involved and are tightly regulated during the metastatic
process (9). Various factors are
involved in tumor recurrence and metastasis, including modification
of cell gene regulation, disorder of cytokine secretion,
enhancement of tumor immunological tolerance, degradation of
extracellular matrix and inhibition of adhesion. CXCR4, a highly
conserved G-protein-coupled receptor (10) of high specificity to SDF-1, is coded
for by a 352-amino acid protein with seven transmembrane domains.
The CXCR4 gene was originally separated and purified from human
monocytes and was found to be located at human chromosome 2q21.
Previous studies (11) have
predominantly focused on its critical function as an essential
coreceptor of the CD4 molecule, which allows the HIV virus to enter
T cells and diffuse in vivo. CXCR4 is expressed in various
tumor cells (12–14), and the CXCR4/SDF-1 axis has a
significant role in malignant tumor genesis, adhesion, infiltration
and metastasis (15–17). The CXCR4/CXCL12-axis has been
demonstrated to exhibit a critical role in the trafficking and
homing of normal stem cells and metastasis of cancer stem cells to
organs that express high levels of CXCL12, including the lymph
nodes, lungs, liver and bone (17).
Smith et al (18) revealed
that metastasis of breast cancer cells in mice lungs was delayed by
intravenous injection at the caudal vein with AMD 3100, a CXCR4
antagonist.
The RNAi technique is a gene therapy method which
uses small interfering double-stranded RNA, originating from the
inside of the cell or transfection to perform gene silencing
following transcription. This process of special homologous mRNA
degradation is mediated by double-stranded RNA (19–21).
Therefore, RNAi may block tumor-associated gene expression and
effectively terminate the translation process of target proteins to
treat tumors (22). In the present
study, CXCR4 mRNA was silenced and CXCR4 protein expression was
decreased using the hairpin structure shRNA technique to
specifically degrade relevant sequences. This allowed the selection
of the most effective interfering CXCR4-shRNA sequence, which had
an inhibitory rate of 81.3%, which was statistically significant
when compared with the blank and negative control groups.
To further investigate the impact of silencing the
CXCR4 gene on the malignant bionomics of breast cancer cells,
MDA-MB-231 cells were transfected with selected sequences of
CXCR4-shRNA, and the CCK-8 kit, cell adhesion assays and
wound-healing assays were used to determine changes prior to and
following shRNA interference. A CCK-8 assay was performed to
determine the proliferation of tumor cells in vitro and the
result revealed that compared with the negative and blank control
groups, the proliferation of CXCR4-shRNA was significantly
inhibited (P<0.05). These results indicated that CXCR4 exhibits
a critical function in tumor genesis and proliferation of breast
cancer cells, and that the selected vector inhibited breast cancer
cell proliferation in vitro, which was consistent with the
results of Lapteva et al (23).
Adhesion, degradation and removal are the three
predominant stages of tumor migration and infiltration. Ueda et
al (24) demonstrated that the
binding between CXCR4 and its ligand, SDF-1, downregulated the
expression of E-cadherin and inhibited adhesion between tumor cells
to promote metastasis, which was inhibited by an anti-CXCR4
monoclonal antibody (25).
Furthermore, Zeelenberg et al (26) revealed that SDF-1/CXCR4 evoked the
aggregation and re-distribution of cytoskeletal proteins to
regulate cell movement and migration. In addition, the migration of
breast cancer cells was inhibited by a CXCR4 antagonist, as well as
infiltration to the lungs and bone (27,28).
In this study, cell-matrix adhesion and wound-healing assays were
performed to determine the impact of CXCR4-shRNA on cancer cell
adhesion and migration. The results indicated that blockade of
CXCR4 expression significantly inhibited tumor cell migration and
adhesion to the matrix (P<0.05), indicating that the
upregulation of CXCR4 causes epithelial cell instability and
promotes tumor cell migration and infiltration.
In conclusion, in this study shRNA eukaryotic
expression vectors targeting CXCR4 were successfully constructed,
and CXCR4-shRNA was found to significantly inhibit the
proliferation, adhesion and migration of breast cancer cells in
vitro. These results may provide a foundation for further study
regarding the mechanisms of CXCR4 involved in breast cancer growth
and metastasis.
Acknowledgements
This study was supported by the Research Innovation
Program of Shanghai Municipal Education Commission (grant no.
09yz79).
References
|
1
|
Sellers TA: Genetic factors in the
pathogenesis of breast cancer: their role and relative importance.
J Nutr. 27(5 Suppl): 929S–932S. 1997.
|
|
2
|
Psaila B and Lyden D: The metastatic
niche: adapting the foreign soil. Nat Rev Cancer. 9:285–293.
2009.
|
|
3
|
Yarden Y: The EGFR family and its ligands
in human cancer. signalling mechanisms and therapeutic
opportunities. Eur J Cancer. 37(Suppl 4): S3–S8. 2001.
|
|
4
|
Ghosh R, Narasanna A, Wang SE, et al:
Trastuzumab has preferential activity against breast cancers driven
by HER2 homodimers. Cancer Res. 71:1871–1882. 2011.
|
|
5
|
Kulbe H, Levinson NR, Balkwill F and
Wilson JL: The chemokine network in cancer - much more than
directing cell movement. Int J Dev Biol. 48:489–496. 2004.
|
|
6
|
Kuhlmann CR, Schaefer CA, Reinhold L,
Tillmanns H and Erdogan A: Signalling mechanisms of SDF-induced
endothelial cell proliferation and migration. Biochem Biophys Res
Commun. 335:1107–1114. 2005.
|
|
7
|
Wang J, Wang J, Sun Y, et al: Diverse
signaling pathways through the SDF-1/CXCR4 chemokine axis in
prostate cancer cell lines leads to altered patterns of cytokine
secretion and angiogenesis. Cell Signal. 17:1578–1592. 2005.
|
|
8
|
Chu H, Zhou H, Liu Y, et al: Functional
expression of CXC chemokine recepter-4 mediates the secretion of
matrix metalloproteinases from mouse hepatocarcinoma cell lines
with different lymphatic metastasis ability. Int J Biochem Cell
Biol. 39:197–205. 2007.
|
|
9
|
Tang J, Zhang L, She X, et al: Inhibiting
CD164 expression in colon cancer cell line HCT116 leads to reduced
cancer cell proliferation, mobility, and metastasis in vitro
and in vivo. Cancer Invest. 30:380–389. 2012.
|
|
10
|
David NB, Sapède D, Saint-Etienne L, et
al: Molecular basis of cell migration in the fish lateral line:
role of the chemokine receptor CXCR4 and of its ligand, SDF1. Proc
Natl Acad Sci USA. 99:16297–16302. 2002.
|
|
11
|
Babcock GJ, Farzan M and Sodroski J:
Ligand-independent dimerization of CXCR4, a principal HIV-1
coreceptor. J Biol Chem. 278:3378–3385. 2003.
|
|
12
|
Sun YX, Wang J, Shelburne CE, et al:
Expression of CXCR4 and CXCL12 (SDF-1) in human prostate cancers
(PCa) in vivo. J Cell Biochem. 89:462–473. 2003.
|
|
13
|
Phillips RJ, Burdick MD, Lutz M, et al:
The stromal derived factor-1/CXCL12-CXC chemokine receptor 4
biological axis in non-small cell lung cancer metastases. Am J
Respir Crit Care Med. 167:1676–1686. 2003.
|
|
14
|
Koshiba T, Hosotani R, Miyamoto Y, et al:
Expression of stromal cell-derived factor 1 and CXCR4 ligand
receptor system in pancreatic cancer: a possible role for tumor
progression. Clin Cancer Res. 6:3530–3535. 2000.
|
|
15
|
Raman D, Baugher PJ, Thu YM and Richmond
A: Role of chemokines in tumor growth. Cancer Lett. 256:137–165.
2007.
|
|
16
|
Sun YX, Fang M, Wang J, et al: Expression
and activation of alpha v beta 3 integrins by SDF-1/CXC12 increases
the aggressiveness of prostate cancer cells. Prostate. 67:61–73.
2007.
|
|
17
|
Sutton A, Friand V, Brulé-Donneger S, et
al: Stromal cell-derived factor-1/chemokine (CXC motif) ligand 12
stimulates human hepatoma cell growth, migration, and invasion. Mol
Cancer Res. 5:21–33. 2007.
|
|
18
|
Smith MC, Luker KE, Garbow JR, et al:
CXCR4 regulates growth of both primary and metastatic breast
cancer. Cancer Res. 64:8604–8612. 2004.
|
|
19
|
Hannon GJ: RNA interference. Nature.
418:244–251. 2002.
|
|
20
|
Elbashir SM, Lendeckel W and Tuschl T: RNA
interference is mediated by 21-and 22-nucleotide RNAs. Genes Dev.
15:188–200. 2001.
|
|
21
|
Aravin AA, Klenov MS, Vagin VV, Rozovskiĭ
IaM and Gvozdev VA: Role of double-stranded RNA in eukaryotic gene
silencing. Mol Biol (Mosk). 36:240–251. 2002.(In Russian).
|
|
22
|
Sanguino A, Lopez-Berestein G and Sood AK:
Strategies for in vivo siRNA delivery in cancer. Mini Rev
Med Chem. 8:248–255. 2008.
|
|
23
|
Lapteva N, Yang AG, Sanders DE, Strube RW
and Chen SY: CXCR4 knockdown by small interfering RNA abrogates
breast tumor growth in vivo. Cancer Gene Ther. 12:84–89.
2005.
|
|
24
|
Ueda Y, Neel NF, Schutyser E, Raman D and
Richmond A: Deletion of the COOH-terminal domain of CXC chemokine
receptor 4 leads to the down-regulation of cell-to-cell contact,
enhanced motility and proliferation in breast carcinoma cells.
Cancer Res. 66:5665–5675. 2006.
|
|
25
|
Yoon Y, Liang Z, Zhang X, et al: CXC
chemokine receptor-4 antagonist blocks both growth of primary tumor
and metastasis of head and neck cancer in xenograft mouse models.
Cancer Res. 67:7518–7524. 2007.
|
|
26
|
Zeelenberg IS, Ruuls-Van Stalle L and Roos
E: The chemokine receptor CXCR4 is required for outgrowth of colon
carcinoma micrometastases. Cancer Res. 63:3833–3839. 2003.
|
|
27
|
Huang EH, Singh B, Cristofanilli M, et al:
A CXCR4 antagonist CTCE-9908 inhibits primary tumor growth and
metastasis of breast cancer. J Surg Res. 155:231–236. 2009.
|
|
28
|
Richert MM, Vaidya KS, Mills CN, et al:
Inhibition of CXCR4 by CTCE-9908 inhibits breast cancer metastasis
to lung and bone. Oncol Rep. 21:761–767. 2009.
|