Introduction
Hepatocellular carcinoma (HCC) represents the fifth
most prevalent malignancy and the second most common cause of
cancer-related mortality worldwide, with ~695,900 mortalities per
year (1). Half of these cases occur
in China, due to high incidence of chronic hepatitis B virus (HBV)
infection (2). Liver surgery,
including liver resection and liver transplantation, are considered
to be curative treatment strategies, as they provide complete
oncologic resection (R0 resection). However, tumor recurrence
following liver surgery remains a critical issue (recurrence rate,
~30–50%), compromising the long-term survival of patients (3–5). The
molecular mechanisms underlying HCC progression remain poorly
understood. Therefore, investigating ideal biomarkers for the
improved prediction of postoperative recurrence may aid surgeons in
selecting patients or adopting preventive strategies for patients
at high risk of recurrence.
Previously, RNA sequencing revealed a novel class of
transcripts termed long non-coding RNAs (lncRNAs). LncRNAs are
>200 nucleotides long and lack protein-coding potential, thus,
they were previously regarded as random transcriptional noises.
However, increasing evidence has implicated lncRNAs in critical
regulatory roles in cancer biology (6–8).
LncRNAs have been demonstrated to control gene expression through
transcriptional and posttranscriptional regulation (9). Various classic lncRNAs have been
characterized in human hepatocarcinogenesis as having oncogenic and
tumor suppressive roles, such as HOX transcript antisense RNA
(HOTAIR) (10,11), metastasis-associated lung
adenocarcinoma transcript 1 (MALAT1) (12), maternally expressed 3 (MEG3)
(13) and H19 (14). Our previous studies have
demonstrated that overexpression of HOTAIR (10) and MALAT1 (12) exhibit oncogenic properties, and may
serve as independent prognostic factors for HCC. Furthermore, MEG3
was frequently downregulated in HCC and inhibited cell growth by
functionally interacting with p53 (13). Additionally, H19 suppressed HCC
metastasis by epigenetic activation of the microRNA (miR)200 family
(14). These data support the
hypothesis that MG3 and H19 act as tumor suppressors in HCC.
PVT1, which maps to chromosome 8q24, is a copy
number amplification-associated lncRNA. Overexpression of PVT1 is a
powerful predictor of tumor progression and patient survival in
colorectal (15), ovarian and
breast cancers (16). Furthermore,
PVT1 exerts regulatory functions in various biological processes,
such as proliferation, apoptosis, mobility and invasion (15,16),
and chromosome 8q24 is a commonly amplified region in HCC (17,18).
However, the association between PVT1 and HCC remains unclear.
The aim of the present study was to examine the
expression pattern of PVT1 and its clinical significance in
HCC.
Materials and methods
Patients and surgical specimens
Fifty-eight snap-frozen HCC tissues and the
corresponding non-cancerous tissues were obtained from patients
undergoing liver resection at the First Affiliated Hospital of
Zhejiang University (Hangzhou, China) between 2009 and 2012 (cohort
one). An additional 214 HCC tissues were collected from patients
undergoing liver transplantation at the First Affiliated Hospital
of Zhejiang University between 2003 and 2012 (cohort two), and were
used for survival analysis and validation. The liver tissue
specimens were immediately frozen in liquid nitrogen following
surgical resection and stored at −80°C prior to the extraction of
total RNA. A postoperative histopathological examination by
experienced pathologists was used to establish a diagnosis of HCC
in these patients. The histological grade of differentiation was
evaluated on hematoxylin and eosin-stained sections according to
the Edmondson-Steiner grading method (19). Complete clinical and laboratory data
were collected in a perspective database. The tumor staging was
defined according to the sixth edition of the tumor-node-metastasis
(TNM) classification system published by the American Joint
Committee on Cancer and the Union for International Cancer Control
(20). The requirements for
selecting transplant candidates varies depending on the criteria,
for example: The Milan criteria defines HCC as a single tumor ≤5 cm
or up to three tumors ≤3 cm (21);
the University of California, San Francisco (UCSF) criteria defines
HCC as a single nodule ≤6.5 cm or up to three nodules ≤4.5 cm, and
total tumor diameter ≤8 cm (22);
and the Hangzhou criteria defines HCC as a total tumor diameter of
≤8 cm, or a total tumor diameter of >8 cm, with a well or
moderately differentiated histopathological grade and a
preoperative serum AFP level of ≤400 ng/ml, simultaneously
(23).
The present study was conducted with the approval of
the Institutional Review Board and Ethics Committee of the First
Affiliated Hospital, Zhejiang University (Hangzhou, China) and in
accordance with the Declaration of Helsinki. Thus, the study
conformed to international and national regulations. Informed
consent was obtained from all of the patients.
Follow-up
Patient follow-up was conducted every 2–3 months
during the first two years following surgery and 3–6 months
thereafter. The endpoint of study was September 3, 2013. During the
follow-up period, all patients were monitored using abdomen
ultrasonography, chest X-ray, emission computed tomography and
serum AFP tests. Following a suspected recurrence, computed
tomography, magnetic resonance imaging or positron emission
tomography-computed tomography, were immediately performed. These
imaging techniques were required for a new lesion to be detected
and recurrence to be diagnosed; an increase in serum AFP levels
alone was not regarded as recurrence. Once recurrent tumors were
diagnosed, treatment was implemented based on the tumor size,
number, location and vascular invasion, as well as the liver
function. The recurrence-free survival (RFS) period was calculated
from the date of surgery to the date of detection of tumor
recurrence, mortality or the most recent observation. All follow-up
examinations were performed by two physicians who were unaware of
the study. The mean follow-up time was 27.58 months (range, 2–124
months).
Cell culture
The healthy liver cell line HL7702, the liver cancer
cell lines Huh7, SK-hep1, SMMC-7721, HepG2, Hep3B, PLC/PRF/5 and
Bel-7402, as well as the metastasis-capable human HCC cell lines
MHCC97L, MHCC97H and HCCLM3, were purchased from the American Type
Culture Collection (Manassas, VA, USA), the Shanghai Institute of
Biochemistry and Cell Biology (Shanghai, China), and the Liver
Cancer Institute of Fudan University (Shanghai, China),
respectively. All of the cell lines were maintained in Dulbecco’s
modified Eagle’s medium with high glucose or RPMI-1640, containing
10% fetal bovine serum, and cultured in a humidified 5%
CO2 incubator at 37°C.
RNA extraction and quantitative reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNAs from the specimens and cell lines were
extracted using TRIzol reagent (Invitrogen Life Technologies,
Carlsbad, CA, USA); and complementary DNA was synthesized using
Moloney murine leukemia virus reverse transcriptase (Promega
Corporation, Madison, WI, USA) according to the manufacturer’s
instructions. The expression of PVT1 was determined using qPCR,
which was performed on an Applied Biosystems 7500 Fast Real-Time
PCR system (Applied Biosystems Life Technologies, Foster City, CA,
USA) and SYBR® Green dye (Takara Biotechnology Co.,
Inc., Dalian, China). All PCRs were performed in triplicate and
GAPDH was used to normalize mRNA expression levels. Relative
quantification was performed using the comparative threshold cycles
(2−ΔΔCt method) as described in the manufacturer’s
instructions. The primer sequences were as follows: Sense,
3′-CATCCGGCGCTCAGCT-5′ and antisense, 3′-TCATGATGGCTGTATGTGCCA-5′
for PVT1; and sense, 3′-ATGGGGAAGGTGAAGGTCG-5′ and antisense,
3′-GGGGTCATTGATGGCAACAATA-5′ for GAPDH.
Statistical analysis
Comparisons of continuous data were analyzed using
the independent t-test between the two groups, whereas categorical
data were analyzed by the χ2 test. Comparisons of
continuous data among multiple groups were calculated using one-way
analysis of variance. A receiver operating characteristic (ROC)
curve was used to determine the cut-off value of PVT1 expression
that yielded the highest combined sensitivity and specificity for
discriminating between patients exhibiting HCC recurrence and those
not exhibiting recurrence. Recurrence-free survival was analyzed
using the Kaplan-Meier method and compared by performing the
log-rank test. Independent prognostic factors were assessed in the
univariate and multivariate analysis using the Cox
proportional-hazards regression model. All statistical analyses
were performed using SPSS for Windows (version 16.0; SPSS, Inc.,
Chicago, IL, USA) and GraphPad Prism (version 5.0; GraphPad
Software, Inc., La Jolla, CA, USA) software. P<0.05 was
considered to indicate a statistically significant difference.
Results
Characteristics of HCC patients
Snap-frozen HCC tissues were obtained from 272 HCC
patients who had undergone liver surgery at the First Affiliated
Hospital of Zhejiang University between 2003 and 2012. The present
study involved two independent cohorts of HCC patients: Cohort one
included 58 HCC patients who had undergone radical resection
between 2009 and 2012; while cohort two included 214 HCC patients
who had received a liver transplant between 2003 and 2012.
The majority of the HCC patients in the two cohorts
were male (91.18%) with a tumor size of >5 cm at the time of
surgery (45.96%), an elevated serum AFP level (47.79%) and with
tumors exceeding the Milan and UCSF criteria (63.60 and 54.41%,
respectively; Table I). In
addition, the patients in cohort two were HCC transplant patients,
thus representing a group of patients with advanced disease.
Furthermore, the patients in cohort two were relatively younger
than those in cohort one, and more patients in cohort two exhibited
HBV infection, liver cirrhosis, multifocal tumor, portal vein tumor
thrombosis (PVTT) and advanced TNM stage.
 | Table IClinical characteristics of HCC
patients. |
Table I
Clinical characteristics of HCC
patients.
| Cohort one
(n=58) | Cohort two
(n=214) | P-value |
|---|
| Age, years | | | 0.000a |
| Median | 60 | 49 | |
| Range | 26–86 | 20–71 | |
| Gender, n (%) | | | 0.951 |
| Female | 5 (8.62) | 19 (8.88) | |
| Male | 53 (91.38) | 195 (91.12) | |
| HBV, n (%) | | | 0.000a |
| Negative | 11 (18.97) | 6 (2.34) | |
| Positive | 47 (81.03) | 209 (97.66) | |
| Cirrhosis, n
(%) | | | 0.000a |
| No | 22 (37.93) | 2 (0.93) | |
| Yes | 36 (62.07) | 212 (99.07) | |
| Tumor size, n
(%) | | | 0.059 |
| ≤5 cm | 25 (43.10) | 122 (57.01) | |
| >5 cm | 33 (56.90) | 92 (42.99) | |
| Tumor number, n
(%) | | | 0.000a |
| =1 | 52 (89.66) | 89 (41.59) | |
| >1 | 6 (10.34) | 125 (58.41) | |
| PVTT, n (%) | | | 0.000a |
| Negative | 54 (93.10) | 142 (66.36) | |
| Positive | 4 (6.90) | 72 (33.64) | |
| AFP, n (%) | | | 0.270 |
| ≤400 ng/ml | 34 (58.62) | 108 (50.47) | |
| >400 ng/ml | 24 (41.38) | 106 (49.53) | |
| Histopathological
grade, n (%) | | | 0.512 |
| Well +
moderately | 27 (46.55) | 110 (51.40) | |
| Poorly | 31 (53.45) | 104 (48.60) | |
| TNM stage, n
(%) | | | 0.000a |
| I + II | 52 (89.66) | 99 (46.26) | |
| III + IV | 6 (10.34) | 115 (53.74) | |
| Milan criteria
(19), n (%) | | | 0.374 |
| Within
criteria | 24 (41.38) | 75 (35.05) | |
| Beyond
criteria | 34 (58.62) | 139 (64.95) | |
| UCSF criteria
(20), n (%) | | | 0.175 |
| Within
criteria | 31 (53.45) | 93 (43.46) | |
| Beyond
criteria | 27 (46.55) | 121 (56.54) | |
| Hangzhou criteria
(21), n (%) | | | 0.000a |
| Within
criteria | 46 (79.31) | 105 (49.07) | |
| Beyond
criteria | 12 (20.69) | 109 (50.93) | |
PVT1 overexpression in the two
independent HCC cohorts and liver cancer cell lines
In the present study, the expression levels of PVT1
in the 272 HCC patients from the two independent cohorts were
determined using RT-qPCR. Compared with the corresponding non-tumor
liver tissues of the HCC cohorts, PVT1 expression was significantly
increased in the cancerous tissues of the patients in cohort one
(P=0.0016; Fig. 1A) and cohort two
(P=0.0274; Fig. 1B). Furthermore,
the expression of PVT1 was defined in 10 liver cancer cell lines
and one healthy liver cell line. Of the 10 liver cancer cell lines,
six (HepG2, SK-Hep1, MHCC-97L, SMMC-7721, MHCC-97H and Bel-7402)
expressed a higher level of PVT1 than the healthy liver cell line
(Fig. 2). Thus, the present study
determined that PVT1 expression was significantly increased in
HCC.
Association between PVT1 and HCC
progression
As PVT1 expression was significantly increased in
HCC, the association between PVT1 expression in HCC and disease
progression was evaluated. First, the expression level of PVT1 in
different TNM-stage patients was assessed, as TNM staging is a
widely accepted system for HCC stratification. In HCC cohort two,
advanced-stage patients (stages III and IV; n=115) exhibited
increased expression levels of PVT1 compared with early-stage
patients (stages I and II; n=99) (P=0.0466; Fig. 3A). Additionally, the patients
exhibiting disease recurrence (n=114) demonstrated higher levels of
PVT1 expression compared with the non-recurrence recipients (n=100)
(P=0.0477;Fig. 3B). However, in HCC
cohort one, it was difficult to analyze the data with sufficient
statistical power due to the limited number of patients with an
advanced disease stage (Fig.
3C).
To investigate the clinicopathological correlation
of PVT1 expression in HCC tissues, the patients were divided into
high and low expression groups according to the cut-off value
obtained from the ROC curve analysis. No significant correlations
with any of the clinicopathological parameters tested were observed
in HCC cohort one (Table II). In
HCC cohort two, no significant correlations were identified between
PVT1 expression and clinicopathological parameters such as age,
gender, tumor number, tumor size, PVTT, histopathological grade or
TNM stage (Table III). However,
high PVT1 expression levels in HCC cohort two were significantly
correlated with a higher AFP level (P=0.011) and higher recurrence
rate (P=0.004) (Table III). Thus,
these data indicate that high PVT1 expression levels in HCC may be
associated with disease progression.
 | Table IIClinicopathological correlation of
PVT1 expression in human HCC (cohort one). |
Table II
Clinicopathological correlation of
PVT1 expression in human HCC (cohort one).
| | Low PVT1
expression | High PVT1
expression | |
|---|
| |
|
| |
|---|
| Factors | Cases, n | n | % | n | % | P-value |
|---|
| Age, years |
| ≤60 | 33 | 5 | 55.56 | 28 | 57.14 | 0.930 |
| >60 | 25 | 4 | 44.44 | 21 | 42.86 | |
| Gender |
| Female | 5 | 0 | 0.00 | 5 | 10.20 | 0.316 |
| Male | 53 | 9 | 100.00 | 44 | 89.80 | |
| HBV |
| Negative | 11 | 1 | 11.11 | 10 | 20.41 | 0.513 |
| Positive | 47 | 8 | 88.89 | 39 | 79.59 | |
| Cirrhosis |
| No | 22 | 4 | 44.44 | 18 | 36.73 | 0.661 |
| Yes | 36 | 5 | 55.56 | 31 | 63.27 | |
| Tumor size, cm |
| ≤5 | 25 | 2 | 22.22 | 23 | 46.94 | 0.169 |
| >5 | 33 | 7 | 77.78 | 26 | 53.06 | |
| Tumor number |
| Single | 52 | 8 | 88.89 | 44 | 89.80 | 0.935 |
| Multiple | 6 | 1 | 11.11 | 5 | 10.20 | |
| PVTT |
| Absent | 54 | 8 | 88.89 | 46 | 93.88 | 0.587 |
| Present | 4 | 1 | 11.11 | 3 | 6.12 | |
| Preoperative AFP
level, ng/ml |
| ≤400 | 34 | 7 | 77.78 | 27 | 55.10 | 0.204 |
| >400 | 24 | 2 | 22.22 | 22 | 44.90 | |
| Histopathological
grade |
| Well +
moderately | 27 | 5 | 55.56 | 22 | 44.90 | 0.556 |
| Poorly | 27 | 4 | 44.44 | 27 | 55.10 | |
| TNM stage |
| I + II | 52 | 7 | 77.78 | 45 | 91.84 | 0.203 |
| III + IV | 6 | 2 | 22.22 | 4 | 8.16 | |
| Milan criteria
(21) |
| Within
criteria | 24 | 2 | 22.22 | 22 | 44.90 | 0.204 |
| Beyond
criteria | 34 | 7 | 77.78 | 27 | 55.10 | |
| UCSF criteria
(22) |
| Within
criteria | 31 | 3 | 33.33 | 28 | 57.14 | 0.188 |
| Beyond
criteria | 27 | 6 | 66.67 | 21 | 42.86 | |
| Hangzhou criteria
(23) |
| Within
criteria | 46 | 6 | 66.67 | 40 | 81.63 | 0.308 |
| Beyond
criteria | 12 | 3 | 33.33 | 9 | 18.37 | |
 | Table IIIClinicopathological correlation of
PVT1 expression in human HCC (cohort two). |
Table III
Clinicopathological correlation of
PVT1 expression in human HCC (cohort two).
| | Low PVT1
expression | High PVT1
expression | |
|---|
| |
|
| |
|---|
| Factor | Cases, n | n | % | n | % | P-value |
|---|
| Age, years |
| ≤60 | 187 | 49 | 85.96 | 138 | 87.90 | 0.707 |
| >60 | 27 | 8 | 14.04 | 19 | 12.10 | |
| Gender |
| Female | 19 | 4 | 7.02 | 15 | 9.55 | 0.564 |
| Male | 195 | 53 | 92.98 | 142 | 90.45 | |
| HBV |
| Negative | 5 | 2 | 3.51 | 3 | 1.91 | 0.494 |
| Positive | 210 | 55 | 96.49 | 154 | 98.09 | |
| Cirrhosis |
| No | 2 | 1 | 1.75 | 1 | 0.64 | 0.453 |
| Yes | 212 | 56 | 98.25 | 156 | 99.36 | |
| Tumor size, cm |
| ≤5 | 122 | 33 | 57.89 | 89 | 56.69 | 0.875 |
| >5 | 92 | 24 | 42.11 | 68 | 43.31 | |
| Tumor number |
| Single | 89 | 28 | 49.12 | 61 | 38.85 | 0.178 |
| Multiple | 125 | 29 | 50.88 | 96 | 61.15 | |
| PVTT |
| Absent | 142 | 40 | 70.18 | 102 | 64.97 | 0.476 |
| Present | 72 | 17 | 29.82 | 55 | 35.03 | |
| Preoperative AFP
level, ng/ml |
| ≤400 | 108 | 37 | 64.91 | 71 | 45.22 | 0.011a |
| >400 | 106 | 20 | 35.09 | 86 | 54.78 | |
| Histopathological
grading |
| Well +
moderately | 110 | 35 | 61.40 | 75 | 47.77 | 0.078 |
| Poorly | 104 | 22 | 38.60 | 82 | 52.23 | |
| TNM stage |
| I + II | 99 | 30 | 52.63 | 69 | 43.95 | 0.260 |
| III + IV | 115 | 27 | 47.37 | 88 | 56.05 | |
| Recurrence |
| No | 100 | 36 | 63.16 | 64 | 40.76 | 0.004a |
| Yes | 114 | 21 | 36.84 | 93 | 59.24 | |
| Milan criteria
(19) |
| Within
criteria | 75 | 24 | 42.11 | 51 | 32.48 | 0.192 |
| Beyond
criteria | 139 | 33 | 57.89 | 106 | 67.52 | |
| UCSF criteria
(20) |
| Within
criteria | 93 | 27 | 47.37 | 66 | 42.04 | 0.487 |
| Beyond
criteria | 121 | 30 | 52.63 | 91 | 57.96 | |
| Hangzhou criteria
(21) |
| Within
criteria | 105 | 31 | 54.39 | 74 | 47.13 | 0.348 |
| Beyond
criteria | 109 | 26 | 45.61 | 83 | 52.87 | |
PVT1 predicts HCC recurrence following
liver transplantation
To determine whether PVT1 could be employed as a
prognostic biomarker for HCC, clinical data of HCC cohort two were
analyzed in detail. Using the cut-off value, the 214 patients were
divided into two groups: A low-expression group (n=57) and a
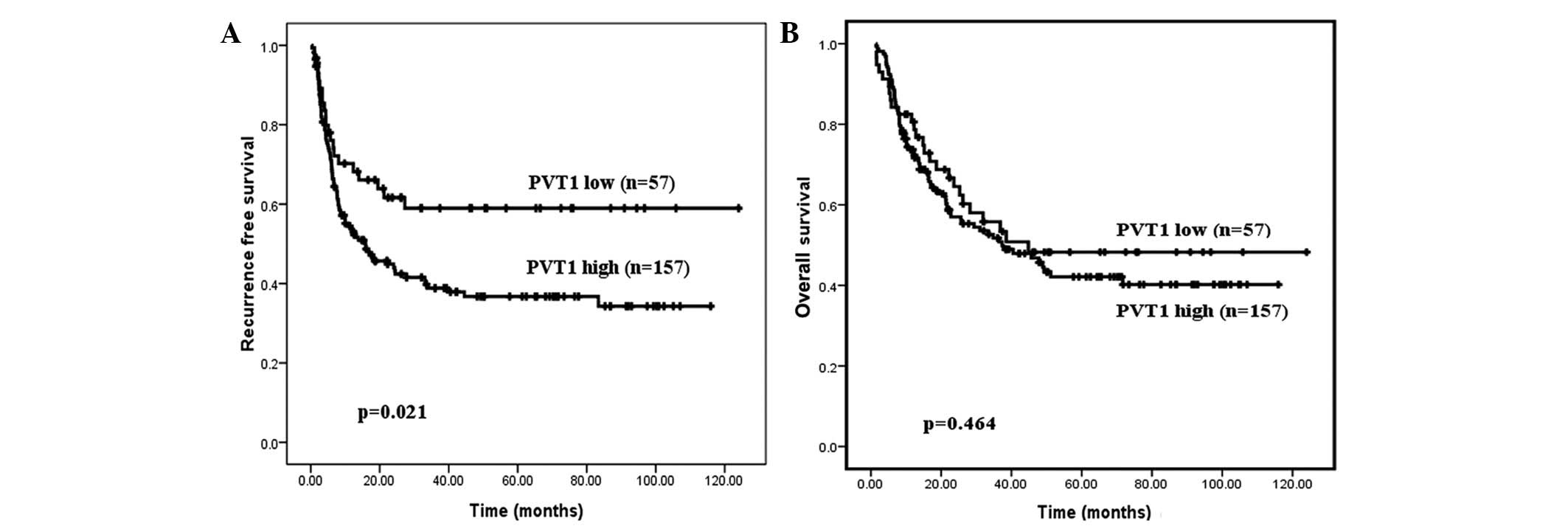
high-expression group (n=157). Kaplan-Meier analysis indicated that
the patients with high PVT1 expression levels had a poor RFS period
(P=0.021; Fig. 4A). Although no
statistical significance of PVT1 expression as a predictor of
overall survival was determined, patients exhibiting high PVT1
expression levels demonstrated a trend for poor prognoses (Fig. 4B; P=0.464).
To identify the risk factors associated with
post-transplant RFS, 11 clinicopathological factors were evaluated
by performing Cox univariate and multivariate analyses. Univariate
analysis demonstrated that the significant prognostic factors for
HCC recurrence were tumor size, tumor number, histopathological
grade, PVTT, preoperative AFP level, TNM stage and PVT1 expression
(all P<0.05). Only tumor size (HR, 2.462; 95% CI, 1.652–3.671;
P<0.001), tumor number (HR, 1.802; 95% CI, 1.194–2.719;
P=0.005), PVTT (HR, 2.075, 95% CI, 1.418–3.037; P<0.001),
preoperative AFP level (HR, 1.539;95% CI, 1.027–2.305; P=0.037) and
PVT1 expression (HR, 1.653; 95% CI, 1.019–2.681; P=0.042) were
identified as independent prognostic factors associated with tumor
recurrence following liver transplantation, as determined by the
Cox multivariate analysis (Table
IV).
 | Table IVCox univariate and multivariate
analysis of predictors of recurrence in hepatocellular carcinoma
patients following liver transplant. |
Table IV
Cox univariate and multivariate
analysis of predictors of recurrence in hepatocellular carcinoma
patients following liver transplant.
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|---|
| Variable for tumor
recurrence | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age, years (>60
vs. ≤60) | 0.669 | 0.349–1.280 | 0.224 | | | |
| Gender (male vs.
female) | 1.577 | 0.768–3.239 | 0.214 | | | |
| HBV | 21.091 | 0.159–2793 | 0.221 | | | |
| Cirrhosis | 1.039 | 0.145–7.443 | 0.970 | | | |
| Tumor size, cm
(>5 vs. ≤5) | 3.431 | 2.340–5.029 | 0.000 | 2.462 | 1.652–3.671 | 0.000a |
| Tumor number
(multiple vs. single) | 2.393 | 1.597–3.582 | 0.000 | 1.802 | 1.194–2.719 | 0.005a |
| Histopathological
grade (poorly vs. well + moderately) | 1.665 | 1.148–2.415 | 0.007 | | | |
| PVTT (present vs.
absent) | 2.826 | 1.947–4.102 | 0.000 | 2.075 | 1.418–3.037 | 0.000a |
| Preoperative AFP
level, ng/ml (>400 vs. ≤400) | 2.380 | 1.622–3.492 | 0.000 | 1.539 | 1.027–2.305 | 0.037a |
| TNM stage (III + IV
vs. I + II) | 4.584 | 2.987–7.034 | 0.000 | | | |
| PVT1 expression
(high vs. low) | 1.738 | 1.082–2.792 | 0.022 | 1.653 | 1.019–2.681 | 0.042a |
Discussion
The complexity of the human transcriptome has been
highlighted by various high-throughput studies (24,25);
≤70% of the genome is transcribed into RNA that does not act as
templates for protein (26). The
non-protein-coding portion of the genome constitutes the vast
majority of genomic information, as well as exhibiting critical
functional roles (27). Improved
understanding of the aberrant expression patterns, cellular
functions and underlying mechanisms of the non-protein-coding
genome may aid in expanding the current understanding of the
complex regulatory network in cancer biology.
Although the majority of previous studies have
focused on short RNAs in cancer research, such as miRNAs, lncRNAs
are gaining prominence. A number of classic lncRNAs have been
implicated in human hepatocarcinogenesis, exhibiting oncogenic or
tumor suppressive roles. One such example of oncogenic lncRNA is
HOTAIR, which was initially identified in foreskin fibroblasts.
HOTAIR resides in the HOX C locus, acting as a modular scaffold to
recruit the polycomb repressive complex 2 to specific target
sequences that ultimately results in the suppression of numerous
genes (28). Our previous study
demonstrated that the expression of HOTAIR is upregulated in HCC
tissues compared with paired non-cancerous tissues, and high
expression levels of HOTAIR were an independent prognostic marker
for HCC recurrence and shorter survival (10). An additional classic oncogenic
lncRNA is MALAT1, which regulated the alternative splicing of a
subset of pre-mRNAs to promote cancer metastasis (29). In our previous study, MALAT1 was
unregulated in HCC and served as a negative prognostic factor for
tumor progression and patient survival (12).
Furthermore, tumor suppressive lncRNAs may affect
various tumor suppressor pathways. For example, MEG3 was identified
to be frequently downregulated in HCC by miR29a-mediated promoter
methylation, and MEG3 inhibited cell growth by functionally
interacting with the p53 signaling pathway (13). Additionally, H19 associated with the
protein complex heterogeneous nuclear ribonucleoprotein
U/P300/cAMP-response element binding protein-associated factor/RNA
polymerase II, epigenetically activating the miR-200 family, and
thus suppressing HCC metastasis (14).
Various reports have presented evidence that PVT1
contributes to cancer pathophysiology. For example, PVT1 was
markedly overexpressed in colorectal (15), ovarian and breast cancers (16). In the current study, PVT1 expression
was initially analyzed in 58 pairs of HCC resection samples (cohort
one), and it was identified that PVT1 was significantly upregulated
in HCC. Subsequently, the PVT1 expression level from an independent
cohort of 214 HCC patients (cohort two) was analyzed. The
upregulation of PVT1 was validated in this independent cohort. The
findings of the present study indicate that PVT1 is predominantly
overexpressed in HCC tissues, regardless of the type of surgical
intervention that the patients undergo. However, the molecular
mechanism underlying the upregulation of PVT1 remains poorly
understood. Guan et al (16)
demonstrated that amplification of chromosome 8q24 increased the
expression of PVT1 in ovarian and breast cancers. Consistent with
this finding, Takahashi et al (15) identified that chromosome 8q24 copy
number gain promoted the expression of PVT1 in colorectal cancer.
Additional studies are required to determine if PVT1 is similarly
regulated in HCC.
Furthermore, the present study identified that PVT1
was more likely to be overexpressed in advanced-stage and
recurrence patients. Correlation analysis indicated that increased
expression of PVT1 was associated with a higher AFP level and a
higher recurrence rate. These data support the hypothesis that PVT1
is associated with disease progression. In addition, survival
analysis demonstrated that the patients with high PVT1 expression
exhibited poor RFS. Multivariate analysis identified that PVT1 was
an independent prognostic factor for RFS. Therefore, data from the
present study indicate that PVT1 may be a novel biomarker for risk
surveillance and adjuvant therapy screening of HCC patients
following liver transplantation. Furthermore, overexpression of
PVT1 may be used by surgeons to identify high-risk patients who may
benefit from preventive strategies as opposed to surgery.
The effects and precise molecular mechanisms
underlying the altered expression of PVT1 in HCC are unclear. Guan
et al (16) demonstrated
that depletion of PVT1 may decrease cell proliferation and increase
cell apoptosis in ovarian and breast cancer cell lines. Similarly,
Takahashi et al (15)
identified that PVT1 knockdown promotes apoptosis in colorectal
cancer cell lines via the TGF-β signaling pathway. Thus, the
detailed mechanism is likely to be complex. Barsotti et al
(30) identified PVT1 as a
p53-inducible target gene. Furthermore, the PVT1 locus produced
numerous spliced non-coding RNAs, as well as six miRNAs, which may
be a molecular switch for cell life and death. Further experiments
are required to elucidate the detailed interplay.
In conclusion, the present study demonstrated that
PVT1 was overexpressed in two independent human HCC cohorts and 10
liver cancer cell lines. Increased expression levels of PVT1 were
associated with a higher AFP level and a higher recurrence rate.
Furthermore, PVT1 served as an independent prognostic factor for
RFS. Thus, the findings of the present study indicate that PVT1 may
act as a novel biomarker for predicting tumor recurrence in HCC
patients and may be a potential therapeutic target.
Acknowledgements
This abstract was presented at the American College
of Surgeons Clinical Congress (October 26–October 30, 2014), San
Francisco, CA, USA, and was published as abstract S23 in the
Journal of the American College of Surgeons 219: S3, 2014. The
present study was supported by grants from the National High
Technology Research and Development Program of China (863 Program),
the Special Fund for Health Research in the Public Welfare, the
Zhejiang Provincial Natural Science Foundation for Young
Distinguished Scholars, the National Science and Technology Major
Project and the Foundation for Innovative Research Groups of the
National Natural Science Foundation of China (grant nos.
2012AA021002, 201302009, R2110125, 2012ZX10002017 and 81121002,
respectively). The authors of the present study would also like to
thank all of the patients enrolled in the study for their
support.
References
|
1
|
Jemal A, Bray F, Center MM, et al: Global
cancer statistics. CA Cancer J Clin. 61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cha C, Fong Y, Jarnagin WR, Blumgart LH
and DeMatteo RP: Predictors and patterns of recurrence after
resection of hepatocellular carcinoma. J Am Coll Surg. 197:753–758.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shah SA, Cleary SP, Wei AC, et al:
Recurrence after liver resection for hepatocellular carcinoma: risk
factors, treatment, and outcomes. Surgery. 141:330–339. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zimmerman MA, Ghobrial RM, Tong MJ, et al:
Recurrence of hepatocellular carcinoma following liver
transplantation: a review of preoperative and postoperative
prognostic indicators. Arch Surg. 143:182–188; discussion 188.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tsai MC, Spitale RC and Chang HY: Long
intergenic noncoding RNAs: new links in cancer progression. Cancer
Res. 71:3–7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang Z, Zhou L, Wu LM, et al:
Overexpression of long non-coding RNA HOTAIR predicts tumor
recurrence in hepatocellular carcinoma patients following liver
transplantation. Ann Surg Oncol. 18:1243–1250. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ishibashi M, Kogo R, Shibata K, et al:
Clinical significance of the expression of long non-coding RNA
HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 29:946–950.
2013.PubMed/NCBI
|
|
12
|
Lai MC, Yang Z, Zhou L, et al: Long
non-coding RNA MALAT-1 overexpression predicts tumor recurrence of
hepatocellular carcinoma after liver transplantation. Med Oncol.
29:1810–1816. 2012. View Article : Google Scholar
|
|
13
|
Braconi C, Kogure T, Valeri N, et al:
microRNA-29 can regulate expression of the long non-coding RNA gene
MEG3 in hepatocellular cancer. Oncogene. 30:4750–4756. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang L, Yang F, Yuan JH, et al:
Epigenetic activation of the MiR-200 family contributes to
H19-mediated metastasis suppression in hepatocellular carcinoma.
Carcinogenesis. 34:577–586. 2013. View Article : Google Scholar
|
|
15
|
Takahashi Y, Sawada G, Kurashige J, et al:
Amplification of PVT-1 is involved in poor prognosis via apoptosis
inhibition in colorectal cancers. Br J Cancer. 110:164–171. 2014.
View Article : Google Scholar :
|
|
16
|
Guan Y, Kuo WL, Stilwell JL, et al:
Amplification of PVT1 contributes to the pathophysiology of ovarian
and breast cancer. Clin Cancer Res. 13:5745–5755. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schlaeger C, Longerich T, Schiller C, et
al: Etiology-dependent molecular mechanisms in human
hepatocarcinogenesis. Hepatology. 47:511–520. 2008. View Article : Google Scholar
|
|
18
|
Ding J, Huang S, Wu S, et al: Gain of
miR-151 on chromosome 8q24.3 facilitates tumour cell migration and
spreading through downregulating RhoGDIA. Nat Cell Biol.
12:390–399. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Edmondson HA and Steiner PE: Primary
carcinoma of the liver: a study of 100 cases among 48,900
necropsies. Cancer. 7:462–503. 1954. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Greene FL, Page DL, Fleming ID, et al:
Liver. AJCC Cancer Staging Manual. 6th edition. Springer; Chicago,
IL: pp. 4352002
|
|
21
|
Mazzaferro V, Regalia E, Doci R, et al:
Liver transplantation for the treatment of small hepatocellular
carcinomas in patients with cirrhosis. N Engl J Med. 334:693–699.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yao FY, Ferrell L, Bass NM, et al: Liver
transplantation for hepatocellular carcinoma: expansion of the
tumor size limits does not adversely impact survival. Hepatology.
33:1394–1403. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zheng SS, Xu X, Wu J, et al: Liver
transplantation for hepatocellular carcinoma: Hangzhou experiences.
Transplantation. 85:1726–1732. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gibb EA, Vucic EA, Enfield KS, et al:
Human cancer long non-coding RNA transcriptomes. PLoS One.
6:e259152011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Prensner JR, Iyer MK, Balbin OA, et al:
Transcriptome sequencing across a prostate cancer cohort identifies
PCAT-1, an unannotated lincRNA implicated in disease progression.
Nat Biotechnol. 29:742–749. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gutschner T and Diederichs S: The
hallmarks of cancer: a long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guttman M and Rinn JL: Modular regulatory
principles of large non-coding RNAs. Nature. 482:339–346. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rinn JL, Kertesz M, Wang JK, et al:
Functional demarcation of active and silent chromatin domains in
human HOX loci by noncoding RNAs. Cell. 129:1311–1323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tripathi V, Ellis JD, Shen Z, et al: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Barsotti AM, Beckerman R, Laptenko O, et
al: p53-dependent induction of PVT1 and miR-1204. J Biol Chem.
287:2509–2519. 2012. View Article : Google Scholar :
|