Introduction
Osteosarcoma is a highly malignant and aggressive
bone tumor, occurring primarily in individuals between 10 and 30
years old (1). This tumor accounts
for ~45% of all the bone sarcomas. Although five-year survival
rates of 50–70% can be achieved through multimodal therapy, the
lack of effective treatment options results in poor prognosis in a
large number of patients (2).
Further investigation of the pathogenesis of osteosarcoma is
required to reduce the morbidity and mortality rates. A previous
study has demonstrated that the established risk factors involved
in the development of osteosarcoma include the following:
Adolescent and young adult age, male gender, previous treatment
with radiotherapy or anticancer drugs, particularly alkylating
agents, and a family history of osteosarcoma (1). However, the etiology of osteosarcoma
is not fully understood based on these risk factors; therefore,
additional risk factors may be involved (3). Molecular biology studies have revealed
strong evidence that genetic factors are important in the
pathogenesis of osteosarcoma (4,5).
Glutathione S-transferases (GSTs) are a family of
phase II enzymes (6). GSTs are
mainly responsible for the detoxification of a wide range of
environmental and nonenvironmental carcinogens (including
polyaromatic hydrocarbons from second-hand cigarette smoke),
chemotherapy agents (including alkylating agents and
anthracyclines), inflammation-associated reactive oxygen species
and metabolism-derived lipid peroxides (7). In addition, GSTs are able to modulate
the induction of other enzymes and proteins that are important for
cellular functions (8). Thus, GSTs
are involved in the protection against various types of cellular
damage and their malfunction may result in carcinogenesis.
In humans, the GST super family consists of numerous
cytosolic, mitochondrial and microsomal proteins. Cytosolic
proteins are divided into eight distinct classes, including the α,
κ, μ, ω, π, σ, θ and ζ (9). The θ
class of GSTs is encoded by the GST θ 1 (GSTT1) gene, which is
located on chromosome 1p13.3 and contains 10 exons (10). In addition, the μ class is encoded
by the GST μ1 (GSTM1) gene on chromosome 22q11.23 and contains six
exons (11). Homozygous deletion
(null genotype) is the most common variant of the GSTM1 and GSTT1
genes. In the Caucasian European populations, the prevalence of the
GSTM1 deletion genotype is 47–58%, while the prevalence of the
GSTT1 null genotype is 13–25% (12). A previous study has indicated that
the null genotype may be associated with the absence of enzyme
activity, increasing vulnerability to cytogenetic damage and
resulting in susceptibility to cancer (13).
A previous study revealed that the null genotypes of
GSTT1 and GSTM1 were associated with increased risk of bladder and
prostate cancer (14). In addition,
a number of studies have investigated the association between GSTM1
and GSTT1 polymorphisms and the risk of developing osteosarcoma
(15,17,18).
However, the results of these studies were inconsistent. A
meta-analysis can be useful in the detection of an association that
may not be identified in sample size studies, particularly studies
evaluating rare allele frequency polymorphisms. The aim of the
present study was to investigate the association between the null
genotypes of GSTM1 and GSTT1 and the development of osteosarcoma by
conducting a meta-analysis investigation of all the eligible
case-control studies published to date.
Materials and methods
Literature search
The literature was screened (titles, abstracts and
full text) by two researchers independently, in order to determine
which studies were eligible for inclusion in this meta-analysis.
The results were compared and disagreements were resolved by
consensus. The PubMed (www.ncbi.nlm.nih.gov/pubmed), Cochrane Library
(www.thecochranelibrary.com) and China
National Knowledge Infrastructure (http://www.cnki.net) databases were examined to
identify all the studies investigating the association between
GSTM1 and GSTT1 polymorphisms and osteosarcoma risk, which were
published prior to March 2014. The following key words were used:
‘glutathione S-transferases’, ‘GST’, ‘osteosarcoma’,
‘polymorphism’, ‘mutation’ and ‘variant’. No publication language
restrictions were imposed. The references of all the studies
identified through the literature search were investigated for
other relevant publications. In the case that sequential or
multiple publications using the same data were identified, the
publication reporting data from the largest or most recent study
was included. The search strategy used in this study is shown in
Table I.
 | Table ICharacteristics of literatures
included in the meta-analysis. |
Table I
Characteristics of literatures
included in the meta-analysis.
| Study included | Year | Country | Ethnicity | Genotyping | Cases/controls | Cases | Controls |
|---|
|
|---|
| Null | Non-null | Null | Non-null |
|---|
| GSTM1 |
| Barnette et
al | 2004 | America | Caucasian | High-throughout
assay | 12/326 | 2 | 10 | 183 | 143 |
| Carolina et
al | 2010 | Brazil | Caucasian | PCR-RFLP | 80/160 | 35 | 45 | 72 | 88 |
| Lu et al | 2011 | China | Asian | TaqMan assay | 110/226 | 61 | 49 | 104 | 122 |
| GSTT1 |
| Barnette et
al | 2004 | America | Caucasian | High-throughout
assay | 12/300 | 2 | 10 | 66 | 234 |
| Carolina et
al | 2010 | Brazil | Caucasian | PCR-RFLP | 80/160 | 26 | 54 | 42 | 118 |
| Lu et al | 2011 | China | Asian | TaqMan assay | 110/226 | 70 | 40 | 111 | 115 |
Inclusion and exclusion criteria
The inclusion criteria for human studies included
the following: i) Case-control studies, addressing osteosarcoma
cases and healthy controls; ii) studies evaluating the association
between GSTM1 and GSTT1 polymorphisms and osteosarcoma risk; and
iii) studies that included sufficient genotype data for extraction.
The exclusion criteria included the following: i) Non-case-control
studies, evaluating the association between GSTM1 and GSTT1
polymorphisms and osteosarcoma risk; ii) case reports, letters,
reviews, meta-analyses and editorial articles; iii) studies
reporting incomplete or insufficient data; iv) studies containing
duplicate data; and v) studies with a family-based design.
Data extraction
The data were independently examined and extracted
by two researchers, based on the aforementioned inclusion and
exclusion criteria. In the case of inconsistency between the
studies selected by the two researchers, consensus was reached
following discussion. The following information was collected from
the eligible studies: First author’s name; year of publication;
country and ethnicity of the studied population; number of cases
and controls; and number of genotyped cases and controls. The cases
and controls were categorized into the Asian or Caucasian ethnic
groups. No minimum number of patients was required to include a
study in this meta-analysis.
Statistical analyses
Odds ratios (ORs) with 95% confidence intervals
(CIs) were used to determine the strength of association between
the GSTM1 and GSTT1 polymorphisms and osteosarcoma risk. Pooled ORs
for the risk associated with the GSTM1 and GSTT1 null genotypes vs.
the non-null genotypes were calculated. Heterogeneities between the
studies were estimated using the I2 test.
I2 values of 25, 50 and 75% were defined as low,
moderate and high estimates, respectively (15). When the I2 value
was >50%, indicating heterogeneity across the studies, the
random effects model was used for meta-analysis; otherwise, the
fixed effects model was used. In addition, subgroup analysis based
on ethnicity was used to investigate and interpret the diversity
among the results of different studies. Sensitivity analysis was
performed by using random effect values compared with the fixed
effect in order to ensure the stability of the findings (16). Publication bias was investigated
using Begg’s funnel plot and P<0.05 was considered to indicate a
statistically significant publication bias. All the analyses were
performed using the STATA version 12.0 software (StataCorp LP,
College Station, TX, USA) and the significance level was set to
0.05.
Results
Identification of eligible studies
Based on the search criteria used in the present
study, 31 individual manuscripts were identified. Of these, eight
full-text publications were preliminarily selected for further
detailed evaluation. According to the exclusion criteria, five of
these publications were then excluded, including one duplicate
study, one meta-analysis and three studies with insufficient data
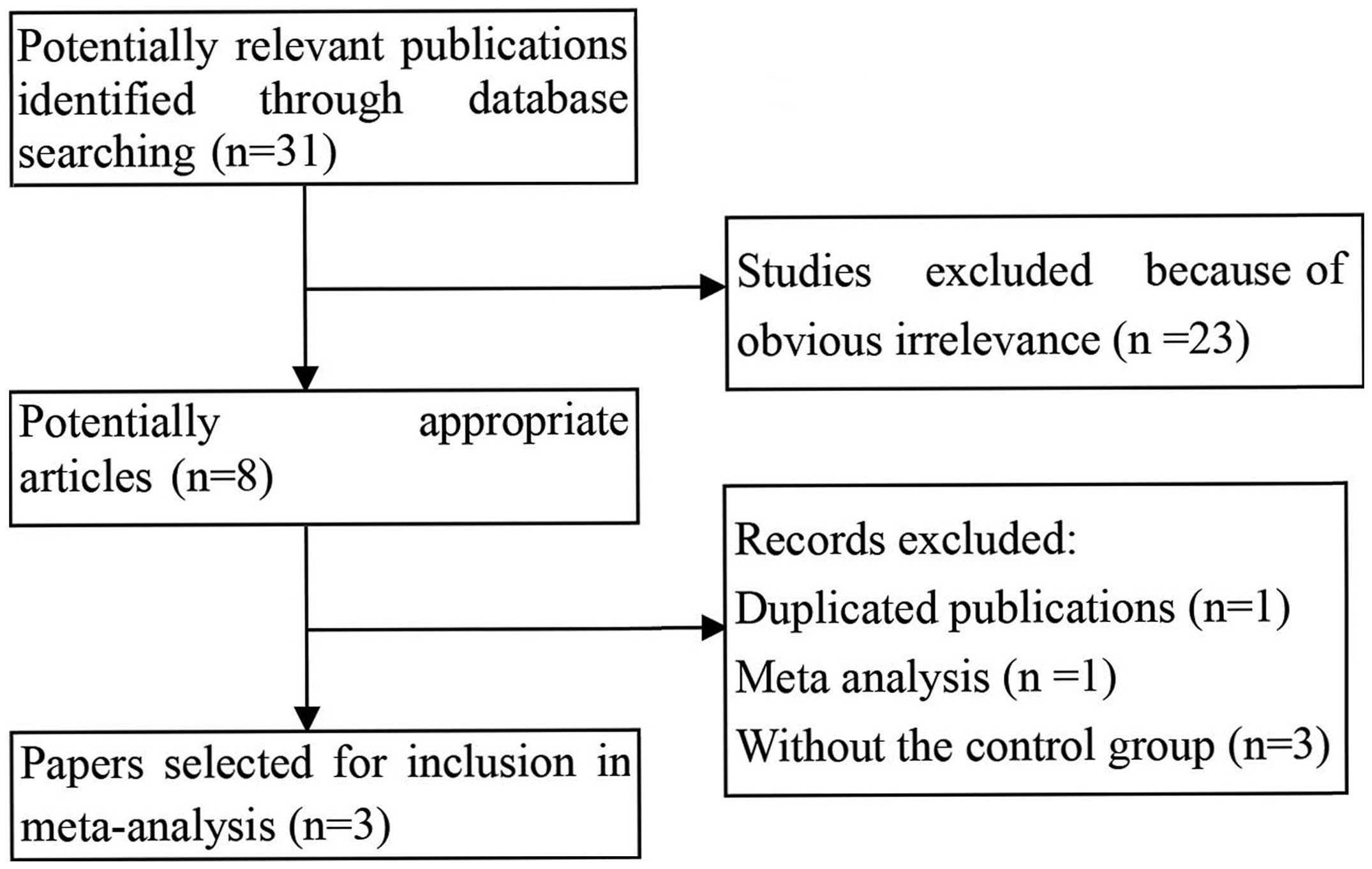
for extraction. Finally, as shown in Fig. 1, three studies with 202 cases and
712 healthy controls were included in the current meta-analysis
(17–19). A flow chart demonstrating the study
selection process is summarized in Fig.
1. All the eligible studies were case-control studies that
evaluated the association of the GSTM1 and GSTT1 null genotypes
with the susceptibility to osteosarcoma. The publication year range
of the included studies was between 2000 and 2014. The main
characteristics of the eligible studies are summarized in Table I.
Meta-analysis
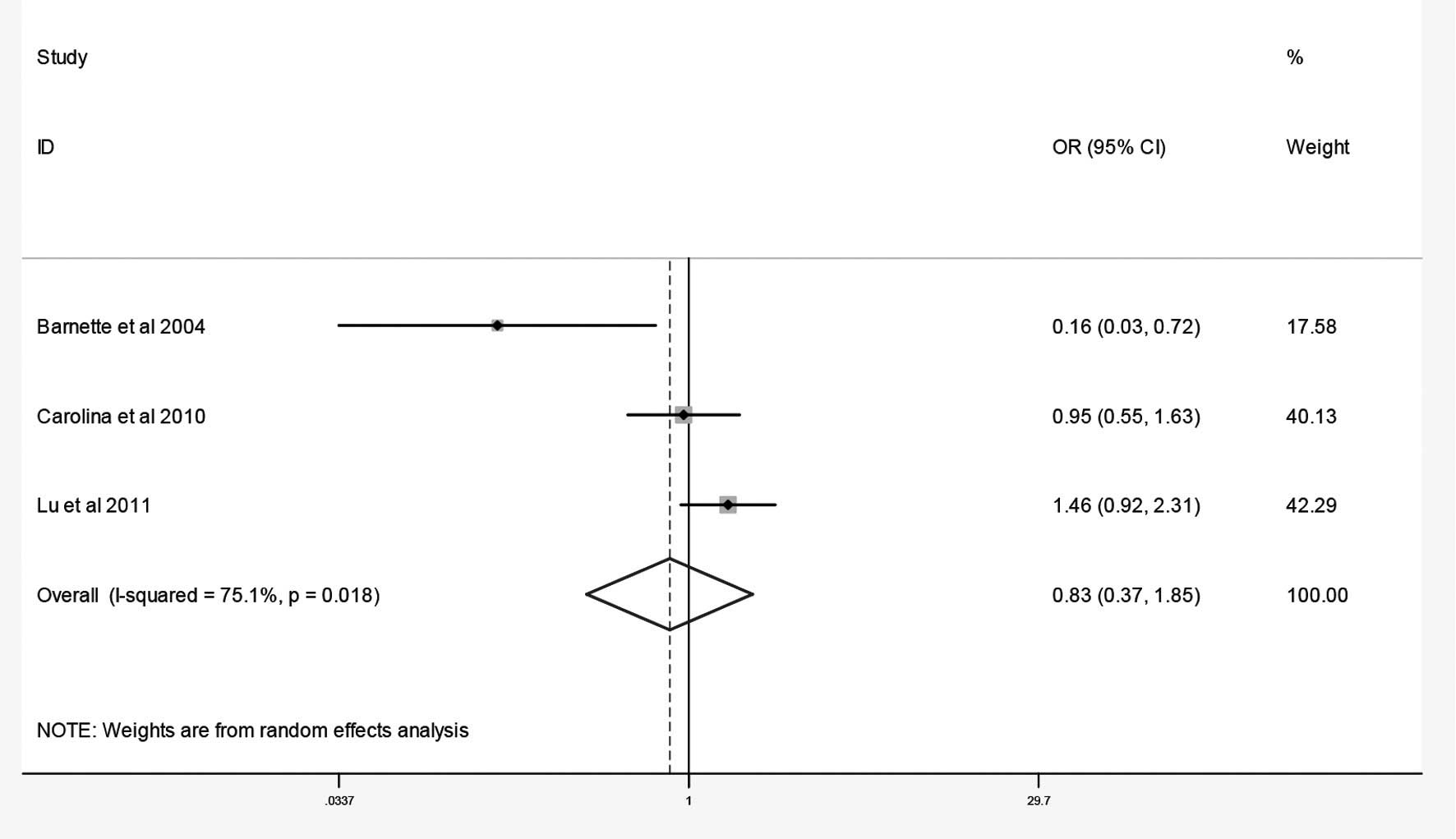
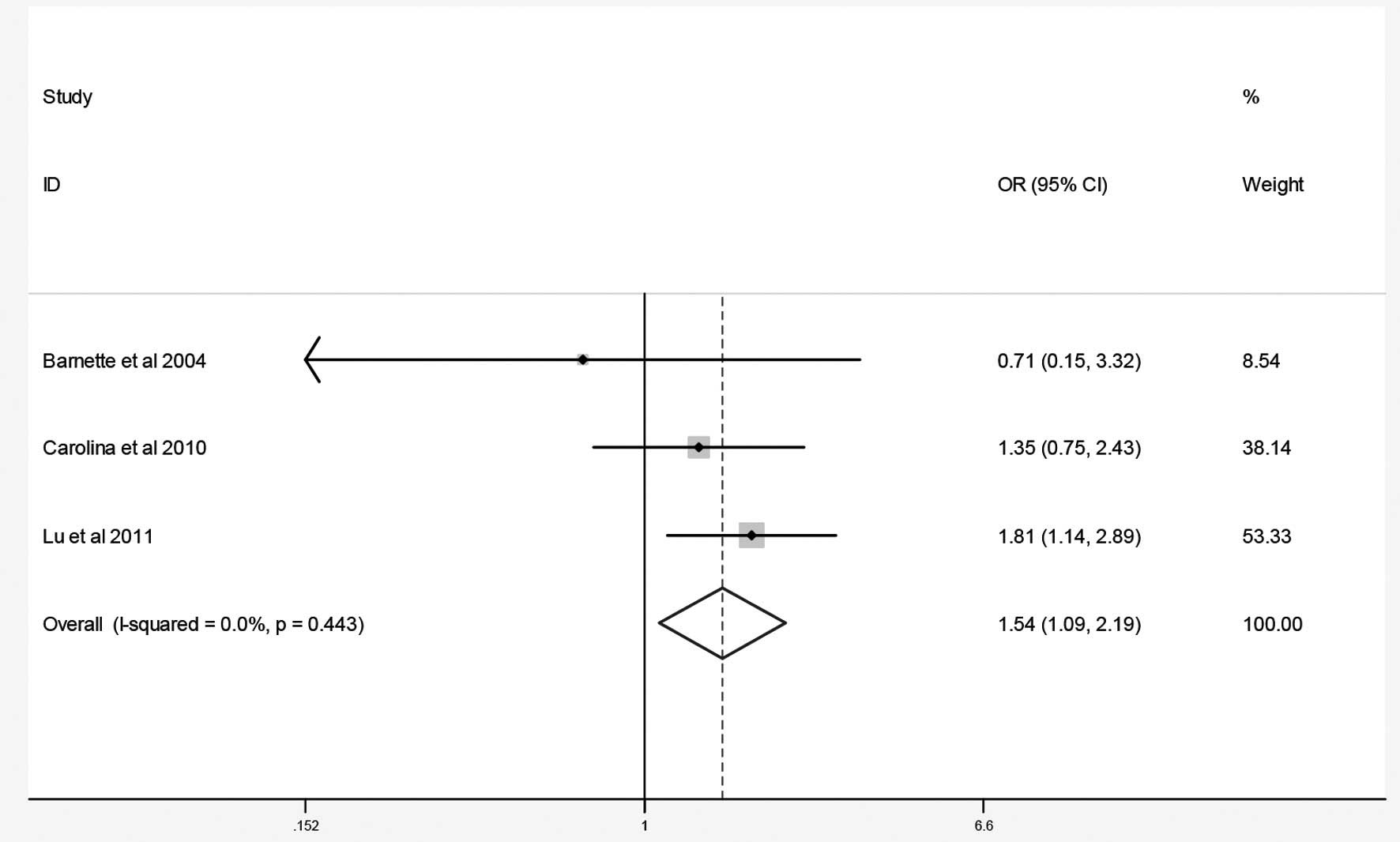
The combined results of the GSTM1 and GSTT1 null
genotypes and osteosarcoma risk are summarized in Figs. 2 and 3 and Table
II. The results of the meta-analysis revealed no association
between the null genotypes of GSTM1 and the risk of osteosarcoma
(OR=0.83; 95% CI, 0.37–1.85). By contrast, the meta-analysis
indicated that the GSTT1 null genotype was associated with an
increased risk of osteosarcoma in the two ethnic groups (OR=1.54;
95% CI, 1.09–2.19). Sensitivity analysis was performed by comparing
the results of the fixed and random effects models. No alterations
were detected in these results, indicating that the data of this
meta-analysis were relatively stable and credible.
 | Table IISummary ORs and 95% CI of null
genotypes of GSTM1 and GSTT1 and osteosarcoma risk. |
Table II
Summary ORs and 95% CI of null
genotypes of GSTM1 and GSTT1 and osteosarcoma risk.
| Subgroup | Contrast | Type of effects
model | Heterogeneity | Association | Publication bias |
|---|
|
|
|
|---|
| I2
(%) | P | OR | 95% CI | z | P |
|---|
| GSTM1 |
| Total | Null vs.
non-null | Random | 75.1 | 0.02 | 0.83 | 0.37–1.85 | 0.00 | 1.00 |
| GSTT1 |
| Total | Null vs.
non-null | Fixed | 0.0 | 0.44 | 1.54 | 1.09–2.19 | 0.00 | 1.00 |
Publication bias
The funnel plot and Begg’s test were used to assess
the publication bias. No evidence of publication bias was detected
in the present meta-analysis (P>0.05; Table II).
Discussion
Even with identical environmental exposure,
different individuals present a varied susceptibility to the same
cancer type. A number of factors, including polymorphisms of genes
involved in carcinogenesis, may account for this susceptibility
variation. Therefore, recent studies have focused on genetic
susceptibility to cancer (4,5). As
important phase II enzymes, the GSTM1 and GSTT1 null genotypes are
known to eliminate enzyme activities; therefore, these null
genotypes have been linked with the increased number of cancer
cases, possibly due to increased susceptibilities to environmental
toxins and carcinogens (13).
The association between GSTM1 and GSTT1 null
genotypes and osteosarcoma risk has been investigated in several
studies (15,17,18);
however, the results of these studies are controversial. The aim of
meta-analyses is to combine similar studies in order to increase
the sample size and statistical potential, obtaining more accurate
results (20). To the best of our
knowledge, the present study is the first systematic meta-analysis
of the association between GSTM1 and GSTT1 null genotypes and
osteosarcoma risk. The current study assessed quantitatively the
association between the GSTM1 and GSTT1 null genotypes and
susceptibility to osteosarcoma. In total, three case-control
studies were found to be eligible and were investigated. These
studies involved a total of 202 osteosarcoma cases and 712 healthy
controls. The results revealed no statistically significant
association between the GSTM1 null genotype and osteosarcoma risk
(OR=0.83; 95%CI, 0.37–1.85). By contrast, the results demonstrated
that the GSTT1 null genotype was significantly associated with the
susceptibility to osteosarcoma (OR=1.54; 95% CI, 1.09–2.19). No
evidence of publication bias was detected in this meta-analysis for
the GSTM1 and GSTT1 null genotypes (P>0.05).
The underlying mechanism of the association between
the GSTT1 null genotype and osteosarcoma risk remains unclear. A
recent study demonstrated that GSTT1 is different compared with
other phase II enzymes (such as GSTM1), which also exhibit phase I
enzyme activity and possess the ability to activate carcinogens
(17). The null genotype of GSTT1
is associated with the absence of enzyme activity and an increased
cancer incidence. As the number of eligible studies selected in the
present meta-analysis was small, these results require further
verification.
The limitations of the current meta-analysis should
be acknowledged. Firstly, the systematic review was based on
unadjusted data, since the genotype information stratified for the
main confounding variables was not available in the original
studies, while the confounding factors addressed across the
different studies were variable. In addition, this meta-analysis
was not able to address all the sources of heterogeneity that
existed among the previous studies for the majority of
polymorphisms, although subgroup stratification analysis may be
possible for the limited number of the included published studies.
Furthermore, gene-gene and gene-environment interactions were not
investigated in the present study, due to the lack of information
from the original studies.
In conclusion, the present meta-analysis indicated
that the GSTT1 null genotype was associated with an increased risk
of developing osteosarcoma. Since only a small number of studies
are available in this field and the current evidence remains
limited, future studies with large sample groups and adequate
methodological quality are required to obtain accurate results.
References
|
1
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: data
from the Surveillance, Epidemiology, and End Results Program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Walkley CR, Qudsi R, Sankaran VG, et al:
Conditional mouse osteosarcoma, dependent on p53 loss and
potentiated by loss of Rb, mimics the human disease. Genes Dev.
22:1662–1676. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Both J, Krijgsman O, Bras J, et al: Focal
chromosomal copy number aberrations identify CMTM8 and GPR177 as
new candidate driver genes in osteosarcoma. PLoS One.
9:e1158352014. View Article : Google Scholar
|
|
4
|
Sadikovic B, Thorner P, Chilton-Macneill
S, et al: Expression analysis of genes associated with human
osteosarcoma tumors shows correlation of RUNX2 overexpression with
poor response to chemotherapy. BMC Cancer. 10:2022010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gokgoz N, Wunder JS, Mousses S, et al:
Comparison of p53 mutations in patients with localized osteosarcoma
and metastatic osteosarcoma. Cancer. 92:2181–2189. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Toh CK, Gao F, Lim WT, et al:
Never-smokers with lung cancer: epidemiologic evidence of a
distinct disease entity. J Clin Oncol. 24:2245–2251. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Krajinovic M, Labuda D, Richer C, Karimi S
and Sinnett D: Susceptibility to childhood acute lymphoblastic
leukemia: influence of CYP1A1, CYP2D6, GSTM1, and GSTT1 genetic
polymorphisms. Blood. 93:1496–1501. 1999.PubMed/NCBI
|
|
8
|
Hayes JD, Flanagan JU and Jowsey IR:
Glutathione transferases. Annu Rev Pharmacol Toxicol. 45:51–88.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Strange RC, Spiteri MA, Ramachandran S and
Fryer AA: Glutathione-S-transferase family of enzymes. Mutat Res.
482:21–26. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Webb G, Vaska V, Coggan M and Board P:
Chromosomal localization of the gene for the human theta class
glutathione transferase (GSTT1). Genomics. 33:121–123. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pearson WR, Vorachek WR, Xu SJ, et al:
Identification of class-mu glutathione transferase genes
GSTM1-GSTM5 on human chromosome 1p13. Am J Hum Genet. 53:220–233.
1993.PubMed/NCBI
|
|
12
|
Rebbeck TR: Molecular epidemiology of the
human glutathione S-transferase genotypes GSTM1 and GSTT1 in cancer
susceptibility. Cancer Epidemiol Biomarkers Prev. 6:733–743.
1997.PubMed/NCBI
|
|
13
|
Mcllwain CC, Townsend DM and Tew KD:
Glutathione S-transferase polymorphisms: cancer incidence and
therapy. Oncogene. 25:1639–1648. 2006. View Article : Google Scholar
|
|
14
|
Steinhoff C, Franke KH, Golka K, et al:
Glutathione transferase isozyme genotypes in patients with prostate
and bladder carcinoma. Arch Toxicol. 74:521–526. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu MS, Huang SP, Chang YT, et al: Tumor
necrosis factor-alpha and interleukin-10 promoter polymorphisms in
Epstein-Barr virus-associated gastric carcinoma. J Infect Dis.
185:106–109. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang YJ and Pan Y: The M235T polymorphism
in the angiotensinogen gene and myocardial infarction risk: a
meta-analysis. J Renin Angiotensin Aldosterone Syst. 15:294–300.
2014. View Article : Google Scholar
|
|
17
|
Barnette P, Scholl R, Blandford M, et al:
High-throughput detection of glutathione s-transferase polymorphic
alleles in a pediatric cancer population. Cancer Epidemiol
Biomarkers Prev. 13:304–313. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Salinas-Souza C, Petrilli AS and de Toledo
SR: Glutathione S-transferase polymorphisms in osteosarcoma
patients. Pharmacogenet Genomics. 20:507–515. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu XF, Yang WL, Wan ZH, et al: Glutathione
S-transferase polymorphisms and bone tumor risk in China. Asian Pac
J Cancer Prev. 12:3357–3360. 2011.PubMed/NCBI
|
|
20
|
Salanti G, Sanderson S and Higgins JP:
Obstacles and opportunities in meta-analysis of genetic association
studies. Genet Med. 7:13–20. 2005. View Article : Google Scholar : PubMed/NCBI
|