Introduction
The retinoblastoma protein-interacting zinc finger
(RIZ) gene was identified by Bird (1) following the application of
retinoblastoma (Rb) probes in combination with Rb protein to screen
for separation-of-function mutants. Fluorescence in situ
hybridization located the gene to human chromosome 1p36. Due to the
presence of various transcription initiation sites, the RIZ
gene encodes two proteins, RIZ1 and RIZ2. RIZ1 contains a positive
regulatory (PR) domain, whereas RIZ2 does not; the sequences of the
two proteins are otherwise identical (2). The PR domain, known as the
PRDI-BF1-RIZ (positive regulatory domain I-binding factor 1-RIZ)
homologous region, contains 100 amino acids that form the protein
binding surface (protein-binding interface), mediating
protein-protein interactions and exerting an important role in
chromosome structure stability and in the regulation of chromatin
gene expression (3). In tumors, the
PR domain gene family expresses various protein products according
to the presence or absence of the PR domain. A preponderance of
either type of protein is indicative of gene inactivation, which is
a predominant mechanism of tumorigenesis (4). Currently, a number of studies have
demonstrated that RIZ1 exerts tumor-inhibiting activity; for
example, the RIZ1 protein may cause tumor cell arrest in the
G2/M phase and induce apoptosis (5,6). In
the present study, human RIZ1 and PR domain eukaryotic
expression vectors were constructed to investigate whether the PR
domain of the tumor suppressor RIZ1 has the ability to induce
apoptosis and reduce cell invasion ability in esophageal carcinoma
cells.
Materials and methods
Cell culture and RNA isolation
The TE13 human esophageal squamous cell carcinoma
(ESCC) cell line was purchased from American Type Culture
Collection (Rockville, MD, USA). The cells were cultured in
RPMI-1640 (Gibco-BRL, Carlsbad, CA, USA) containing 4.76 g HEPES,
2.0 g NaCO3, 10.4 g RPMI 1640 and 1,000 ml
double-distilled (dd) H2O supplemented with 10% fetal
bovine serum (FBS; Gibco-BRL), 1X L-glutamine (2 mm), 100 U/ml
penicillin and 100 μg/ml streptomycin. The cells were incubated at
37°C in a 5% CO2 humidified incubator.
RNA was isolated from the cells using TRIzol
(Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s
instructions, and 1 ml TRIzol was added to
5×106-1×107 cells. The RNA pellets were
resuspended in diethylpyrocarbonate-treated H2O. The
total RNA concentrations were quantified using an ultraviolet (UV)
spectrophotometer (Beckman Coulter, Miami, FL, USA).
Reverse transcription (RT) amplification
of mRNA
RT reactions were performed to generate cDNA using 2
μg RNA, Moloney murine leukemia virus reverse transcriptase,
ribonuclease inhibitor and a dNTP mixture (Takara Bio, Inc., Shiga,
Japan), according to the manufacturer’s instructions.
Semi-quantitative, RT-polymerase chain reaction (PCR) was conducted
using the cDNA templates.
According to the RIZ1 mRNA sequence published
by the National Center for Biotechnology Information (NCBI), the
5,157 bp protein-coding region is located between base pairs 857
and 6,013. Due to the amplicon size, the open reading frame was
divided into five sections, termed A603, A1200, B, C and D. The
following primers were designed for the five RIZ1 sections,
hereafter referred to as amplicons, using Primer Premier 5.0
software (Premier Biosoft, Palo Alto, CA, USA): A603 forward,
5′-GTGGCTAGCATGAATCAGAACACTACTG-3′ and reverse,
5′-TTGGCTAGCAGAGGTGAAATCTGGCTC-3′; A1200 forward,
5′-TGGCTGCGATATGTGAATTG-3′ and reverse, 5′-CTCTACGCTGATGCCGTCTC-3′;
B forward, 5′-GCTGATGGCAAAGCATCTG-3′ and reverse,
5′-AATTCCTTGCCTTCAGAGTCAC-3′; C forward,
5′-TCAAAGAAAGTCATTCAGTGC-3′ and reverse, 5′-CGGTGATGGTACTGAAATG-3′;
and D forward, 5′-GCCTCAATCAGCATTACC-3′ and reverse,
5′-GTCTACTCTTTGAAGAATGGTC-3′. PCR was conducted in 50-μl reactions
containing 5 μl 10X KOD buffer, 5 μl 2 mm dNTPs, 3 μl 25 mm
MgSO4, 2 μl of each forward and reverse primer, 1 μl
cDNA, 1 μl KOD-Plus-Ver. 2 polymerase (Toyobo Corporation, Osaka,
Japan) and ddH2O. Each reaction required the following
specific conditions in accordance with the melting temperature and
size of each amplicon: Initial denaturation at 94°C for 2 min, 35
cycles of denaturation at 98°C for 10 sec, annealing (A603U, 60°C
at 30 sec; A1200, 57°C at 30 sec; B, 55°C at 30 sec; and C and D,
50°C at 30 sec) and extension at 72°C for 1 min, and a final
extension at 72°C for 10 min.
The PR domain primers were as follows: Forward,
5′-GTGGCTAGCATGAATCAGAACACTACTG-3′ and reverse,
5′-TTGGGATCCTCAAGAGGTGAAATCTG-3′. The 5′ terminal of the forward
and reverse primers contained NheI and BamHI
endonuclease sites, respectively, and three protective base pairs.
The downstream primer also contained a termination codon. Initial
denaturation was performed at 94°C for 3 min, followed by 35 cycles
of denaturation at 98°C for 10 sec, annealing at 60°C for 1 min and
final extension at 72°C for 10 min. Subsequently, 0.3 μl Easy Taq
(Tiangen, Beijing, China) was added to append a polyA tail at the
end of the PCR products, turning the blunt end into a sticky end.
This was followed by a final extension step at 72°C for 30 min.
The quality of the amplified products was analyzed
on 12 g/l agarose gels using a UV spectrophotometer and the RT-PCR
products were sequenced.
Construction and transfection of the
pcDNA3.1(+)/RIZ1 and pcDNA3.1(+)/PR domain
The amplicons were extracted from the agarose gels
using the Tiangel Midi Purification kit (Tiangen, Beijing, China)
according to the manufacturer’s instructions. The five RIZ1
and PR domain amplicons were inserted into Trans1-T1 Phage
Resistant vectors (Promega Corporation, Madison, WI, USA) that were
subsequently transformed into Trans1-T1 Phage Resistant competent
cells, and plated on agar containing ampicillin and X-gal. White
colonies were selected for further analysis. Subsequent to
expansion of the selected bacterial colonies, plasmid DNA was
extracted by alkaline lysis (5).
Restriction enzyme digests were employed to validate successful
recombination, with confirmation provided by sequencing. The
sequences of each plasmid were compared with the sequences listed
by NCBI using the Basic Local Alignment Search Tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The
RIZ1 and PR domain amplicons were digested from plasmids
containing the correct insert and were ligated into the pcDNA3.1(+)
eukaryotic expression vector (Invitrogen Life Technologies).
Insertion was verified by restriction enzyme digestion followed by
sequencing.
The TE13 cells were seeded in six-well culture
plates at a density of 2×105 cells/well in 2 ml media,
and then incubated at 37°C to 90–95% confluence. After 24 h, the
media was replaced with complete serum- or antibiotic-free
RPMI-1640 in preparation for transfection. Ultra pure
pcDNA3.1(+)/RIZ1 and pcDNA3.1(+)/PR domain plasmid DNA was
extracted using a HighPure Mini Plasmid kit (Tiangen). A
liposome-mediated method (6) was
employed to transfect the TE13 cells with either the
pcDNA3.1(+)/RIZ1 or the pcDNA3.1(+)/PR domain, with empty
vector-transfected and untransfected cells serving as negative
controls. Subsequent to 6 h of incubation with media containing the
recombinant plasmids and the transfection reagents, the media was
replaced with antibiotic-free RPMI-1640 containing 10% FBS. The
transfected cells were incubated for a further 48 h, before being
harvested for further analysis.
Quantitative PCR
For the RNA isolation and reverse transcription
reaction, 2μl of cDNA was mixed with 2X SYBR real-time PCR
premixture (BioTeke, Beijing, China). The primers for the genes of
interest (10μM) were as follows: RIZ1/PR domain forward,
5′-AATCAGAACACTACTGAGCCTGT-3′ and reverse,
5′-ACCAATCCGGGTCTTGTCAAC-3′; and glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) forward, 5′-GAAGGTGAAGGTCGGAGTC-3′ and
reverse, 5′-GGGTGGAATCATATTGGAAC-3′. The reactions were conducted
using an CFX96 Real-time PCR System (Bio-Rad, Hercules, CA, USA)
according to the manufacturer’s instructions. Briefly, initial
denaturation was performed at 95°C for 2 min, followed by 45 cycles
of denaturation at 95°C for 15 sec, annealing for the RIZ1/PR
domain at 63°C for 15 sec or for GAPDH at 60°C for 15 sec, and
extension at 72°C for 40 sec, followed by the production of thermal
melting curves. Each sample for each gene was conducted in
triplicate.
Western blotting
The TE13 cells transfected with pcDNA3.1(+)/RIZ1
were homogenized in radioimmunoprecipitation buffer (containing 50
mm Tris-HCl, pH 7.4; 150 mm NaCl; 1% Nonidet P-40; 0.5% sodium
deoxycholate; 0.1% SDS; 1 mm EDTA; 1 mm phenylmethylsulfonyl
fluoride and 1 mg/ml aprotinin) and the protein concentrations were
determined using a bicinchoninic acid protein assay kit (Pierce,
Rockford, IL, USA). Cell lysates (30 μg) were separated by 8%
SDS-PAGE, transferred to nitrocellulose membranes (Amersham
Biosciences, Chalfont St. Giles, UK) and immunoblotted with the
indicated antibodies overnight in the Orbital Shaker (Thermo Fisher
Scientific, Waltham, MA, USA) at 4°C. The monoclonal mouse
anti-human RIZ1/PR domain antibodies, monoclonal mouse anti-human
β-actin primary antibody and secondary polyclonal goat anti-mouse
polyclonal antibody were all obtained from Abcam (Cambridge,
UK).
Bands were visualized using a PowerLook scanner
(UMAX Technologies, Hsinchu, Taiwan) and quantified using
ImageQuant software (GE Healthcare, Little Chalfont, UK). The
relative expression levels of RIZ1 and the PR domain were
calculated as the gray values of RIZ1, the PR domain and β-actin.
Untransfected and empty vector-transfected TE13 cells served as
negative controls.
Flow cytometric analysis
To investigate the effect of overexpression of RIZ1
or the PR domain on apoptosis, TE13 cells were seeded in six-well
plates at a density of 2×105 cells/well and allowed to
attach for 12 h. The cells were then transfected with either
pcDNA3.1(+)/RIZ1 or pcDNA3.1(+)/PR domain and harvested after 24 h.
A total of ~1×105 cells were washed with cold
phosphate-buffered saline (PBS; Solomen, Tianjin, China) for each
test. Cells were suspended in 1 ml 1X binding buffer using the
Annexin V-fluorescein isothiocyanate (FITC) Apoptosis Detection
Analysis Kit (Tianjin Sungene Biotech Co., Ltd., Tianjin, China).
Next, the cells were centrifuged (5415D; Ruicong Co., Ltd.,
Shanghai, China) at 300 × g for 10 min at room temperature and the
supernatant was removed. The cells were then resuspended in 1 ml 1X
binding buffer and the cell concentration was adjusted to
1×106 cells/ml. A total of 100 μl cell suspension was
used for analysis. Next, 5 μl Annexin V-FITC staining solution was
added and the tube was kept in the dark at room temperature for 10
min. A total of 5 μl propidium iodide staining solution (Tianjin
Sungene Biotech Co., Ltd.) was added and the tube was kept in the
dark at room temperature for 5 min. Finally, 500 μl PBS was added
to the cells and vortexed gently for 1 h. The cells were then
analyzed using a BD FACSAria II cell sorter (BD Biosciences,
Franklin Lakes, NJ, USA). Untransfected and empty
vector-transfected TE13 cells served as negative controls.
Cell Invasion Assay
To study the invasion ability using a cell invasion
assay kit (ECM550; Millipore, Billerica, MA, USA). The invasion
chamber was allowed to adjust to room temperature in a tissue
culture hood. Media containing 10% FBS (500 μl) was added to the
lower chamber and 300 μl of cell suspension, containing
1.0×106 cells/ml in serum-free media, was added to each
insert. The invasion assay was then incubated for 24 h in a tissue
culture incubator. Using a cotton-tipped swab, the non-invading
cells were then gently removed, and the ECMatrix gel was also
removed from the interior of the inserts. The invasive cells on the
lower surface of the membrane were stained by dipping the inserts
in the crystal violet staining solution for 20 minutes and then
dipping the inserts in a beaker of water three times to rinse. The
cells were counted by capturing images of the membrane through the
microscope (DFC480, Leica, Wetzlar, Germany).
Statistical analysis
Statistical analysis was conducted using SPSS 18.0
software (SPSS, Inc., Chicago, IL, USA). The data are presented as
the mean ± SD. The quantitative PCR results are shown as
2−averageΔΔCT × 100%. Student’s t-test and one-way
analysis of variance (ANOVA) were employed to examine parametric
data. The χ2 test was used for statistical analysis of
the group comparisons and to compare enumerated data. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression levels of RIZ1 and PR domain
following pcDNA3.1(+)/RIZ1 and pcDNA3.1(+)/PR domain
transfection
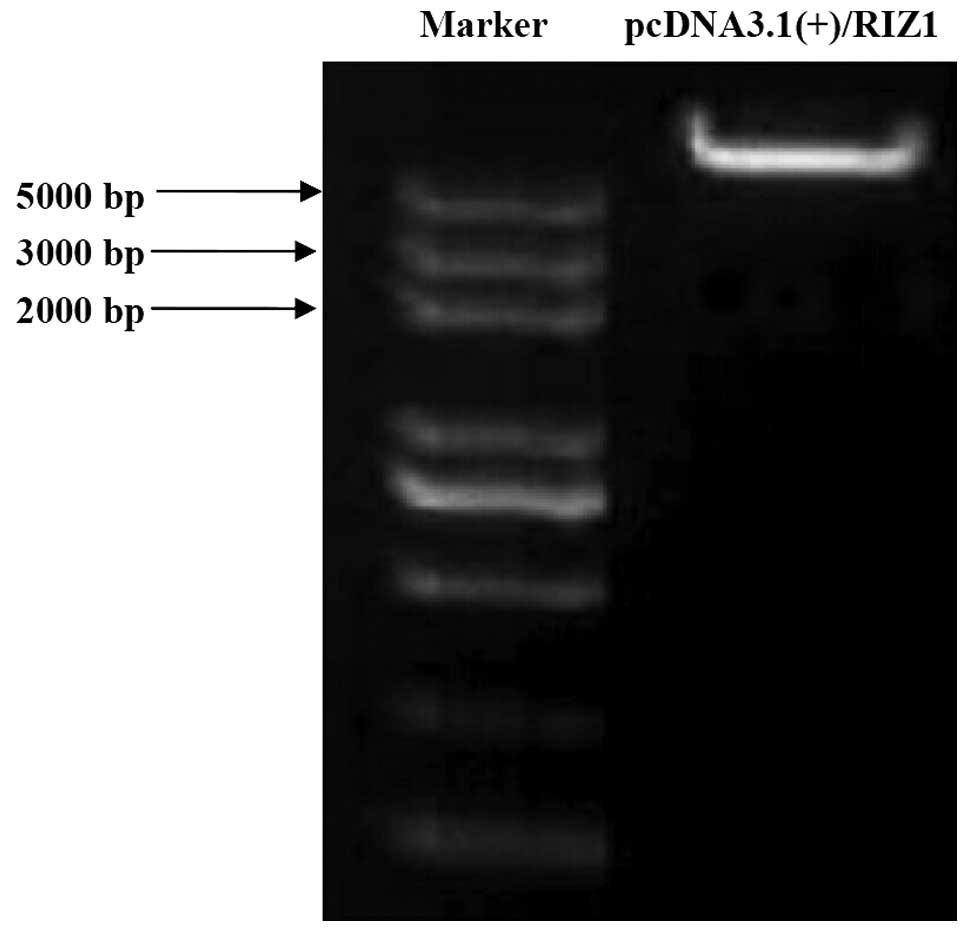
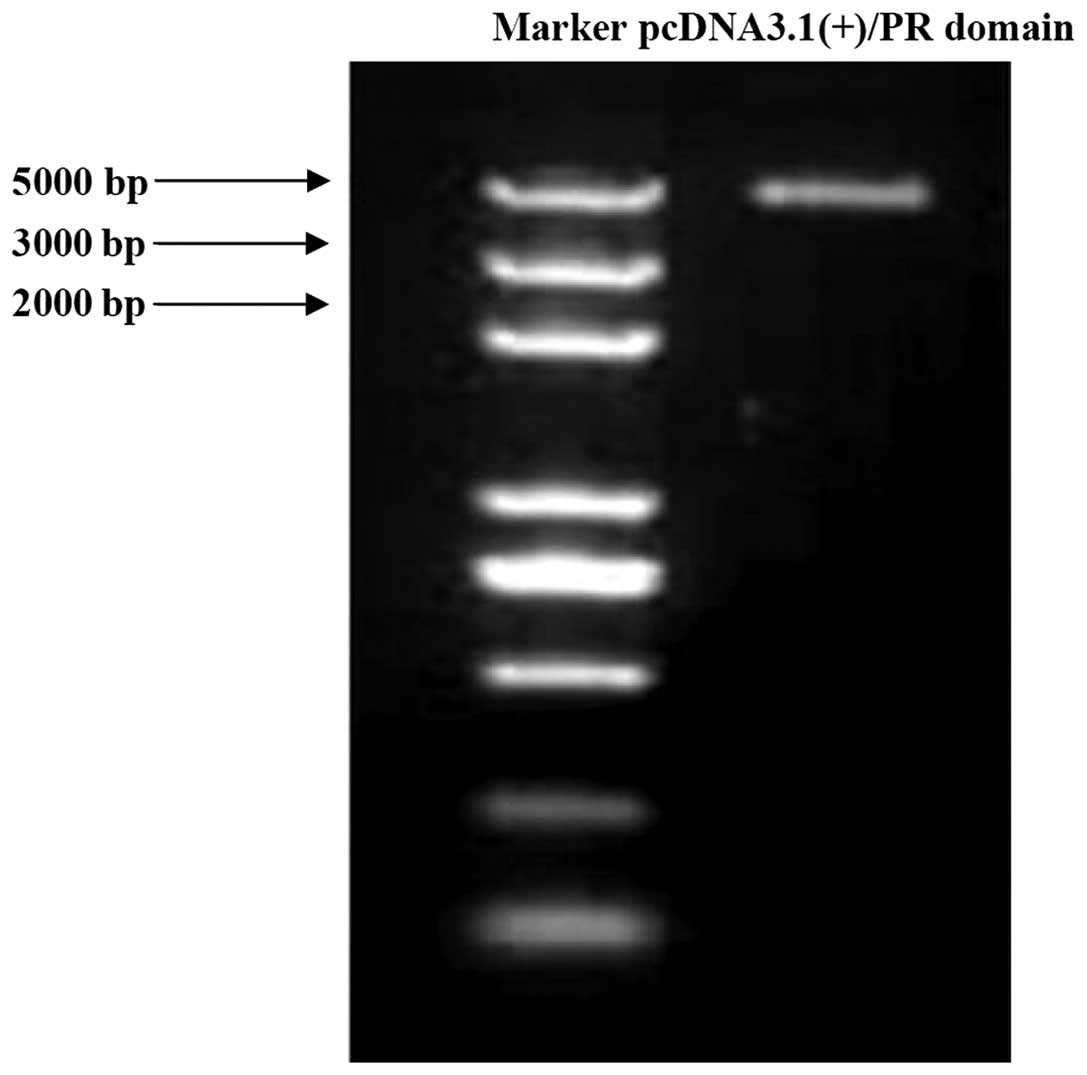
To overexpress RIZ1 and the PR domain,
recombinant plasmids were generated to enable ectopic
overexpression of RIZ1 (Fig.
1) and the PR domain (Fig. 2)
in TE13 cells.
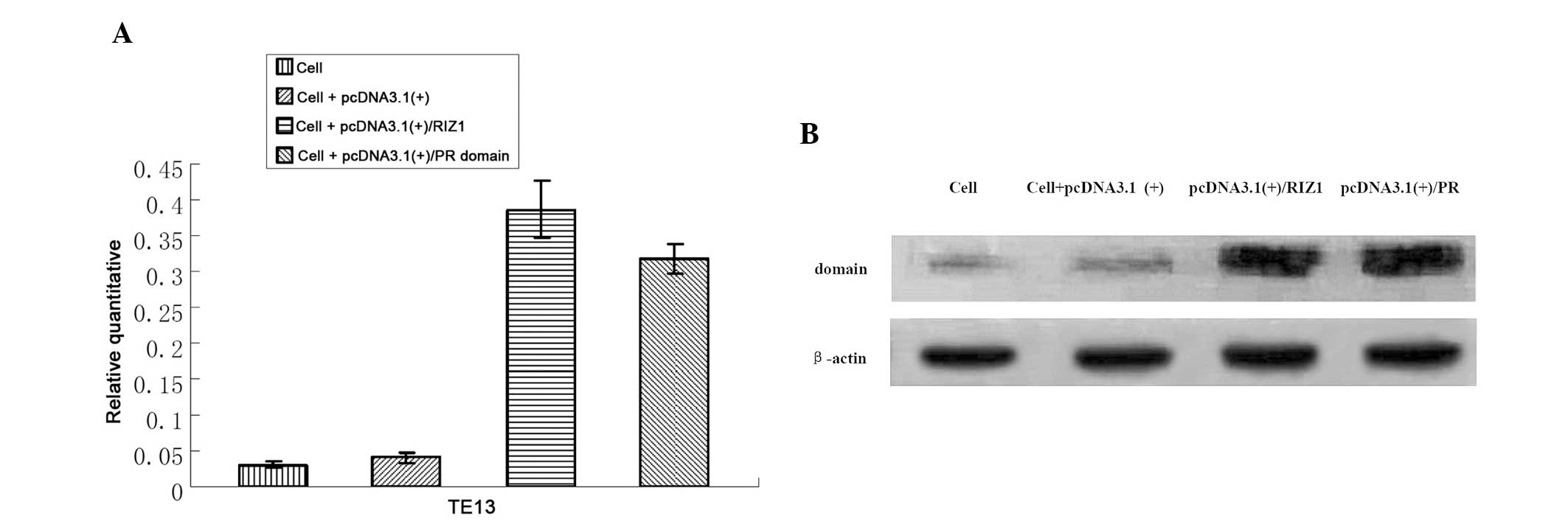
Quantitative RT-PCR and western
blotting
The RIZ1 and PR domain mRNA and protein expression
levels in TE13 cells transfected with pcDNA3.1(+)/RIZ1 or
pcDNA3.1(+)/PR were significantly higher compared with
untransfected and empty vector-transfected cells (negative control
groups) (P<0.05; Fig. 3). No
statistically significant differences were identified between the
negative control groups (P>0.05).
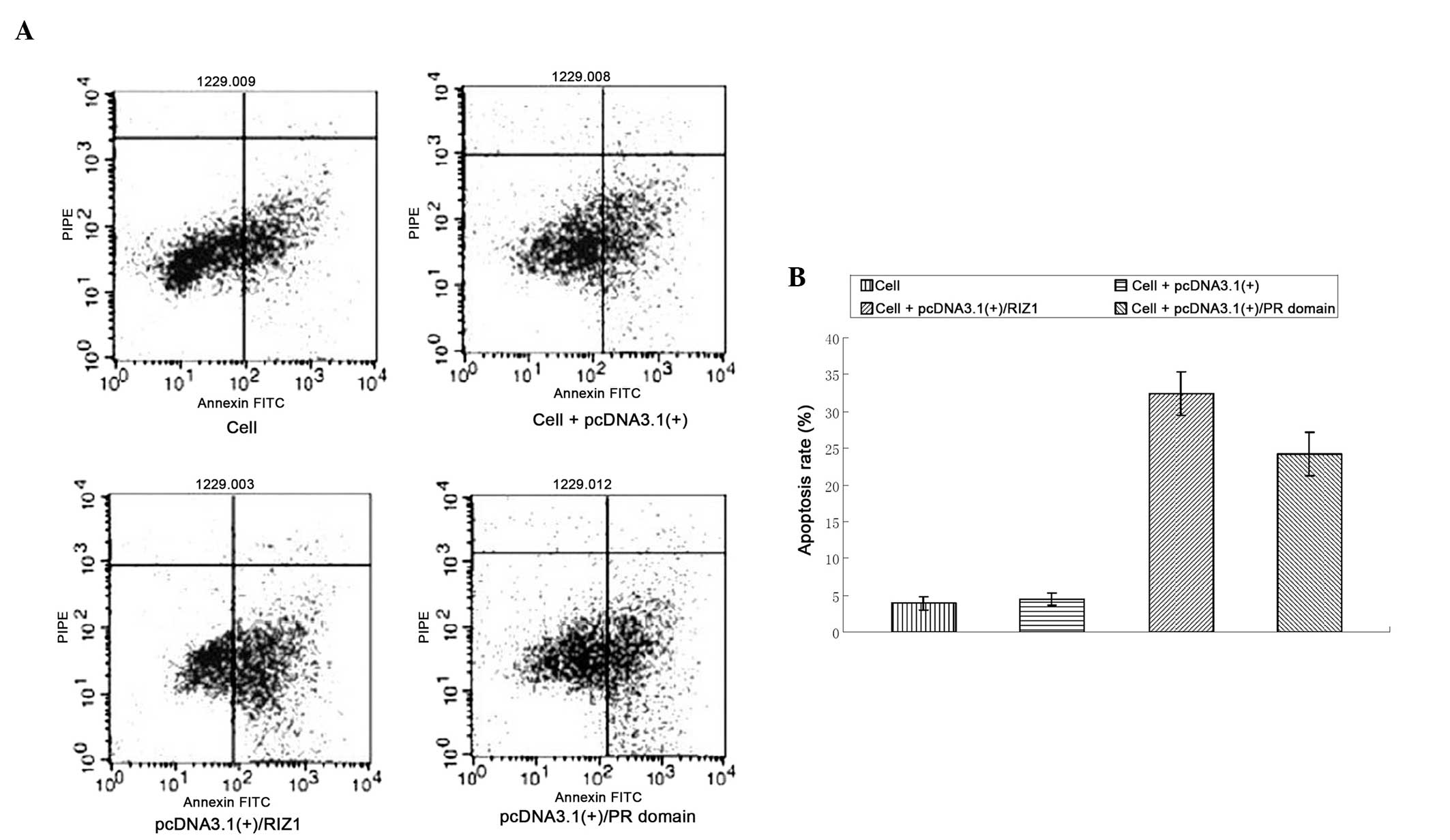
Flow cytometric analysis of the apoptotic
rate in pcDNA3.1(+)/RIZ1 or pcDNA3.1(+)/PR domain-transfected TE13
cells
The apoptotic rates were significantly higher in the
cells transfected with pcDNA3.1(+)/RIZ1 or the pcDNA3.1(+)/PR
domain compared with the untransfected and empty vector-transfected
TE13 cells (P<0.01; Fig. 4).
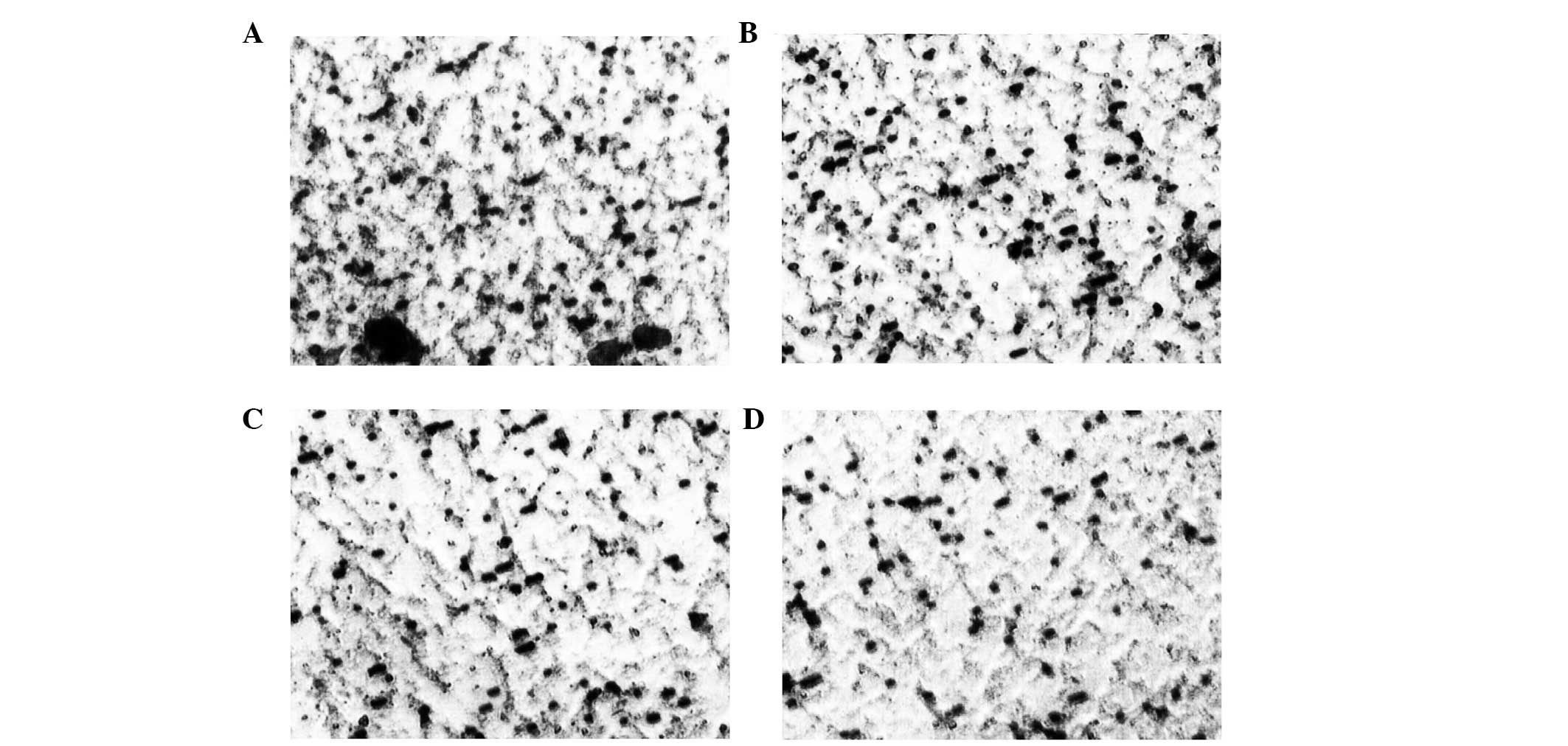
Cell Invasion Assay
The Matrigel invasion ability of the TE13 cells
transfected with pcDNA3.1(+)/RIZ1 or pcDNA3.1(+)/PR domain was
significantly reduced compared with the invasion ability of the
negative controls (P<0.05; Fig.
5).
Discussion
The silencing of tumor suppressor genes (TSGs) by
genetic and epigenetic pathways is recognized as important in human
carcinogenesis (7,8). In addition to gross chromosomal
instability and the instability of small repetitive DNA sequences,
epigenetic silencing is also considered to contribute significantly
to human carcinogenesis. TSG silencing by the methylation of
CpG-rich promoter regions has been reported in numerous types of
human cancer. The distal region of the short arm of human
chromosome 1 (1p36) is commonly deleted in a variety of human tumor
types. This region is known to harbor several TSGs. One candidate
TSG in this region is RIZ1 (9).
The RIZ gene encodes two protein products of
different lengths, RIZ1 and RIZ2. As a TSG, RIZ1 contains
the PR or Suvar3–9, Enhancer-of-zeste, Trithorax domain, but RIZ2
lacks this domain. A number of studies have investigated the
underlying mechanism of RIZ1 gene inactivation, which has
been shown to include genetic and epigenetic changes (10–12).
Chromosomal and microsatellite instability deactivates the RIZ1
gene, resulting in frameshift mutation, point mutations and
heterozygous deficiency (13).
Silenced or reduced expression levels of the RIZ1 gene have been
observed in numerous types of human tumor and tumor cell lines;
however, to the best of our knowledge, no study of the RIZ1 gene in
esophageal cancer has been reported.
Previously, quantitative RT-PCR was performed and
the RIZ1 mRNA expression levels in esophageal cancer were found to
be significantly lower compared with those in normal esophageal
tissue, and this was associated with CpG island methylation
(14,15). This indicated that inactivation of
RIZ1 may be important in the progression of esophageal cancer.
The ability of the PR domain alone to exert any
anticancer activity was examined, as the PR domain is the only
structural difference between the two protein products of the RIZ
gene. Compared with RIZ2, RIZ1 possesses an additional 100 amino
acid residues at the amino terminus, and the PR domain represents
the main functional motif within the amino terminus of RIZ1. In the
present study, TE13 cells were transfected with pcDNA3.1(+)/PR
domain to investigate whether the domain could be expressed
independently and whether it influenced apoptosis or invasion.
Quantitative PCR and western blotting determined that cells
transfected with the recombinant plasmid successfully expressed
RIZ1 and the PR domain. Flow cytometry revealed that transfection
with either the PR domain or RIZ1 inhibited cell proliferation. The
cell invasion assay revealed that the invasion ability was
significantly reduced in the TE13 cells transfected with
pcDNA3.1(+)/RIZ1 or pcDNA3.1(+)/PR-domain (P<0.05). These
findings indicate that the PR domain of RIZ1 exerts anticancer
activity in ESCC. In conclusion, further study regarding the
mechanism of action of the RIZ1 tumor suppressor gene and the PR
domain may reveal the underlying mechanism of anticancer function,
and thus may lead to the development of novel biomarkers for early
diagnosis and prognostic evaluation in ESCC.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant no. 81201945), the Science
Foundation of Tianjin Medical University (grant no. 2011KY08), the
Doctoral Program of Higher Education Research Fund (grant no.
20091202110009) and the Natural Science Foundation of Tianjin
(grant no. 10JCYBJC11300).
References
|
1
|
Bird A: Perceptions of epigenetics.
Nature. 447:396–398. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jaenisch R and Bird A: Epigenetic
regulation of gene expression: how the genome integrates intrinsic
and environmental signals. Nat Genet. 33(Suppl): 245–254. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Du Y, Carling T, Fang W, Piao Z, Sheu JC
and Huang S: Hypermethylation in human cancers of the RIZ1 tumor
suppressor gene, a member of a histone/protein methyltransferase
superfamily. Cancer Res. 61:8094–8099. 2001.PubMed/NCBI
|
|
4
|
Braig M and Schmitt CA: Oncogene-induced
senescence: putting the brakes on tumor development. Cancer Res.
66:2881–2884. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pors K and Patterson LH: DNA mismatch
repair deficiency, resistance to cancer chemotherapy and the
development of hypersensitive agents. Curr Top Med Chem.
5:1133–1149. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Oliveira AM, Ross JS and Fletcher JA:
Tumor suppressor genes in breast cancer: the gatekeepers and the
caretakers. Am J Clin Pathol. 124(Suppl): S16–S28. 2005.
|
|
7
|
Dong SW, Ma L, Xu N, Yan HQ, Liu HY, et
al: Research on the reactivation of Syk expression caused by the
inhibition of DNA promoter methylation in the lung cancer.
Neoplasma. 58:89–95. 2011. View Article : Google Scholar
|
|
8
|
Ma L, Dong S, Zhang P, Xu N, Yan H, et al:
The relationship between methylation of the Syk gene in the
promoter region and the genesis of lung cancer. Clin Lab.
56:407–416. 2010.PubMed/NCBI
|
|
9
|
Buyse IM, Shao G and Huang S: The
retinoblastoma protein binds to RIZ. a zinc-finger protein that
shares an epitope with the adenovirus EIA protein. Proc Natl Acad
Sci USA. 92:4467–4471. 1995. View Article : Google Scholar
|
|
10
|
Costello JF, Frünwald MC, Smiraglia DJ, et
al: Aberrant CpG-island methylation has non-random and
tumour-type-specific patterns. Nat Genet. 24:132–138. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dong SW, Li D, Xu C, Sun P, Wang YG and
Zhang P: Alteration in gene expression profile and oncogenicity of
esophageal squamous cell carcinoma by RIZ1 upregulation. World J
Gastroenterol. 19:6170–6177. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dong SW, Zhang H, Wang BL, Sun P, Wang YG
and Zhang P: Effect of the downregulation of SMYD3 expression by
RNAi on RIZ1 expression and proliferation of esophageal squamous
cell carcinoma. Oncol Rep. 32:1064–1070. 2014.PubMed/NCBI
|
|
13
|
Rountree MR, Bachman KE, Herman JG and
Baylin SB: DNA methylation, chromatin inheritance, and cancer.
Oncogene. 20:3156–3165. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dong SW, Cui YT, Zhong RR, Liang DC, Liu
YM, et al: Decreased expression of retinoblastoma
protein-interacting zinc-finger gene 1 in human esophageal squamous
cell cancer by DNA methylation. Clin Lab. 58:41–51. 2012.PubMed/NCBI
|
|
15
|
Dong SW, Zhang P, Liu YM, Cui YT, Wang S,
et al: Study on RIZ1 gene promoter methylation status in human
esophageal squamous cell carcinoma. World J Gastroenterol.
18:576–582. 2012. View Article : Google Scholar : PubMed/NCBI
|