Introduction
Breast cancer is the leading cause of cancer-related
mortality in females, worldwide (1). In the previous two decades, the
incidence rate of breast cancer has increased at an average rate of
3.1% per year and the mortality rate has increased at an average
rate of 1.8% per year (2).
Chemotherapy is an important adjuvant systemic therapeutic approach
for the successful treatment of breast cancer (3) and, during early-stage breast cancer,
has been demonstrated to improve survival rate (4).
MicroRNAs (miRNAs) have previously been identified
as important regulators of a number of key genes associated with
chemoresistance (5,6). miRNAs are a class of endogenous, 18 to
25-nucleotide long, non-coding RNAs, which regulate gene expression
at the post-transcription level (7,8). As a
recognition mechanism, miRNAs complementarily pair to the
3′-untranslated region (UTR) of their target mRNAs, resulting in
decreased translational efficiency and/or decreased mRNA expression
levels (9–11). It has previously been reported that
miRNAs commonly deregulate gene expression levels in specific types
of human cancer, and may serve as oncogenes or tumor suppressors
(12,13). However, dysregulated miRNAs appear
to be associated with every aspect of the cancer-related biological
process, including tumor progression, invasion and metastasis, as
well as the acquisition of resistance to various chemotherapeutic
agents (14,15). Previous studies have indicated that
single nucleotide polymorphisms (SNPs) occurring in or near miRNA
binding sites may be associated with tumor susceptibility and
chemotherapeutic response in humans (6,17–19).
B-cell lymphoma-extra large (Bcl-xL) belongs to the
Bcl-2 protein family and appears to confer resistance to apoptosis,
thereby reducing the effectiveness of chemotherapy (20). It has previously been reported that
overexpression of Bcl-2 and Bcl-xL increases resistance to totaxol
and etoposide administration in MCF-7 cells (21), whereas their downregulation
sensitizes MCF-7 and MDA-MB-231 cells to doxorubicin, paclitaxel
and cyclophosphamide administration (22). In the present study, we hypothesized
that SNPs located in let-7b binding sites of the Bcl-xL gene 3′-UTR
may regulate Bcl-xL expression, thus, increasing cellular
resistance to chemotherapeutic agents in breast cancer cells. To
investigate this hypothesis, bioinformatic analyses were performed
to identify SNPs in the 3′-UTR of the Bcl-xL gene. We then
functionally validated SNP rs3208684 A>C, which is located in
the let-7b binding site in the 3′UTR of the Bcl-xL gene.
Materials and methods
SNP selection
To predict putative miRNA binding sites in the
Bcl-xL 3′-UTR, microrna.org (http://www.microrna.org/microrna/home.do), PicTar
(http://pictar.mdc-berlin.de/cgi-bin/PicTar_vertebrate.cgi)
and TargetScan version 6.2 (http://www.targetscan.org/) were used. Furthermore,
the National Center for Biotechnology Information SNP database
(dbSNP; http://www.ncbi.nlm.nih.gov/SNP) was used to identify
SNPs within putative miRNA target sites in the 3′-UTR of Bcl-xL.
The search was focused on the miRNA seed region, as the seed
sequence nucleates interaction between the miRNA and the
complementary Bcl-xL mRNA target region, and is the predominant
determinant for successful miRNA targeting.
Cell culture and transfections
The human breast cancer cell line, MCF-7 (Cell Bank
of the Chinese Academy of Sciences, Shanghai, China) was cultured
in Dulbecco’s modified Eagle medium (HyClone Laboratories, Inc.,
Logan, UT, USA) with 10% fetal bovine serum (HyClone Laboratories,
Inc.) at 37°C and 5% CO2. Upon reaching 70% confluence,
the MCF-7 cells (5×103 cells per well) were transfected
with let-7b mimics or control miRNA mimics [normal control (NC)]
(GenePharma Co., Ltd, Shanghai, China), or wild-type Bcl-xL
(WT-Bcl-xL), mutant Bcl-xL (Mut-Bcl-xL) or vector alone (pcDNA3.1)
(Invitrogen Life Technologies, Shanghai, China) using
Lipofectamine® 2000 transfection reagent (Invitrogen
Life Technologies, Carlsbad, CA, USA), according to the
manufacturer’s instructions.
Luciferase reporter assay
To conduct the luciferase reporter assay, HEK293T
cells (Land Co., Ltd, Guangzhou, China) were seeded onto 24-well
plates (2×104 cells per well). In each well, HEK293T
cells were transfected with 0.5 μg Bcl-xL 3′-UTR luciferase
reporter plasmids containing A or C alleles (Land Co., Ltd) and 100
nM let-7b mimics, let-7b inhibitor or NC inhibitor using
Lipofectamine 2000 (Invitrogen Life Technologies), according to the
manufacturer’s instructions. At 48 h post-transfection, the cell
lysates were collected and a dual luciferase reporter assay system
was used to measure firefly and Renilla luciferase activity
(Promega Corporation, Madison, WI, USA); relative luciferase
activity was calculated by normalizing the Renilla luciferase
activity to the firefly luciferase activity.
Western blot analysis
The cell lysates were separated on 10% SDS-PAGE and
transferred to polyvinylidene fluoride membranes (EMD Millipore
Co., Hayward, CA, USA). The membranes were blocked with 5% skimmed
milk for 1 h and incubated with rabbit monoclonal anti-Bcl-xL (cat.
no. 2764), anti-Bcl-2-associated X protein (Bax) (cat. no. 5023)
and anti-β-actin (cat. no. 4970) (Cell Signaling Technology, Inc.,
Danvers, MA, USA) antibodies at dilutions of 1:1,000 overnight at
4°C, respectively. Subsequently, the membranes were incubated with
a polyclonal goat anti-rabbit horseradish peroxidase-conjugated
secondary antibody (dilution, 1:4,000; cat. no. 7074; Cell
Signaling Technology, Inc.) for 1 h. Finally, enhanced
chemiluminescence (Enhanced Chemiluminescence Western Blotting kit;
Amersham Biosciences, Piscataway, NJ, USA) was used to visualize
the results and β-actin was used as internal control.
3-(4,
5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
Cell viability was detected by performing an MTT
(Sigma Aldrich, St. Louis, MO, USA) assay. MCF-7 cells were
transfected prior to treatment with 0, 6, 60, 600, 6,000 and 60,000
μM 5-fluorouracil (5-FU) (Sigma-Aldrich) or 0.0, 0.25, 0.5, 1.0,
2.0 and 4.0 μM doxorubicin (ADM) (Sigma-Aldrich) for 48 h.
Following a 4-h re-incubation with 10% MTT solution, 150 μl DMSO
(Sigma-Aldrich) was added to solubilize the resultant formazan
crystals. The absorbance of the plate was measured in a microplate
reader reader (ELX-800, Bio-Tek Instruments, Inc., Winooski, VT,
USA) at a wavelength of 570 nm, with a reference wavelength of 650
nm, and the results were expressed as the percentage of absorbance
relative to the untreated controls.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism software (version 5.0; GraphPad Software Inc., La Jolla, CA,
USA) and Student’s t-test. Data are expressed as the mean ±
standard deviation and P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification of let-7b SNP binding
sites in the Bcl-xL 3′-UTR
To identify possible miRNA binding sites in the
3′-UTR of the Bcl-xL gene, bioinformatic analysis was performed
using three online prediction programs (PicTar, TargetScan and
microrna.org). According to the putatively identified miRNA binding
sites combined with information from the dbSNP database, it was
identified that the Bcl-xL 3′-UTR SNP rs3208684 A>C is located
within a predicted miRNA binding site for let-7b (Fig. 1). These results indicate that the C
allele forms a non-perfect pairing with the let-7b miRNA seed and,
thus, may escape let-7b-mediated regulation.
Let-7b negatively regulates the protein
expression levels of Bcl-xL and sensitizes MCF-7 cells to 5-FU and
ADM
Bioinformatic analysis identified a potential
binding site of let-7b in the 3′-UTR of the Bcl-xL gene and a
dual-luciferase reporter was performed to determine whether let-7b
binds at this putative binding site. The full length Bcl-xL 3′-UTR
was cloned into the psiCHECK-2 vector (Fig. 2A) and this psiCHECK2-WT-Bcl-xL
3′-UTR vector was subsequently cotransfected into HEK293T cells
with let-7b mimics, NC, let-7b inhibitor or inhibitor NC. As
indicated in Fig. 2B, luciferase
activity was significantly suppressed in the presence of let-7b
mimics compared with the NC (P<0.01), whereas the luciferase
activity displayed no significant difference in cotransfection rate
between the let-7b inhibitor and NC inhibitor groups
(P>0.05).
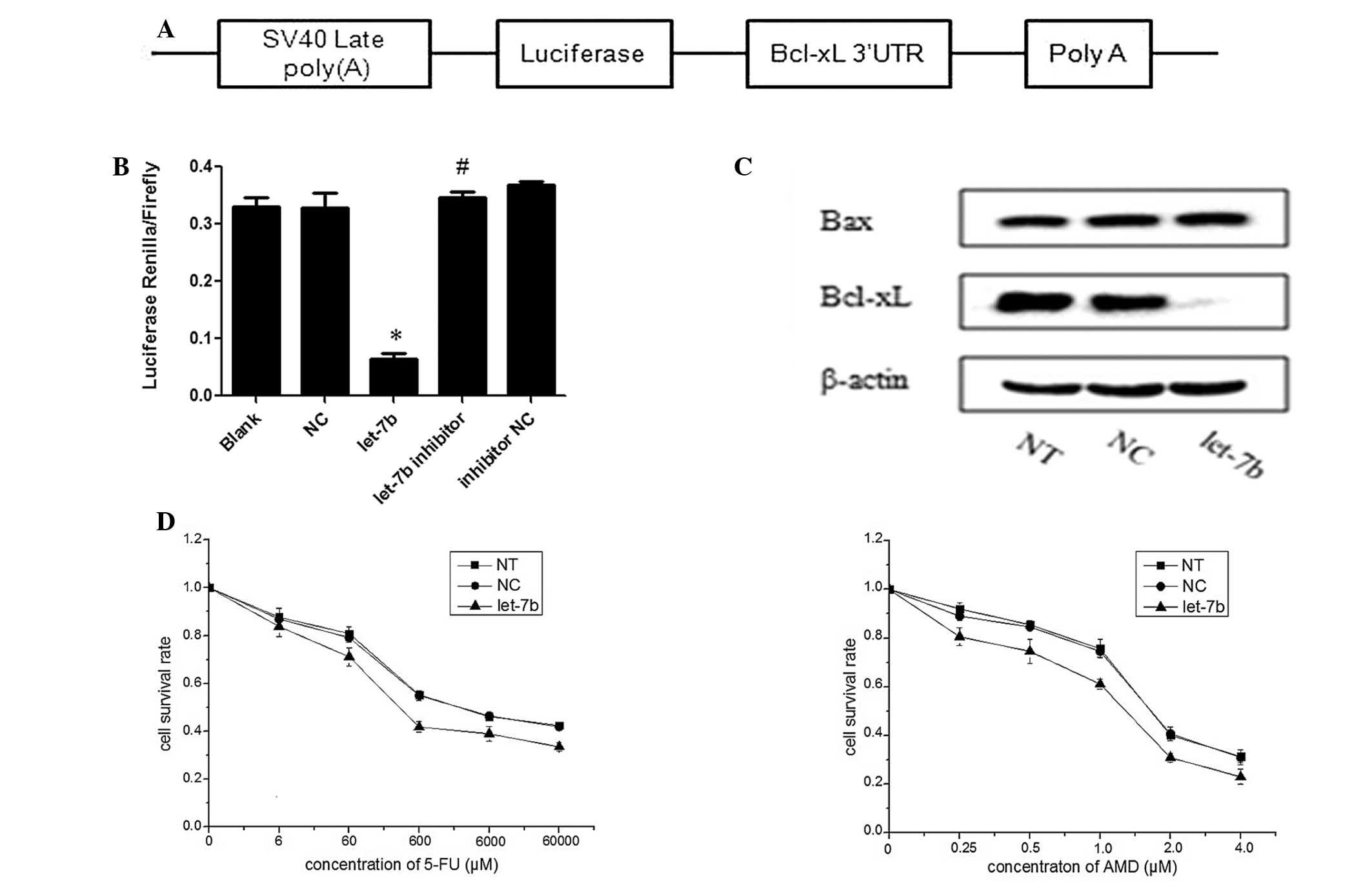 | Figure 2Let-7b negatively regulates the
expression of Bcl-xL and sensitizes MCF-7 cells to 5-FU and ADM.
(A) Bcl-xL 3′-UTR was cloned into the psiCHECK-2 vector. (B)
Luciferase reporter plasmids containing the wild-type Bcl-xL 3′-UTR
were transfected in HEK293T cells with 100 nM let-7b, NC, let-7b
inhibitor or inhibitor NC and luciferase expression was measured 48
h after transfection. The data are represented as the mean ±
standard deviation (SD) of a minimum of three independent
transfections analyses. *P<0.01 vs. NC;
#P>0.05 vs. inhibitor NC. (C) Let-7b mimics were
transfected into MCF-7 cells, and, 48 h after transfection, cells
lysates were prepared and subjected to western blot analysis. NT
and NC were used as negative controls, and the data are represented
as the mean ± SD of three independent analyses. (D) MCF-7 cells
were transfected with let-7b mimics or NC and treated with
different concentrations of 5-FU or ADM. *P<0.05 vs.
NC. The cell survival rate was determined by performing an MTT
assay and the data are represented as the mean ± SD of a minimum of
three independent transfections analyses. ADM, doxorubicin; Bcl,
B-cell lymphoma-extra large; FU, fluorouracil; let-7b, let-7b
mimic; MTT, 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
bromide; NC, normal control (microRNA mimic); NT, non-transfected
cells; SV40 late poly(A), Simian virus 40 late polyadenylation (A);
UTR, untranslated region. |
To exert their function, miRNAs suppress the
expression of their target genes (23). To verify whether Bcl-xL is a target
of the miRNA let-7b, Bcl-xL protein expression levels were analyzed
in response to enforced expression of let-7b in MCF-7 cells.
Following transfection with let-7b mimics or NC, MCF-7 cells were
analyzed by performing western blot analysis and it was determined
that overexpression of let-7b significantly inhibited endogenous
Bcl-xL protein expression levels. However, overexpression of let-7b
had no effect on Bax protein expression levels (Fig. 2C). These biochemical findings
indicate that let-7b is a posttranscriptional regulator of Bcl-xL
expression in breast cancer cells.
To evaluate the effect of let-7b on the response of
MCF-7 cells to 5-FU and ADM treatment, let-7b mimics or NC were
transfected into MCF-7 cells and the sensitivity of these
mimic-transfected cells to different concentrations of 5-FU or ADM
was determined. The MTT assay indicated that MCF-7 cells
overexpressing let-7b were significantly more sensitive to 5-FU and
ADM, compared with the control cells (P<0.05; Fig. 2D).
SNP rs3208684 A>C increases Bcl-xL
expression levels by interfering with let-7b function
As SNP rs3208684 A>C is located within the let-7b
seed binding site, we hypothesized that this SNP may result in
differential regulation of Bcl-xL induced by let-7b due to the
differential binding affinity of let-7b for the two Bcl-xL 3′-UTR
genotypes. To investigate this hypothesis, the wild-type
(containing the A allele) and mutant (containing the C allele)
Bcl-xL 3′-UTRs were cloned into the dual-luciferase psiCHECK-2
reporter vector, and HEK293T cells were co-transfected with let-7b
mimics or NC. It was identified that luciferase activity
significantly decreased in the presence of psiCHECK2-WT-Bcl-xL
3′-UTR plasmids (P<0.01) but did not change in the presence of
psiCHECK2-Mut-Bcl-xL 3′-UTR plasmids (P>0.05). These data
indicate that the SNP rs3208684 A>C may affect let-7b binding to
the Bcl-xL 3′-UTR (Fig. 3A).
To determine the effect of the SNP rs3208684 A>C
on let-7b-mediated regulation of Bcl-xL expression, Bcl-xL gene
expression constructs containing WT-Bcl-xL and Mut-Bcl-xL were
generated and co-transfected into MCF-7 cells with let-7b mimics.
As indicated in Fig. 3B,
overexpression of let-7b caused a decrease in Bcl-xL gene
expression in the presence of WT-Bcl-xL compared with Mut-Bcl-xL;
however, the SNP rs3208684 A>C did not change Bax protein
expression levels. Thus, the present study proposes that variation
in the SNP rs3208684 A>C may mediate the upregulation of Bcl-xL
protein expression by interfering with the binding of let-7b to the
3′-UTR of Bcl-xL in breast cancer cells.
SNP rs3208684 A>C enhances the
resistance of MCF-7 cells to 5-FU and ADM
The current study investigated whether the presence
of an SNP in the let-7b binding site of the Bcl-xL 3′-UTR results
in resistance to chemotherapeutic agents. WT-Bcl-xL or Mut-Bcl-xL
were co-transfected into MCF-7 cells with let-7b mimic and, after
24 h, these transfected MCF-7 cells were treated with various
concentrations of 5-FU or ADM for an additional 48 h. As indicated
in Fig. 4, the survival rate of
MCF-7 cells transfected with Mut-Bcl-xL and let-7b was
significantly higher than that in MCF-7 cells transfected with
WT-Bcl-xL and let-7b (P<0.05). Thus, the SNP rs3208684 A>C in
the let-7b binding site of Bcl-xL 3′-UTR appears to result in
let-7b-associated 5-FU and ADM resistance in MCF-7 cells.
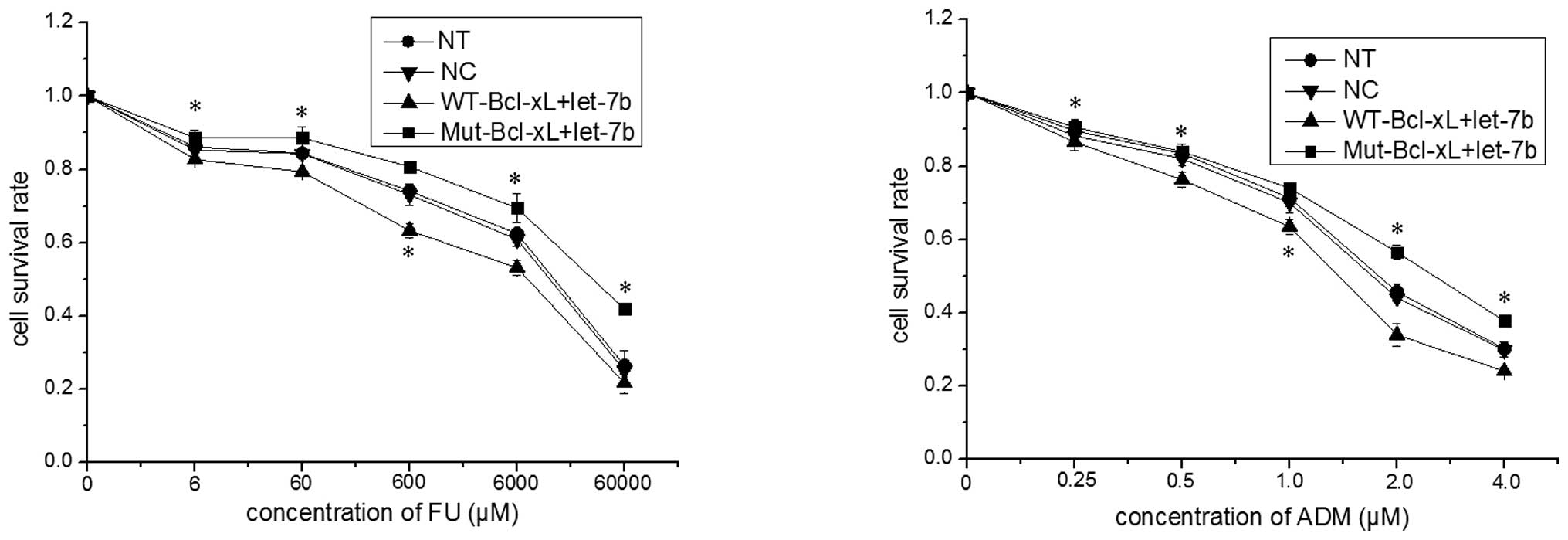 | Figure 4Single nucleotide polymorphism
rs3208684 A>C enhances the resistance of MCF-7 cells to 5-FU and
ADM. The expression constructs of the full-length Bcl-xL gene
containing A (WT-Bcl-xL) or C (Mut-Bcl-xL) were co-transfected with
let-7b into MCF-7 cells and, 24 h after transfection, the MCF-7
cells were treated with various concentrations of 5-FU or ADM for
48 h. The cell survival rate was determined by performing an MTT
assay and the data are expressed as the mean ± standard deviation
of a minimum of three independent transfection analyses.
*P<0.05 vs. WT-Bcl-xL+let-7b. ADM, doxorubicin; Bcl,
B-cell lymphoma-extra large; FU, fluorouracil; MTT, 3-(4,
5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; mut,
mutant; NT, non-transfected cells; WT wild-type; NC, normal control
(miRNA mimic). |
Discussion
Bcl-xL is one of two protein products of the Bcl2l1
gene (24) and is a primary
antiapoptotic factor, which has been recognized to mediate
chemotherapeutic agent resistance. Previously, Bcl-xL and Bax were
identified as key factors in the regulation of apoptosis by
homodimerization and heterodimerization (25). The human let-7 family is classified
as a tumor suppressor family in human cancer (26) and a number of previous studies have
indicated that the expression of members of the let-7 family are
significantly downregulated in various types of cancer (27), including breast cancer; furthermore,
this downregulation is associated with a poorer clinical outcome
(28). In the present study, let-7b
was demonstrated to target Bcl-xL, resulting in its downregulation
in MCF-7 cells. In addition, the present study indicated that
let-7b enhances the sensitivity of MCF-7 cells to ADM and 5-FU.
These results indicate that let-7b overexpression may enhance
cellular sensitivity to 5-FU and ADM via the repression Bcl-xL
expression in MCF-7 cells.
In a number of critical genes, SNPs at or adjacent
to miRNA binding sites are associated with the chemotherapeutic
response of a tumor via the disturbance or obstruction of miRNA
binding (29–32). The possible causes of this are the
SNPs located in the ‘seed’ regions at the 3′-UTRs of human genes
involved in multiple pathways such as cell proliferation, cell
death, stress resistance which are likely to affect miRNA-target
interaction and target expression accordingly (31). The dual-luciferase reporter assay
conducted in the current study revealed that the presence of
rs3208684 A-3′-UTR in the let-7b binding site of the Bcl-xL 3′-UTR
significantly reduces the expression of luciferase compared with
the presence of rs3208684 C-3′-UTR. This is consistent with the
initial bioinformatic analysis, which indicated a functional
interaction between let-7b and Bcl-xL mRNA. Additionally, the
transfection of MCF-7 cells with rs3208684 A-3′-UTR and let-7b
mimic demonstrated significant inhibition of Bcl-xL expression, and
the SNP rs3208684 A>C was identified to cause 5-FU and ADM
resistance in MCF-7 cells. Thus, these results indicate that the
occurrence of an SNP in rs3208684 A-3′-UTR of Bcl-xL may contribute
to the alteration of cellular resistance to 5-FU and ADM.
In conclusion, the present study, demonstrated that
let-7b may enhance the sensitivity of MCF-7 cells to 5-FU and ADM
by regulating Bcl-xL expression. The SNP rs3208684 A to C may
inhibit the interaction between let-7b and Bcl-xL 3′-UTR, resulting
in higher Bcl-xL expression, as well as cellular resistance to 5-FU
and ADM. Thus, we propose that the SNP rs3208684 A>C may be a
potential marker for personalized therapeutic approaches.
Furthermore, these results provide insight into a potential novel
chemotherapeutic strategy for breast cancer by combining let-7b
with currently used chemotherapeutic agents.
Acknowledgements
This present study was supported by the National
Natural Science Foundation of China (grant nos. 81372579, 31371277
and 30900625), the Colleges and Universities Innovation Platform
Open Fund Project of Hunan Province of China (grant no. 13K084),
the Science and Technology Program fund of Hunan Province of China
(grant no. 212FJ2016) and Provincial Key Discipline for Pharmacy of
Hunan Province of China during the Twelfth Five-year Plan
Period.
References
|
1
|
Jemal A, Bray F, Center M M, Ferlay J,
Ward E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Forouzanfar MH, Foreman KJ, Delossantos
AM, et al: Breast and cervical cancer in 187 countries between 1980
and 2010: a systematic analysis. Lancet. 378:1461–1484. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Oostendorp LJ, Stalmeier PF, Donders AR,
et al: Efficacy and safety of palliative chemotherapy for patients
with advanced breast cancer pretreated with anthracyclines and
taxanes: a systematic review. Lancet Oncol. 12:1053–1061. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Perez E and Muss HB: Optimizing adjuvant
chemotherapy in early-stage breast cancer. Oncology (Williston
Park). 19:1759–1767; discussion 1768, 1772–1774, 1777–1778.
2005.
|
|
5
|
Sarkar FH, Li Y, Wang Z, Kong D and Ali S:
Implication of microRNAs in drug resistance for designing novel
cancer therapy. Drug Resist Updat. 13:57–66. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Majumder S and Jacob ST: Emerging role of
microRNAs in drug-resistant breast cancer. Gene Expr. 15:141–151.
2011. View Article : Google Scholar
|
|
7
|
Garzon R, Marcucci G and Croce CM:
Targeting microRNAs in cancer: rationale, strategies and
challenges. Nat Rev Drug Discov. 9:775–789. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee S and Vasudevan S:
Post-transcriptional stimulation of gene expression by microRNAs.
Adv Exp Med Biol. 768:97–126. 2013. View Article : Google Scholar
|
|
9
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo H, Ingolia NT, Weissman JS and Bartel
DP: Mammalian microRNAs predominantly act to decrease target mRNA
levels. Nature. 466:835–840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Djuranovic S, Nahvi A and Green R:
miRNA-mediated gene silencing by translational repression followed
by mRNA deadenylation and decay. Science. 336:237–240. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lujambio A and Lowe SW: The microcosmos of
cancer. Nature. 482:347–355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Singh R and Mo YY: Role of microRNAs in
breast cancer. Cancer Biol Ther. 14:201–212. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sandhu S and Garzon R: Potential
applications of microRNAs in cancer diagnosis, prognosis, and
treatment. Semin Oncol. 38:781–787. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cho WC: MicroRNAs as therapeutic targets
and their potential applications in cancer therapy. Expert Opin
Ther Targets. 16:747–759. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mishra PJ, Humeniuk R, Mishra PJ, et al: A
miR-24 microRNA binding-site polymorphism in dihydrofolate
reductase gene leads to methotrexate resistance. Proc Natl Acad Sci
USA. 104:13513–13518. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Saetrom P, Biesinger J, Li SM, et al: A
risk variant in an miR-125b binding site in BMPR1B is associated
with breast cancer pathogenesis. Cancer Res. 69:7459–7465. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu Y, Xiao Y, Ding X, et al: A
miR-200b/200c/429-binding site polymorphism in the 3′ untranslated
region of the AP-2α gene is associated with cisplatin resistance.
PLoS One. 6:e290432011. View Article : Google Scholar
|
|
19
|
Brendle A, Lei H, Brandt A, et al:
Polymorphisms in predicted microRNA-binding sites in integrin genes
and breast cancer: ITGB4 as prognostic marker. Carcinogenesis.
29:1394–1399. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Leber B, Geng F, Kale J and Andrews DW:
Drugs targeting Bcl-2 family members as an emerging strategy in
cancer. Expert Rev Mol Med. 12:e282010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Thomadaki H, Talieri M and Scorilas A:
Treatment of MCF-7 cells with taxol and etoposide induces distinct
alterations in the expression of apoptosis-related genes BCL2,
BCL2L12, BAX, CASPASE-9 and FAS. Biol Chem. 387:1081–1086. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Simões-Wüst AP, Schürpf T, Hall J, et al:
Bcl-2/bcl-xL bispecific antisense treatment sensitizes breast
carcinoma cells to doxorubicin, paclitaxel and cyclophosphamide.
Breast Cancer Res Treat. 76:157–166. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Baek D, Villén J, Shin C, et al: The
impact of microRNAs on protein output. Nature. 455:64–71. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Michels J, Kepp O, Senovilla L, et al:
Functions of BCL-X L at the interface between cell death and
metabolism. Int J Cell Biol. 2013:7052942013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Billen LP, Kokoski CL, Lovell JF, et al:
Bcl-XL inhibits membrane permeabilization by competing with Bax.
PLoS Biol. 6:e1472008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Roush S and Slack FJ: The let-7 family of
microRNAs. Trends Cell Biol. 18:505–516. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Boyerinas B, Park SM, Hau A, et al: The
role of let-7 in cell differentiation and cancer. Endocr Relat
Cancer. 17:F19–F36. 2010. View Article : Google Scholar
|
|
28
|
O’Day E and Lal A: MicroRNAs and their
target gene networks in breast cancer. Breast Cancer Res.
12:2012010. View
Article : Google Scholar
|
|
29
|
Mishra PJ, Mishra PJ, Banerjee D and
Bertino JR: MiRSNPs or MiR-polymorphisms, new players in microRNA
mediated regulation of the cell: Introducing microRNA
pharmacogenomics. Cell Cycle. 7:853–858. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Blitzblau RC and Weidhaas JB: MicroRNA
binding-site polymorphisms as potential biomarkers of cancer risk.
Mol Diagn Ther. 14:335–342. 2010. View Article : Google Scholar
|
|
31
|
Yu Z, Li Z, Jolicoeur N, et al: Aberrant
allele frequencies of the SNPs located in microRNA target sites are
potentially associated with human cancers. Nucleic Acids Res.
35:4535–4541. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen K, Song F, Calin GA, et al:
Polymorphisms in microRNA targets: a gold mine for molecular
epidemiology. Carcinogenesis. 29:1306–1311. 2008. View Article : Google Scholar : PubMed/NCBI
|


















