Introduction
Cancer is a worldwide public health problem, which
results from a complex interaction between environmental and
genetic factors (1). Several
polymorphic genes that are directly involved in tumorigenesis have
also been proposed to contribute to the individual susceptibility
to cancer (2).
The host immune defense has been shown to play a
vital role in modulating human carcinogenesis (3). Regulatory T cells aid in keeping the
balance between immunity and autotolerance, and are mainly
characterized by CD4+/FOXP3+ or
CD4+/CD25+/FOXP3+ expression.
FOXP3 belongs to the forkhead family of transcription factors, and
is involved in the regulation, activation and differentiation of T
cells (4). In fact, the absence of
a functional FOXP3 gene product has been revealed to cause an
abnormal production of regulatory T cells (5). In addition, the loss of expression and
somatic mutation of the human FOXP3 gene has been identified in
human prostate and breast cancers. This suggests that FOXP3 may be
a tumor suppressor and that inactivation of the FOXP3 gene may
contribute to the development of cancer in humans (6,7).
The FOXP3 gene is positioned at the Xp11.23 locus on
the X chromosome and encodes the FOXP3 protein, which is expressed
in epithelial cells from various organs, such as the lungs and the
thymus (8–11). The promoter polymorphisms in the
FOXP3 gene are considered to affect FOXP3 production and activity.
The FOXP3 gene rs3761549 (C>T) and rs3761548 (C>A)
polymorphisms, located on the promoter region of the FOXP3 gene,
are two of the most common single nucleotide polymorphisms.
Previous studies have investigated the association between the
FOXP3 rs3761549 and rs3761548 polymorphisms and the cancer risk,
however, they have yielded conflicting results (12–16).
Therefore, the present meta-analysis was performed to evaluate the
role of these two polymorphisms and their association with the risk
of cancer.
Materials and methods
Publication search and inclusion
criteria
A comprehensive literature search, using the
keywords ‘FOXP3’, ‘polymorphism’ and ‘tumor or cancer’, was
performed using the PubMed, EMBASE and Chinese Wanfang databases
(last search updated in February 10, 2014). Additional eligible
studies were identified by manually searching the reference lists
of reviews and original articles. In the event that data were
published in more than one article, only studies with the largest
sample size were selected for. The selection criteria to identify
an eligible study were as follows: i) Investigation of the
rs3761549 (C>T) and rs3761548 (C>A) polymorphisms of the
FOXP3 gene and cancer risk; ii) the use of a case-control design,
based on unrelated individuals; and iii) sufficient genotype
distributions for cases and controls, so that an odds ratio (OR)
with a 95% confidence interval (CI) could be assessed.
Data extraction
The two authors independently reviewed and extracted
the required data. Disagreements were resolved through discussion
among the authors to achieve a consensus. The following information
was recorded for each study: First author, year of publication,
country, ethnicity, cancer type and number of genotypes (Table I).
 | Table ICharacteristics of studies included in
the present meta-analysis. |
Table I
Characteristics of studies included in
the present meta-analysis.
| | | | | rs3761549 | rs3761548 |
|---|
| | | | |
|
|
|---|
| First author
(ref.) | Year | Country | Ethnicity | Cancer type | Case (TT/CT/CC) | Control
(TT/CT/CC) | Case (AA/AC/CC) | Control
(AA/AC/CC) |
|---|
| Chen et al
(12) | 2013 | China | Asian | Hepatocellular
carcinoma | 59/28/301 | 41/88/233 | - | - |
| He et al
(13) | 2013 | China | Asian | Non-small cell lung
cancer | - | - | 37/80/75 | 18/80/161 |
| Jahan et al
(14) | 2013 | India | Asian | Breast cancer | 0/198/4 | 0/128/2 | 27/160/15 | 20/106/4 |
| Raskin et al
(15) | 2009 | Israel | Asian | Breast cancer | - | - | 320/722/402 | 303/763/392 |
| Zheng et al
(16) | 2013 | China | Asian | Breast cancer | 32/283/734 | 34/290/767 | 38/338/673 | 30/342/719 |
Statistical analysis
The OR corresponding to the 95% CI was used to
assess the association between the FOXP3 polymorphisms and the risk
of cancer. In addition to this comparison among all subjects, a
stratified analysis by cancer type was also performed. The
statistical heterogeneity among studies was assessed using
I2 statistics and the Q-test (17). In the absence of any obvious
heterogeneity, the fixed-effects model (the Mantel-Haenszel method)
was applied to estimate the summary OR. Otherwise, the
random-effects model (the DerSimonian and Laird method) was used
(18,19). Sensitivity analysis was performed to
identify the effect that the data from each study had on the pooled
OR. Finally, any publication bias was evaluated using a funnel
plot. All of the statistical tests were performed using RevMan 5.0
software (The Cochrane Collaboration, Oxford, UK).
Results
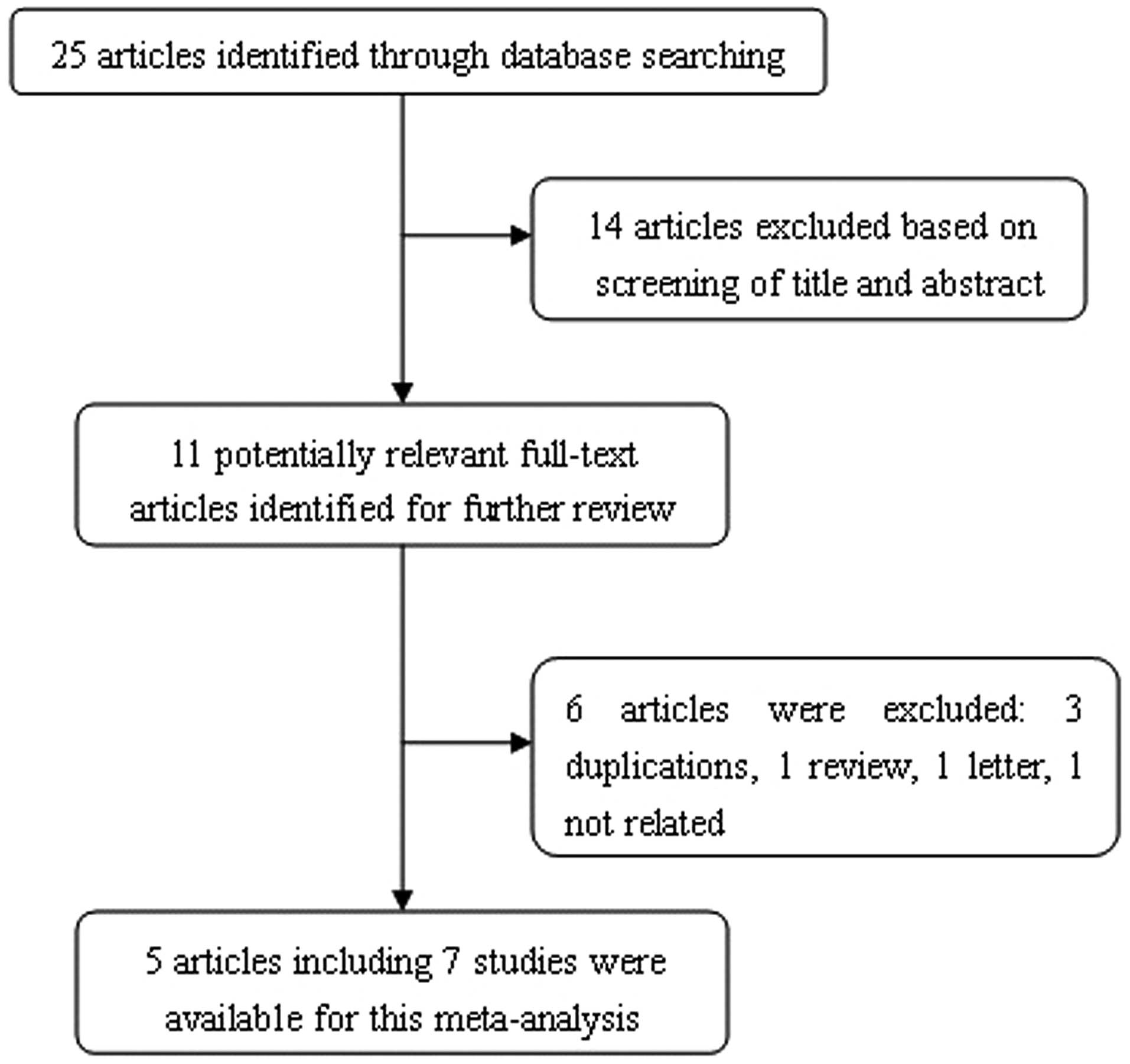
The process of identifying suitable studies is shown
in Fig. 1. A total of five studies
(12–16), including 3,275 cases and 3,300
controls, were included in the present meta-analysis. All of the
selected studies were based on Asian populations (Table I). The results of the pooled
analysis revealed no significant association between the FOXP3 gene
polymorphisms and the cancer risk (for rs3761549: TT vs. CT+CC OR,
1.20, 95% CI, 0.87–1.66; TT+CT vs. CC OR, 0.74, 95% CI, 0.41–1.33;
TT vs. CC OR, 1.06, 95% CI, 0.76–1.46; TC vs. CC OR, 0.56, 95% CI,
0.17–1.80; T vs. C OR, 0.94, 95% CI, 0.83–1.06. For rs3761548: AA
vs. AC+CC OR, 1.37, 95% CI, 0.87–2.16; AA+AC vs. CC OR, 1.18, 95%
CI, 0.79–1.78; AA vs. CC OR, 1.36, 95% CI, 0.67–2.77; AC vs. CC OR,
1.11, 95% CI, 0.79–1.58; A vs. C OR, 1.21, 95% CI, 0.90–1.62).
Further subgroup analysis was conducted based on cancer type,
however, no association between the FOXP3 gene polymorphisms and
the risk of breast cancer was revealed (for rs3761549: TT vs. CT+CC
OR, 0.98, 95% CI, 0.60–1.60; TT+CT vs. CC OR, 1.01, 95% CI,
0.84–1.22; TT vs. CC OR, 0.98, 95% CI, 0.60–1.61; TC vs. CC OR,
1.02, 95% CI, 0.84–1.23; T vs. C OR, 1.01, 95% CI, 0.87–1.16. For
rs3761548: AA vs. AC+CC OR, 1.09, 95% CI, 0.93–1.28; AA+AC vs. CC
OR, 1.00, 95% CI, 0.88–1.12; AA vs. CC OR, 1.04, 95% CI, 0.86–1.26;
AC vs. CC OR, 0.97, 95% CI, 0.86–1.10; A vs. C OR, 1.02, 95% CI,
0.94–1.11). However, statistical associations were observed with
respect to hepatocellular carcinoma (HCC) and non-small cell lung
cancer (NSCLC) (for rs3761549: TT+CT vs. CC OR, 0.52, 95% CI,
0.38–0.72; TC vs. CC OR, 0.25, 95% CI, 0.16–0.39; T vs. C: OR,
0.76, 95% CI, 0.59–0.97. For rs3761548: AA vs. AC+CC OR, 3.20, 95%
CI, 1.76–5.81; AA+AC vs. CC OR, 2.56, 95% CI, 1.75–3.76; AA vs. CC
OR, 4.41, 95% CI, 2.36–8.25; AC vs. CC OR, 2.15, 95% CI, 1.42–3.25;
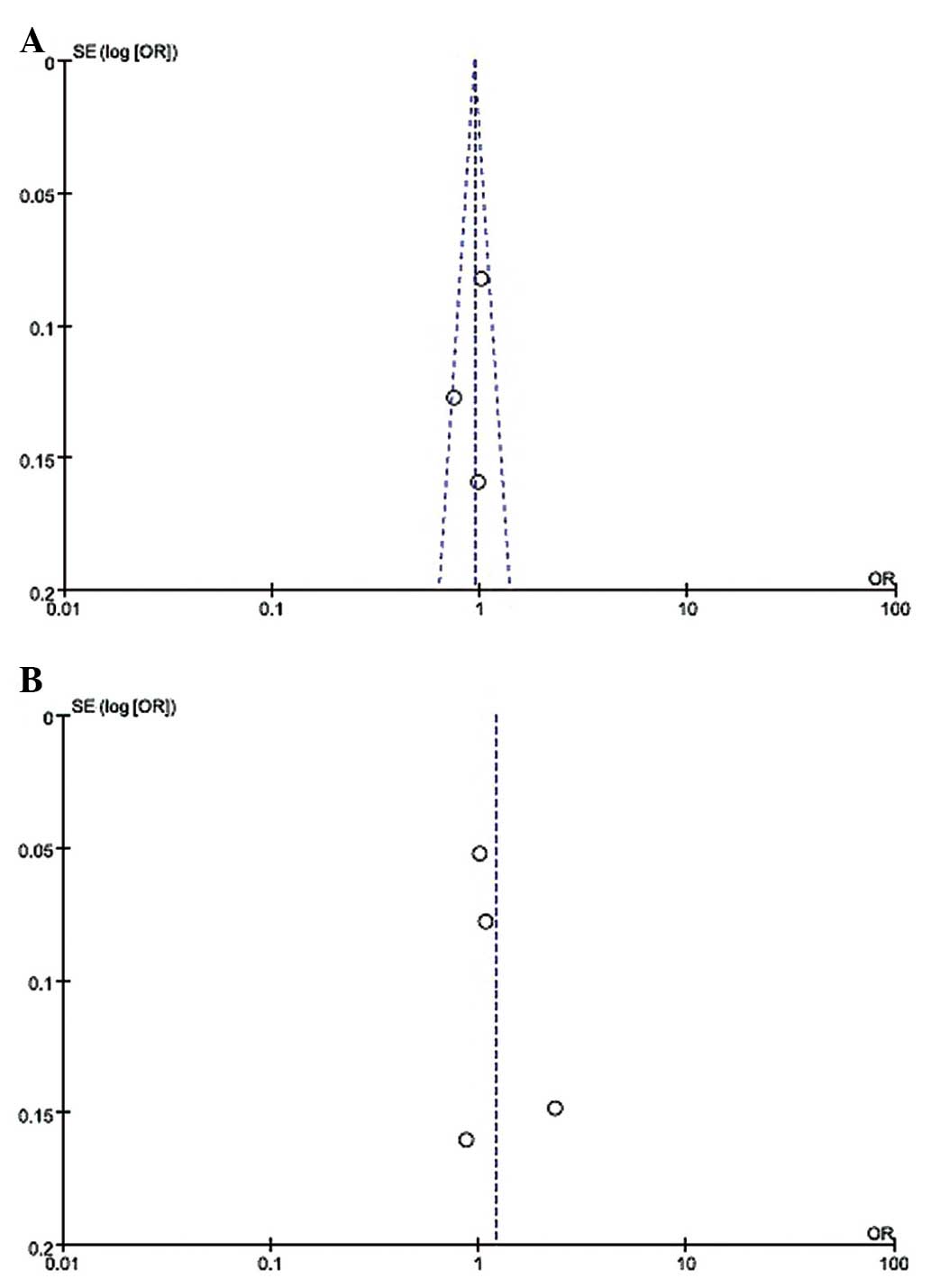
A vs. C OR, 2.32, 95% CI, 1.74–3.10) (Table II). The funnel plot, which assessed
publication bias of the literature, appeared symmetrical in all of
the genetic models (Fig. 2).
 | Table IIMeta-analysis data of the associations
between the FOXP3 promoter polymorphisms and the cancer risk in all
genetic models. |
Table II
Meta-analysis data of the associations
between the FOXP3 promoter polymorphisms and the cancer risk in all
genetic models.
| A, rs3761549
polymorphism |
|---|
|
|---|
| TT vs. CT+CC | TT+CT vs. CC | TT vs. CC | TC vs. CC | T vs. C |
|---|
|
|
|
|
|
|
|---|
| Variables | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value |
|---|
| Total | 1.20 (0.87–1.66) | 0.28 | 0.74 (0.41–1.33) | 0.01 | 1.06 (0.76–1.46) | 0.71 | 0.56 (0.17–1.80) | 0.01 | 0.94 (0.83–1.06) | 0.15 |
| Breast cancer | 0.98 (0.60–1.60) | - | 1.01 (0.84–1.22) | 0.76 | 0.98 (0.60–1.61) | - | 1.02 (0.84–1.23) | 0.75 | 1.01 (0.87–1.16) | 0.92 |
| HCC | 1.40 (0.92–2.15) | - | 0.52
(0.38–0.72) | - | 1.11 (0.72–1.72) | - | 0.25
(0.16–0.39) | - | 0.76
(0.59–0.97) | - |
|
| B, rs3761548
polymorphism |
|
| AA vs. AC+CC | AA+AC vs. CC | AA vs. CC | AC vs. CC | A vs. C |
|
|
|
|
|
|
| Variables | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value |
|
| Total | 1.37 (0.87–2.16) | 0.01 | 1.18 (0.79–1.78) | 0.01 | 1.36 (0.67–2.77) | 0.01 | 1.11 (0.79–1.58) | 0.01 | 1.21 (0.90–1.62) | 0.01 |
| Breast cancer | 1.09 (0.93–1.28) | 0.53 | 1.00 (0.88–1.12) | 0.16 | 1.04 (0.86–1.26) | 0.14 | 0.97 (0.86–1.10) | 0.18 | 1.02 (0.94–1.11) | 0.45 |
| NSCLC | 3.20
(1.76–5.81) | - | 2.56
(1.75–3.76) | - | 4.41
(2.36–8.25) | - | 2.15
(1.42–3.25) | - | 2.32
(1.74–3.10) | - |
Discussion
The characterization and identification of genes
involved in the genetic predisposition and progression of cancer
are critical for clinical practice and basic medical research.
FOXP3 is an immunological regulator, and is able to repress
oncogenes whilst activating additional tumor suppressor genes
(6,20–22).
FOXP3-mediated gene regulation follows the histone code of gene
activation and suppression and alters histone modifications by
binding to gene promoters (23,24).
Epidemiological studies suggest that the FOXP3 promoter
polymorphisms, rs3761549 and rs376154, are associated with the
cancer risk. However, the results from these studies are
conflicting. To provide a more detailed overview of the
association, five genetic models were used in the current
meta-analysis.
To the best of our knowledge, this was the first
meta-analysis to provide comprehensive insight into the association
between the FOXP3 polymorphisms and the risk of cancer. It was
identified that the FOXP3 rs3761549 (C>T) and rs3761548 (C>A)
polymorphisms were not associated with the risk of cancer among an
Asian population. In addition, subgroup analysis revealed that the
FOXP3 gene rs3761549 (C>T) and rs3761548 (C>A) polymorphisms
were not associated with the risk of breast cancer. However, the
rs3761549 (C>T) and rs3761548 (C>A) polymorphisms were linked
with the risk of HCC and NSCLC, respectively. The results therefore
indicated that the rs3761549 (C>T) and rs3761548 (C>A)
polymorphisms may have a varying effect on carcinogenesis within
different organs. However, these findings must be viewed with
caution, since studies on HCC and NSCLC are rare. Therefore, the
results from the present study may be due to chance.
There were certain limitations of this
meta-analysis. Firstly, a relatively small number of studies and
subjects were included, which could reduce the statistical power of
the analysis. Secondly, the results were based on unadjusted
estimates. A more precise analysis could be conducted if individual
data were available. Thirdly, all published studies were based on
Asian populations. Therefore, the results of this meta-analysis may
be applicable to the specified ethnicity alone.
In conclusion, the present study demonstrated that
the rs3761549 (C>T) and rs3761548 (C>A) polymorphisms in the
promoter region of the FOXP3 gene were not associated with breast
cancer, but instead were associated with HCC and NSCLC. Therefore,
a future study that consists of a larger sample size is required to
further evaluate this association.
References
|
1
|
Gao X, Huang M, Liu L, et al:
Insertion/deletion polymorphisms in the promoter region of BRM
contribute to risk of hepatocellular carcinoma in Chinese
populations. PLoS One. 8:e551692013.
|
|
2
|
Ying H, Wang J and Gao X: CCL5-403,
CCR5-59029, and Delta32 polymorphisms and cancer risk: a
meta-analysis based on 20,625 subjects. Tumour Biol. 35:5895–5904.
2014.
|
|
3
|
Schneeberger A, Koszik F and Stingl G:
Immunologic host defense in melanoma: delineation of effector
mechanisms involved and of strategies for the augmentation of their
efficacy. J Invest Dermatol. 105(Suppl 1): S110–S116. 1995.
|
|
4
|
Hori S, Nomura T and Sakaguchi S: Control
of regulatory T cell development by the transcription factor Foxp3.
Science. 299:1057–1061. 2003.
|
|
5
|
Roncador G, Garcia JF, Garcia JF, et al:
FOXP3, a selective marker for a subset of adult T-cell
leukemia/lymphoma. Leukemia. 19:2247–2253. 2005.
|
|
6
|
Wang L, Liu R, Li W, et al: Somatic single
hits inactivate the X-linked tumor suppressor FOXP3 in the
prostate. Cancer Cell. 16:336–346. 2009.
|
|
7
|
Zuo T, Liu R, Zhang H, et al: FOXP3 is a
novel transcriptional repressor for the breast cancer oncogene
SKP2. J Clin Invest. 117:3765–3773. 2007.
|
|
8
|
Bennett CL, Brunkow ME, Ramsdell F, et al:
A rare polyadenylation signal mutation of the FOXP3 gene
(AAUAAA→AAUGAA) leads to the IPEX syndrome. Immunogenetics.
53:435–439. 2001.
|
|
9
|
Gupta S, Joshi K, Wig JD and Arora SK:
Intratumoral FOXP3 expression in infiltrating breast carcinoma: Its
association with clinicopathologic parameters and angiogenesis.
Acta Oncol. 46:792–797. 2007.
|
|
10
|
Karanikas V, Speletas M, Zamanakou M, et
al: FOXP3 expression in human cancer cells. J Transl Med.
6:192008.
|
|
11
|
Katoh H, Zheng P and Liu Y: Signalling
through FoxP3 as an X-linked tumor suppressor. Int J Biochem Cell
Biol. 42:1784–1787. 2010.
|
|
12
|
Chen Y, Zhang H, Liao W, et al: FOXP3 gene
polymorphism is associated with hepatitis B-related hepatocellular
carcinoma in China. J Exp Clin Cancer Res. 32:392013.
|
|
13
|
He YQ, Bo Q, Yong W, Qiu ZX, Li YL and Li
WM: FoxP3 genetic variants and risk of non-small cell lung cancer
in the Chinese Han population. Gene. 531:422–425. 2013.
|
|
14
|
Jahan P, Ramachander VR, Maruthi G, Nalini
S, Latha KP and Murthy TS: Foxp3 promoter polymorphism (rs3761548)
in breast cancer progression: a study from India. Tumour Biol.
35:3785–3791. 2014.
|
|
15
|
Raskin L, Rennert G and Gruber SB: FOXP3
germline polymorphisms are not associated with risk of breast
cancer. Cancer Genet Cytogenet. 190:40–42. 2009.
|
|
16
|
Zheng J, Deng J, Jiang L, et al:
Heterozygous genetic variations of FOXP3 in Xp11.23 elevate breast
cancer risk in Chinese population via skewed X-chromosome
inactivation. Hum Mutat. 34:619–628. 2013.
|
|
17
|
Higgins JP and Thompson SG: Quantifying
heterogeneity in a meta-analysis. Stat Med. 21:1539–1558. 2002.
|
|
18
|
Mantel N and Haenszel W: Statistical
aspects of the analysis of data from retrospective studies of
disease. J Natl Cancer Inst. 22:719–748. 1959.
|
|
19
|
DerSimonian R and Laird N: Meta-analysis
in clinical trials. Control Clin Trials. 7:177–188. 1986.
|
|
20
|
Zuo T, Wang L, Morrison C, et al: FOXP3 is
an X-linked breast cancer suppressor gene and an important
repressor of the HER-2/ErbB2 oncogene. Cell. 129:1275–1286.
2007.
|
|
21
|
Liu R, Wang L, Chen G, et al: FOXP3
up-regulates p21 expression by site-specific inhibition of histone
deacetylase 2/histone deacetylase 4 association to the locus.
Cancer Res. 69:2252–2259. 2009.
|
|
22
|
Li W, Wang L, Katoh H, et al:
Identification of a tumor suppressor relay between the FOXP3 and
the Hippo pathways in breast and prostate cancers. Cancer Res.
71:2162–2171. 2011.
|
|
23
|
Katoh H, Qin ZS, Liu R, et al: FOXP3
orchestrates H4K16 acetylation and H3K4 trimethylation for
activation of multiple genes by recruiting MOF and causing
displacement of PLU-1. Mol Cell. 44:770–784. 2011.
|
|
24
|
Marson A, Kretschmer K, Frampton GM, et
al: FoxP3 occupancy and regulation of key target genes during
T-cell stimulation. Nature. 445:931–935. 2007.
|