Introduction
Transitional cell carcinoma (TCC) of the urinary
bladder is the fourth most common solid tumor in males in the
United States, with ~68,810 new cases reported annually in this
region (1), and is the ninth most
common solid cancer worldwide (2).
Smoking tobacco and occupational exposure to chemical carcinogens
have been established as the strongest risk factors for developing
bladder cancer (3). Although
numerous individuals are exposed to these risk factors, only a
proportion develops bladder cancer, indicating that genetic
susceptibility to bladder carcinogenesis may vary in the general
population.
Previously, a genome-wide association study (GWAS)
identified a significant association between the PSCA rs2294008
(C>T) polymorphism and the risk of bladder cancer in US and
European populations (4). A
subsequent study confirmed that this single-nucleotide polymorphism
(SNP) was also associated with bladder cancer risk in the Chinese
population (5). PSCA has been
reported to be highly expressed in bladder cancer and, thus, has
been considered as a useful marker in the diagnosis and progression
of bladder cancer (6). Furthermore,
the clinical use of PSCA in immunotherapeutic strategies has been
evaluated (7,8).
PSCA is a cell surface antigen that belongs to the
Ly-6/Thy-1 family of glycosylphosphatidylinositol-anchored cell
surface antigens (9).
Predominantly, PSCA is prostate-specific and expressed in a subset
of basal and secretary cells of the healthy prostate. PSCA
overexpression has been associated with an increase in tumor grade,
stage, metastasis and recurrence in prostate cancer patients
(10). The PSCA gene, located on
chromosome 8q24.2, encodes a 123-amino acid protein with 30%
homology to the immature lymphocyte cell surface marker SCA type 2.
Furthermore, high PSCA gene expression has been identified in
healthy extra-prostatic tissues, such as the bladder, esophagus,
stomach, pancreas and kidney, as well as in other non-prostatic
malignancies analogous to prostate cancer, including bladder, clear
renal cell, gestational trophoblastic, pancreatic and non-small
cell lung cancer (9).
PSCA protein expression has been reported in the
transitional epithelium of the healthy bladder, and the majority of
superficial and muscle-invasive TCC specimens exhibit high levels
of PSCA expression (6,11). Furthermore, a previous study
revealed that the T allele reduced the transcriptional activity of
the PSCA promoter in vitro (12). Paradoxically, the T risk allele
reduces PSCA transcription, whereas PSCA is overexpressed in
bladder tumors. However, Fu et al (13) confirmed that the T risk allele of
rs2294008 was associated with increased PSCA mRNA expression in
normal and tumorous bladder tissue samples. On the basis of this,
we conducted the present study.
Quantitative polymerase chain reaction (qPCR) is
sensitive enough to detect low-level gene expression and accurate
enough to quantify the full range of expression. The aim of the
present study was to use this method to evaluate PSCA mRNA
expression levels in TCC of the bladder and normal urothelium
specimens, and to determine whether the rs2294008 polymorphism
influences PSCA mRNA expression levels.
Patients and methods
Patients and tissue samples
The present study included 80 specimens of TCC of
the urinary bladder and 38 specimens of normal urothelium from 80
patients who underwent surgery at the Beijing Friendship Hospital
(Beijing, China) from September 2010 to May 2011. All patients were
diagnosed with TCC of the urinary bladder. TCC tumor tissue was
obtained from patients who underwent transurethral resection or
radical cystectomy for bladder cancer, while normal urothelium
samples were obtained from those who underwent radical cystectomy.
Primary TCC tissue samples were obtained from tissue that grossly
and clearly appeared to comprise a tumor of the urinary bladder.
Tumor tissue samples from the same areas of tumor specimens and
normal mucosa samples from patients undergoing radical cystectomy
were stored in liquid nitrogen and confirmed by a senior
pathologist of the Beijing Friendship Hospital. Tumor samples
exhibiting marked inflammation or necrosis were excluded from
further analysis. Tumors were staged according to International
Union Against Cancer (14) and
graded histologically according to the World Health Organization
classification system (15). None
of the patients had received previous intravenous chemotherapy or
radiation. Patient and tumor characteristics, and the clinical
outcome, are summarized in Table I.
Informed consent for this study was obtained from all patients, and
the study was approved by the Research Ethics Committee of the
Capital Medical University.
 | Table IPatient and tumor characterics, and
clinical outcome (mean patient age at surgery, 68.9 years; range,
48–92 years). |
Table I
Patient and tumor characterics, and
clinical outcome (mean patient age at surgery, 68.9 years; range,
48–92 years).
| Variable | Patients, n (%)
(n=80) |
|---|
| Gender |
| Male | 66 (82.5%) |
| Female | 14 (17.5%) |
| Tumor stage |
| pT1 | 54 (67.5%) |
| ≥pT2 | 26 (32.5%) |
| Histological
grade |
| G1-2 | 56 (70.0%) |
| G3 | 24 (30.0%) |
| Surgical
intervention |
| Transurethral
resection | 42 (52.5%) |
| Radical
cystectomy | 38 (47.5%) |
DNA extraction and genotyping
Genomic DNA was isolated from tumor tissue using a
DNA extraction kit (QIAamp DNA mini kit; Qiagen, Valencia, CA, USA)
according to the manufacturer’s instructions. PCR was performed to
amplify the DNA, using PSCA rs2294008-specific primers, synthesized
at the Biosune Biotechnology Co., Ltd. (Shanghai, China). The
primer sequences were as follows: Sense,
5′-AAACCCGCTGGTGTTGACTGTG-3′ and anti-sense,
5′-CATCTCTGCCCATCCATCCGT-3′, producing a 459-bp product. The
samples were amplified using the PCR Amplification kit (Takara
Biotechnology Co., Ltd., Dalian, China), and each sample was
prepared to a final volume of 20 μl, containing 1X PCR buffer, 4.0
mM MgCl2, 4.0 mM each deoxynucleotide, 0.4 μl each
primer, 0.1 μl Takara Ex Taq polymerase (Takara Biotechnology Co.,
Ltd.) and 1–10 ng genomic DNA. PCR was performed under the
following conditions: Denaturation at 94°C for 5 min; 35 cycles of
denaturation at 95°C for 30s, annealing at 55°C for 30s and
extension at 72°C for 30s; and a final extension at 72°C for 2 min
in a Mastercycler® ep gradient thermocycler (Eppendorf,
Fujian, China). Amplicons of 459 bp were analyzed by
electrophoresis in 3% agarose gel and visualized using the
Molecular Imager® Gel Doc™ XR+ system with Quantity
One® 1-D analysis software (Bio-Rad Laboratories,
Hercules, CA, USA) following staining with ethidium bromide. The
rs2294008 C>T polymorphism of PSCA was genotyped using
sequencing by Biosune Biotechnology Co., Ltd.
RNA isolation and RT reaction
Total cellular RNA was extracted from the tissue
samples using an RNeasy Mini kit (Qiagen) according to the
manufacturer’s instruction. Purified total RNA was assessed for
purity by measuring the absorbance at 260 and 280 nm using the
DU730 Nucleic Acid/Protein Analyzer (Beckman Coulter, Inc., Brea,
CA, USA), and stored at −80°C prior to analysis. Total RNA was
reverse-transcribed using a SuperScript® III First
Strand kit (Invitrogen, Carlsbad, CA, USA), according to the
manufacturer’s instructions.
qPCR for PSCA mRNA
To examine the mRNA expression levels of the target
gene (PSCA) and the housekeeping gene (GAPDH), qPCR was performed
using the Mastercycler ep realplex (Eppendorf) and SYBR®
Green I dye (Toyobo Co., Ltd., Osaka, Japan). The primer sequences
were as follows: Sense, 5′-AAAGCCCAGGTGAGCAACGAG-3′ and anti-sense,
5′-CTGTGAGTCATCCACGCAGTTC-3 for PSCA, producing a 147-bp product;
and sense, 5′-TCAAGATCATCAGCAATGCC-3′ and anti-sense
5′-TGTGGTCATGAGTCCTTCCA-3′ for GAPDH, producing a 100-bp product.
Each PCR reaction (final volume, 10 μl) contained 1 μl
complementary DNA, 0.6 μl primers and 4 mM MgCl2. The
thermal cycling conditions for the PCR were as followed:
Denaturation at 95°C for 5 min; and 40 cycles of denaturation at
95°C for 15 sec, annealing at 56.4°C for 20 sec and extension at
72°C for 20 sec. The PSCA and GAPDH sequences were amplified in
duplicate from the tissue samples. To ensure that the correct
product was amplified, all of the samples were separated by 3%
agarose gel electrophoresis. The quantity of the target was
calculated as the ratio of target gene to reference gene copies, to
obtain a normalized value. A separate standard melting point curve
for PSCA and GAPDH was constructed using a serial dilution gradient
(range, 1/41–1/45) to determine the
amplification efficiency. To verify that the melting curve results
were correct, representative PCR products were sequenced.
Statistical analysis
The PSCA rs2294008 allele frequency was calculated
as: [1 × (h + 2H)]/2n, where ‘h’ represents the heterozygous
genotype, ‘H’ the homozygous genotype and ‘n’ the sample size for
each population. The differences between PSCA mRNA expression and
various clinicopathological variables, such as stage and grade,
were assessed using the Mann-Whitney U test. Values were presented
as the median and the upper and lower quartiles (P75 and P25,
respectively), and P<0.05 was considered to indicate a
statistically significant difference. All statistical analyses were
performed using SPSS software (version 16; SPSS, Inc., Chicago, IL,
USA).
Results
Genotypes of rs2294008 and identification
of the amplified product
The allele frequencies of rs2294008 C>T are
demonstrated in Table II. Gel
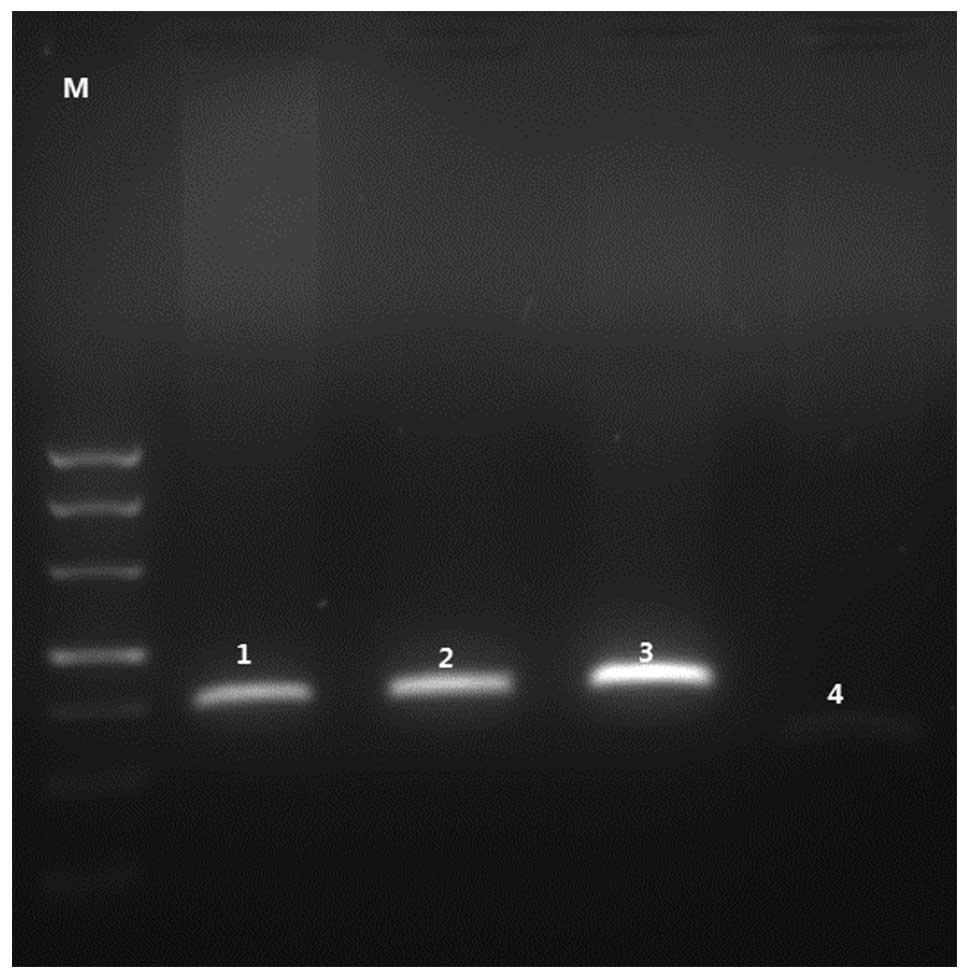
electrophoresis showed 147-bp PSCA and 100-bp GAPDH products
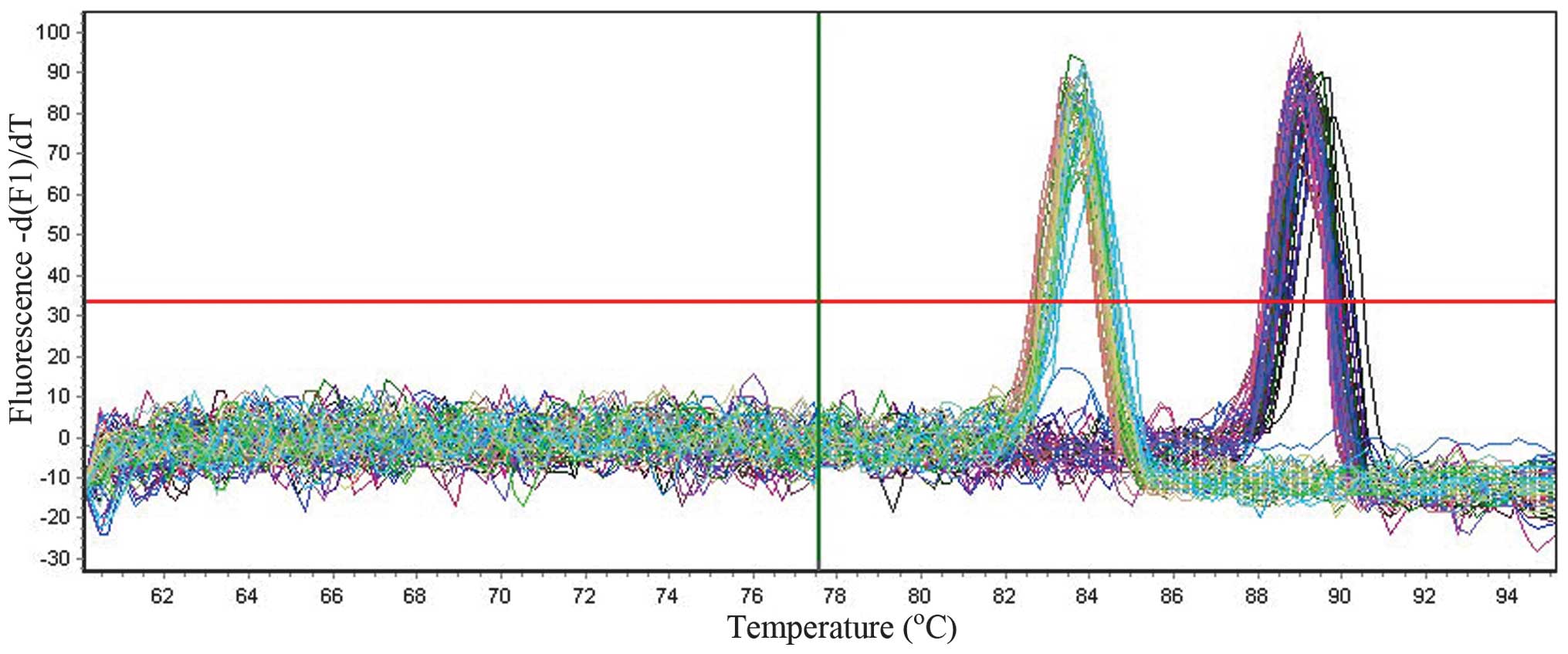
(Fig. 1), and the melting curves
for all of the PCR products only produced one sharp peak for PSCA
and GAPDH, respectively (Fig. 2),
indicating that the specific PCR products were amplified.
Representative samples were sequenced to verify the correct
products of PSCA and GAPDH.
 | Table IIGenotypic and allelic frequencies for
the prostate stem cell antigen rs2294008 allele T and C in
transitional cell carcinoma of the 80 urinary bladder patients. |
Table II
Genotypic and allelic frequencies for
the prostate stem cell antigen rs2294008 allele T and C in
transitional cell carcinoma of the 80 urinary bladder patients.
| Variable | Patients, n (%) |
|---|
| Genotype |
| CC | 57.50 |
| TC | 32.50 |
| TT | 10.00 |
| Allele |
| C | 73.75 |
| T | 26.25 |
Amplification efficiency of PSCA and
GAPDH
PSCA mRNA expression was detected in all samples
(100%). The amplification products of PSCA and GAPDH were diluted
in concentration gradients (range, 1/41–1/45)
and amplified. The Ct values obtained were used to construct the
standard melting curves. The Pfaffl method was used to calculate
the amplification efficiencies of PSCA and GAPDH, which were 106.85
and 109.87%, respectively (16).
Differences in PSCA mRNA expression
The PSCA mRNA expression levels in bladder cancer
and normal urothelium tissues were normalized using GAPDH mRNA.
RT-qPCR data was analyzed using the comparative Ct method. The
following formula was applied: ΔCt = CtPSCA -
CtGAPDH, and the resulting 2−ΔCt data
exhibited a non-normal distribution. Therefore, the nonparametric,
two independent samples, Mann-Whitney U test was performed.
Quantification of PSCA mRNA expression revealed a
significantly greater expression of PSCA in TCC specimens compared
with normal urothelium (P=0.038), and a significantly greater
expression in G3 compared with G1-2 tumors of the bladder
(P=0.001). No significant difference was identified in PSCA mRNA
expression between patients exhibiting T1 and ≥pT2 tumors
(P=0.250). Furthermore, T allele carriers demonstrated higher
levels of PSCA mRNA expression compared with CC homozygous patients
(P=0.001) (Table III).
 | Table IIIRelative prostate stem cell antigen
mRNA expression levels in various samples from 80 patients
diagnosed with TCC of the urinary bladder. |
Table III
Relative prostate stem cell antigen
mRNA expression levels in various samples from 80 patients
diagnosed with TCC of the urinary bladder.
| Category | Samples, n | Median (P25,
P75) | P-value |
|---|
| Tissue type | | | 0.038 |
| Healthy | 38 | 0.12 (0.03,
0.54) | |
| TCC | 80 | 0.22 (0.11,
0.84) | |
| Pathological
grade | | | 0.001 |
| G1-2 | 56 | 0.25 (0.10,
0.47) | |
| G3 | 24 | 1.35 (0.69,
5.04) | |
| Pathological
stage | | | 0.250 |
| T1 | 54 | 0.22 (0.10,
0.47) | |
| ≥pT2 | 26 | 1.29 (0.36,
4.28) | |
| Genotype | | | 0.001 |
| CC homozygous | 46 | 0.20 (0.10,
0.26) | |
| T carrier | 34 | 0.48 (0.17,
1.59) | |
Discussion
Various studies have reported the potential
importance of PSCA as a cell-surface antigen in the diagnosis and
treatment of prostate, bladder, gastric and pancreatic cancers
(6,17–22).
Elsamman et al (6) reported
that PSCA mRNA was expressed less in superficial TCC of the bladder
in patients with disease recurrence compared with patients
exhibiting no recurrence. Cheng et al (23) measured PSCA protein expression in
urine samples, and demonstrated its possible application as a
cytological marker of urothelial carcinoma via immunocytochemical
analysis of urine. Subsequently, Cheng et al (11) used quantum dot-based technology to
detect the levels of PSCA protein expression in human TCC, and
revealed that PSCA expression correlated with tumor stage and
grade. Furthermore, Kohaar et al (21) proposed that anti-PSCA immunotherapy
may be beneficial for the treatment of bladder cancer patients with
high tumor PSCA expression.
Numerous GWAS studies have been conducted to
identify susceptibility loci of bladder cancer (4,24,25).
One such study identified a significant association between the
PSCA rs2294008 (C>T) polymorphism and the risk of bladder cancer
in US and European populations (4).
Ma et al (5) identified that
this SNP was also associated with bladder cancer risk in the
Chinese population. Notably, HapMap data indicates that the T
allele is present at a higher frequency in individuals of European
[minor allele frequency (MAF), 0.46] and Korean (MAF, 0.46)
ancestry compared with Chinese (MAF, 0.26) and African (MAF, 0.25)
populations (4). However, the T
allele is a major allele in Japanese populations (MAF, 0.62). The
population history of this SNP and the reason for only the Japanese
population possessing a different minor allele is yet to be
elucidated (4). rs2294008 is a
missense SNP located in exon 1 of the PSCA gene (4). Linkage disequilibrium (LD) analysis of
all HapMap SNPs in the vicinity of rs2294008 showed that it maps to
an 11-kb LD block on chromosome 8q24 (4). Previously, an in vitro analysis
of gastric cell lines determined that the T risk allele of
rs2294008 was associated with a significant reduction in the
transcriptional activity of the PSCA promoter (12). However, more recently, Fu et
al (13) determined that the T
risk allele was associated with increased PSCA mRNA expression
levels in bladder tumor samples and normal bladder samples.
PSCA is overexpressed in bladder tumors. However, to
date, few studies have been conducted to explain the association
between the PSCA rs2294008 (C>T) polymorphism and mRNA
expression in tumor tissue from TCC of the urinary bladder. The
predominant aim of the present study was to evaluate the
association between PSCA mRNA expression levels and rs2294008
(C>T) polymorphism and various clinicopathological features,
including tumor stage and grade, and to determine whether the
rs2294008 (C>T) polymorphism influences PSCA mRNA expression
levels.
In the present study, PSCA mRNA was expressed at a
significantly higher level in the tumor tissue of T allele carriers
compared with CC homozygous patients (P=0.038). A previous study
demonstrated that rs2294008 altered the length of the PSCA
N-terminal signal peptide, which may affect protein folding,
intracellular modifications and/or trafficking of PSCA proteins
(4). However, the physiological
function of PSCA, the impact of different N-terminal signal lengths
on protein function and the in vivo functional consequence
of the T risk allele remain unclear. Rs2294008 was considered to be
an independent bladder cancer susceptibility locus on 8q24
(4). Additional studies are
required to fully elucidate the physiological function of PSCA and
the biological mechanisms underlying the association of the PSCA
rs2294008 (C>T) polymorphism with bladder carcinogenesis.
In agreement with Amara et al (26), the present study identified a higher
level of PSCA mRNA expression in TCC compared with normal
urothelium. However, Bahrenberg et al (27) determined that PSCA expression levels
were reduced in bladder cancer tumors and identified that PSCA
expression was greater in confluent RT112 cells compared with
non-confluent cells. In addition, Bahrenberg et al (27) reported that, in RT112 cells, PSCA
expression is dependent on cell-cell contact and surface adhesion.
In agreement with this, Elsamman et al (6) identified that confluent HT1376 TCC
cells exhibited three-fold higher PSCA expression levels compared
with the scattered (non-confluent) HT1376 cells. Therefore, the
function of PSCA may be associated with the adhesion of cells,
indicating that the expression level of PSCA correlates with tumor
invasiveness. However, Amara et al (26) and Elsamman et al (6) reported that PSCA expression was
inversely correlated with tumor stage. Instead, the greatest mean
level of PSCA expression occurred in cases of superficial (Ta or
T1) tumors, while lower PSCA expression levels occurred in cases of
muscle-invasive (≥pT2) tumors. In the present study, no significant
difference was identified between superficial (T1) and
muscle-invasive (≥pT2) tumors (P=0.250), and patients with G3
tumors exhibited significantly higher PSCA mRNA expression levels
compared with patients exhibiting G1-2 tumors (P=0.001). Research
conducted by Kohaar et al (21) demonstrated a similar level of PSCA
expression in non-muscle-invasive tumors, stages Ta and T1, and
muscle-invasive tumors, stages T2 and T3/4. Furthermore, Elsamman
et al (6) proposed that PSCA
expression level is a predictor of disease recurrence in patients
exhibiting superficial TCC of the urinary bladder, independent of
tumor stage or grade.
In conclusion, the present study demonstrated that
PSCA mRNA was more highly expressed in T allele carriers compared
with CC homozygous patients, and PSCA mRNA expression was
associated with TCC and tumor histological grade. Additional
studies are required to define the physiological role of PSCA and
the biological mechanisms underlying the association of the PSCA
rs2294008 (C>T) polymorphism with bladder carcinogenesis.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81241079) and the
Beijing Natural Scientific Foundation (grant no.7122043).
References
|
1
|
Jemal A, Siegel R, Ward E, et al: Cancer
statistics, 2008. CA Cancer J Clin. 58:71–96. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Murray T, Ward E, et al: Cancer
statistics, 2005. CA Cancer J Clin. 55:10–30. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Corral R, Lewinger JP, Van Den Berg D, et
al: Comprehensive analyses of DNA repair pathways, smoking, and
bladder cancer risk in Los Angeles and Shanghai. Int J Cancer.
135:335–347. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu X, Ye Y, Kiemeney LA, et al: Genetic
variation in the prostate stem cell antigen gene PSCA confers
susceptibility to urinary bladder cancer. Nat Genet. 41:991–995.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ma Z, Hu Q, Chen Z, et al: Systematic
evaluation of bladder cancer risk-associated single-nucleotide
polymorphisms in a Chinese population. Mol Carcinog. 52:916–921.
2013. View
Article : Google Scholar
|
|
6
|
Elsamman E, Fukumori T, Kasai T, et al:
Prostate stem cell antigen predicts tumour recurrence in
superficial transitional cell carcinoma of the urinary bladder. BJU
Int. 97:1202–1207. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang X, Guo Z, Liu Y, et al: Prostate stem
cell antigen and cancer risk, mechanisms and therapeutic
implications. Expert Rev Anticancer Ther. 14:31–37. 2014.
View Article : Google Scholar
|
|
8
|
Marra E, Uva P, Viti V, et al: Growth
delay of human bladder cancer cells by Prostate Stem Cell Antigen
downregulation is associated with activation of immune signaling
pathways. BMC Cancer. 10:1292010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Saeki N, Gu J, Yoshida T and Wu X:
Prostate stem cell antigen: a Jekyll and Hyde molecule? Clin Cancer
Res. 16:3533–3538. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lam JS, Yamashiro J, Shintaku IP, et al:
Prostate stem cell antigen is overexpressed in prostate cancer
metastases. Clin Cancer Res. 11:2591–2596. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cheng F, Yu W, Zhang X and Ruan Y:
Quantum-dot-based technology for sensitive and stable detection of
prostate stem cell antigen expression in human transitional cell
carcinoma. Int J Biol Markers. 24:271–276. 2009.
|
|
12
|
Study Group of Millennium Genome Project
for Cancer. Sakamoto H, Yoshimura K, Saeki N, et al: Genetic
variation in PSCA is associated with susceptibility to diffuse-type
gastric cancer. Nat Genet. 40:730–740. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fu YP, Kohaar I, Rothman N, et al: Common
genetic variants in the PSCA gene influence gene expression and
bladder cancer risk. Proc Natl Acad Sci USA. 109:4974–4979. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sobin LH, Gospodariwicz M and Wittekind C:
TNM Classification of Malignant Tumors. UICC International Union
Against Cancer. 7th edition. Wiley-Blackwell; Hoboken, NJ: pp.
262–265. 2005
|
|
15
|
Sauter G, Algaba F, Amin M, et al: Tumours
of the urinary system: non-invasive urothelial neoplasias. WHO
Classification of Tumours: Pathology and Genetics of Tumours of the
Urinary System and Male Genital Organs. Eble JN, Sauter G, Epstein
JI and Sesterhenn I: IARCC Press; Lyon: 2004
|
|
16
|
Chini V, Foka A, Dimitracopoulos G and
Spiliopoulou I: Absolute and relative real-time PCR in the
quantification of tst gene expression among methicillin-resistant
Staphylococcus aureus: evaluation by two mathematical models. Lett
Appl Microbiol. 45:479–484. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Barbisan F, Mazzucchelli R, Santinelli A,
et al: Expression of prostate stem cell antigen in high-grade
prostatic intraepithelial neoplasia and prostate cancer.
Histopathology. 57:572–579. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yagn WB, Cai F, Cheng CT, et al:
Expression of prostate stem cell antigen and Claudin-4 in human
pancreatic carcinoma. Zhongguo Yi Xue Ke Xue Yuan Xue Bao.
30:728–731. 2008.(In Chinese).
|
|
19
|
Antonarakis ES, Carducci MA, Eisenberger
MA, et al: Phase I rapid dose-escalation study of AGS-1C4D4, a
human anti-PSCA (prostate stem cell antigen) monoclonal antibody,
in patients with castration-resistant prostate cancer: a PCCTC
trial. Cancer Chemother Pharmacol. 69:763–771. 2012. View Article : Google Scholar
|
|
20
|
Krupa M, Canamero M, Gomez CE, Najera JL,
Gil J and Esteban M: Immunization with recombinant DNA and modified
vaccinia virus Ankara (MVA) vectors delivering PSCA and STEAP1
antigens inhibits prostate cancer progression. Vaccine.
29:1504–1513. 2011. View Article : Google Scholar
|
|
21
|
Kohaar I, Porter-Gill P, Lenz P, et al:
Genetic variant as a selection marker for anti-prostate stem cell
antigen immunotherapy of bladder cancer. J Natl Cancer Inst.
105:69–73. 2013. View Article : Google Scholar :
|
|
22
|
Sala N, Muñoz X, Travier N, et al:
Prostate stem-cell antigen gene is associated with diffuse and
intestinal gastric cancer in Caucasians: results from the
EPIC-EURGAST study. Int J Cancer. 130:2417–2427. 2012. View Article : Google Scholar
|
|
23
|
Cheng L, Reiter RE, Jin Y, et al:
Immunocytochemical analysis of prostate stem cell antigen as
adjunct marker for detection of urothelial transitional cell
carcinoma in voided urine specimens. J Urol. 169:2094–2100. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kiemeney LA, Thorlacius S, Sulem P, et al:
Sequence variant on 8q24 confers susceptibility to urinary bladder
cancer. Nat Genet. 40:1307–1312. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rothman N, Garcia-Closas M, Chatterjee N,
et al: A multi-stage genome-wide association study of bladder
cancer identifies multiple susceptibility loci. Nat Genet.
42:978–984. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Amara N, Palapattu GS, Schrage M, et al:
Prostate stem cell antigen is overexpressed in human transitional
cell carcinoma. Cancer Res. 61:4660–4665. 2001.PubMed/NCBI
|
|
27
|
Bahrenberg G, Brauers A, Joost HG and
Jakse G: Reduced expression of PSCA, a member of the LY-6 family of
cell surface antigens, in bladder, esophagus, and stomach tumors.
Biochem Biophys Res Commun. 275:783–788. 2000. View Article : Google Scholar : PubMed/NCBI
|