Introduction
The hedgehog (Hh) signaling pathway is important in
embryonic cell differentiation, tissue development and organ
formation (1–4). In mammals, sonic Hh (Shh), the
glycoprotein ligand of Hh, binds to the transmembrane receptors
Patched (Ptch) 1 and 2 to activate the Hh signaling pathway and
relieve its suppression of the transmembrane protein Smoothened
(Smo). Subsequently, the activated Smo protein induces nuclear
translocation of a family of transcription factors, including
glioma-associated oncogene homolog (Gli) 1, 2 and 3, to activate
specific downstream target genes (2,5,6). Following maturation, Smo proteins are
suppressed and the pathway is inactivated; however, if excessive
activation mutations in the Smo gene and loss of function
mutations in the ptch gene occur, Smo activity is not
suppressed, and full-length Gli proteins are translocated to the
nucleus. In the nucleus, Gli proteins activate downstream genes,
such as c-myc and vascular endothelial growth factor (VEGF),
resulting in excessive cell proliferation or tumorigenesis.
Previous studies have identified that the Hh signaling pathway is
involved in inducing cancer, including skin cancer (7), medulloblastoma (8), and lung (9,10),
gastrointestinal (11–13), breast (14), prostate (15), ovarian (16) and endometrial cancer (17), in various mammalian systems. In
addition, it has been demonstrated that inhibiting the Hh signaling
pathway with a ligand-blocking antibody or Smo inhibitor, such as
cyclopamine, may lead to the inhibition of the growth of tumor
tissue (18,19).
Previous studies have indicated that the
proliferation, migration and invasion of gastric cancer cells are
associated with excessive Hh signaling. A study conducted in 90
gastric cancer patients identified that 70% of the collected
gastric samples exhibited high Shh, Ptch1 and Gli1 expression
levels (63/90 samples) (20).
Additionally, excessive overexpression of Shh has been detected in
intestinal metaplasia and stomach adenoma (21). A number of studies have also
determined that the Hh signaling pathway appears to directly
participate in cell proliferation and migration in the majority of
gastric cancer cell lines, including the AGS, MKN1, MKN7, MKN45 and
MKN74 cell lines (22,23).
Although the Hh signaling pathway is critical in
inducing gastric tumorigenesis, the underlying cellular and
molecular mechanisms are largely unknown. In the present study,
cyclopamine was used to specifically block the Hh signaling pathway
in the human gastric cancer AGS cell line, and its effect on cell
proliferation, migration and invasion were evaluated in a dose- and
time-dependent manner. Furthermore, the mechanism of this
inhibition was investigated by examining the protein and RNA
expression levels of key factors associated with the Hh signaling
pathway, Gli1, C-X-C chemokine receptor type (CXCR) 4 and
transforming growth factor (TGF)-β1, as well as determining the
rate of TGF-β1 protein secretion in the AGS cells.
Materials and methods
Cell culture and treatment
The human gastric carcinoma AGS cell line was
obtained from the Shanghai Institute of Biological Sciences,
Chinese Academy of Science (Shanghai, China). The cells were
maintained in RPMI 1640 medium supplemented with 10% fetal bovine
serum (Invitrogen Life Technologies, Carlsbad, CA, USA) and 100
U/ml penicillin/streptomycin. Various concentrations of cyclopamine
(2.5, 5, 10, 20, 40 and 80 µM; EMD Millipore, Billerica, MA, USA)
were added to the medium and the cells were maintained at 37°C in a
humidified atmosphere containing 5% CO2 for 24, 48 or 72
h.
Cell proliferation assay
The AGS cells were plated at a concentration of
2.5×104 cells/ml culture medium in 96-well plates and
treated with the abovementioned concentrations of cyclopamine, in
triplicate. After 24, 48 and 72 h, the number of viable cells were
determined by performing an MTT assay (Sigma-Aldrich, St. Louis,
MO, USA), according to the manufacturer's instructions. Briefly,
cells were seeded at 1×104 cells/well in 96-well plates
overnight, then the cells were treated with cyclopamine for 24, 48
or 72 h. Subsequently, 20 µl MTT solution was added and after 4 h
the medium was gently aspirated and 150 µl DMSO (Sigma-Aldrich) was
added to each well to dissolve any formazan crystals. The plate was
shaken for 10 min to allow for complete solubilization. Cell
viability was determined spectrophotometrically by measuring the
absorbance at 490 nm using a 96-well plate reader (Multiskan MK3,
Thermo Fisher Scientific, Waltham, MA, USA) and the results were
calculated as the mean of eight wells per group. Each experiment
was performed in 8 wells a minimum of three times
independently.
Apoptosis assay
After 24 h in culture with 0, 40 and 80µM
cyclopamine, 1×106 gastric cancer cells were washed
twice with phosphate-buffered saline (PBS) and resuspended in
binding buffer (10 mM HEPES/NaOH, 140 mM NaCl and 2.5 mM
CaCl2). Fluorescein isothiocyanate Annexin V (BD
Biosciences, Franklin Lakes, NJ, USA) was added at a final
concentration of 1 mg/ml, followed by 10 mg/ml propidium iodide.
The mixture was incubated for 10 min in the dark at room
temperature and subsequent cell counting was conducted using a
FACScan™ flow cytometer with CellQuest™ software (BD
Biosciences).
Matrigel invasion assay
A migration assay was performed using a quantitative
cell migration assay kit (ECM500; EMD Millipore), according to the
manufacturer's instructions. Briefly, serum-free RPMI-1640 medium
(200 µl) was added to the extracellular matrix layer in the upper
chamber and allowed to hydrate for 1–2 h at ambient temperature.
The cells were dislodged following brief trypsinization, dispersed
into homogeneous single-cell suspensions, washed and resuspended in
serum-free medium at a concentration of 5×105 cells/ml.
The cell suspension (100 µl) was applied to the surface and allowed
to adhere for 1 h at 37°C, and 500 µl migration medium containing
0, 2.5, 5 or 10 µM cyclopamine was added to the bottom chamber.
After 24 h of incubation at 37°C in an atmosphere of 5%
CO2 in air, cells within the inserts were removed from
the upper membrane surface using a moist cotton-tipped swab.
Invasive cells on the lower membrane surface, which had migrated
through the polycarbonate membrane with a precoated thin layer of
basement membrane matrix, were fixed in 100% ethanol and were rinsed
with PBS. After being air-dried and photographed, the cells in the
upper chamber were stained with crystal violet (AppliChem GmbHm,
Darmstadt, Germany) for 20 min and dissolved in 10% acetic acid.
Finally, the optical density was read at an absorbance of 560 nm on
a standard microplate reader (Multiskan MK3, Thermo Fisher
Scientific).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from 2×106 AGS
cells treated with 0, 2.5, 5 or 10 µM cyclopamine for 24 h using
TRIzol® reagent (Roche Diagnostics, Basel, Switzerland), according
to the manufacturer's instructions. First strand complementary
(c)DNA synthesis and amplification were performed using a Revert
Aid First Strand cDNA Synthesis kit (Thermo Fisher Scientific), and
the qPCR was performed using an iQ5 Multicolor Real-Time PCR
Detection system (Bio-Rad Laboratories, Hercules, CA, USA) in
96-well plates. The PCR was run in a 20-µl reaction containing 1 µl
DNA template, 0.2 µl Taq polymerase, 2 µl dNTPs, 0.2 µl each primer
and 2 µl 10X Taq buffer. The mixture was incubated at 95°C for 5
min, followed by 25 cycles at 95°C for 40 sec, 58°C for 40 sec and
72°C for 1 min, with a final extension at 72°C for 10 min. Cycle
threshold values were obtained using ABI PRISM® 7000 software
(Applied Biosystems, Foster City, CA, USA) and the fold change of
relative mRNA expression levels were determined using the
2−ΔΔCt method. The primer sequences were as follows:
Forward, 5′-TCCTTTGGGGTCCAGCCT TG-3′ and reverse,
5′-ATGCCTGTGGAGTTGGGGCT-3′ for Gli1; forward,
5′-TGGAGCTGGTGAAGCGGAAG-3′ and reverse, 5′-TTTCCACCATTAGCACGCGG-3′
for TGF-β1; forward, 5′-TCAGTCTGGACCGCTACCTG-3′ and reverse,
5′-CCACCCACAAGTCATTGGGG-3′ for CXCR4; and forward,
5′-AGGTCGGAGTCAACGGATTTG-3′ and reverse, 5′-GTGATGGCATGGACTGTGGT-3′
for GAPDH.
Western blot analysis
Whole-cell collection of AGS cells treated with 0,
2.5, 5 or 10 µM cyclopamine for 24 h was conducted using
radioimmunoprecipitation assay buffer [50 mM Tris, 150 mM NaCl, 1%
Triton X-100, 0.1% sodium dodecyl sulfate and 1% sodium
deoxycholate (pH 7.4)] supplemented with protease inhibitor.
Following protein concentration determination using a Bio-Rad
protein assay kit (Bio-Rad Laboratories), the protein lysates were
resolved by sodium dodecyl sulfate-polyacrylamide gel
electrophoresis and transferred onto nitrocellulose membranes
(Hybond™-P; GE Healthcare Life Sciences, Chalfont, UK). The
membranes were blocked with PBS containing 0.2% Tween 20 and 5%
skimmed dry milk, and incubated with primary rabbit anti-human
polyclonal antibodies against Gli1 (1:1,000, cat. no. AB3444,
Millipore) and CXCR4 (1:500, cat. no. AB1846, Millipore), rabbit
monoclonal antibody against TGF-β1 (1:1,000, cat. no. 3709, Cell
Signaling Technology, inc., Beverly, MA, USA) and β-actin (1:500,
cat. no. sc-130656, Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA) followed by a horseradish peroxidase-labeled goat anti-rabbit
secondary antibody IgG-HRP (1:5,000, cat. no. sc-2004, Santa Cruz
Biotechnology, Inc.). Finally, X-ray film was used to image the
western blots and determine protein expression levels.
TGF-β1 quantification
After 24 h of cell culture in various concentrations
of cyclopamine, the quantity of TGF-β1 released into the culture
supernatant was measured using an ELISA kit (Fujirebio Diagnostics,
Inc., Malvern, PA, USA), according to manufacturer's instructions.
Absorbance was determined at a wavelength of 490 nm.
Statistical analysis
Data were analyzed using SPSS software, version 13.0
(SPSS, Inc., Chicago, IL, USA). All the results were obtained in
triplicate and are presented as the mean ± standard error of the
mean. Comparisons were made by one-way analysis of variance or
Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Cyclopamine inhibits the proliferation
of AGS cells
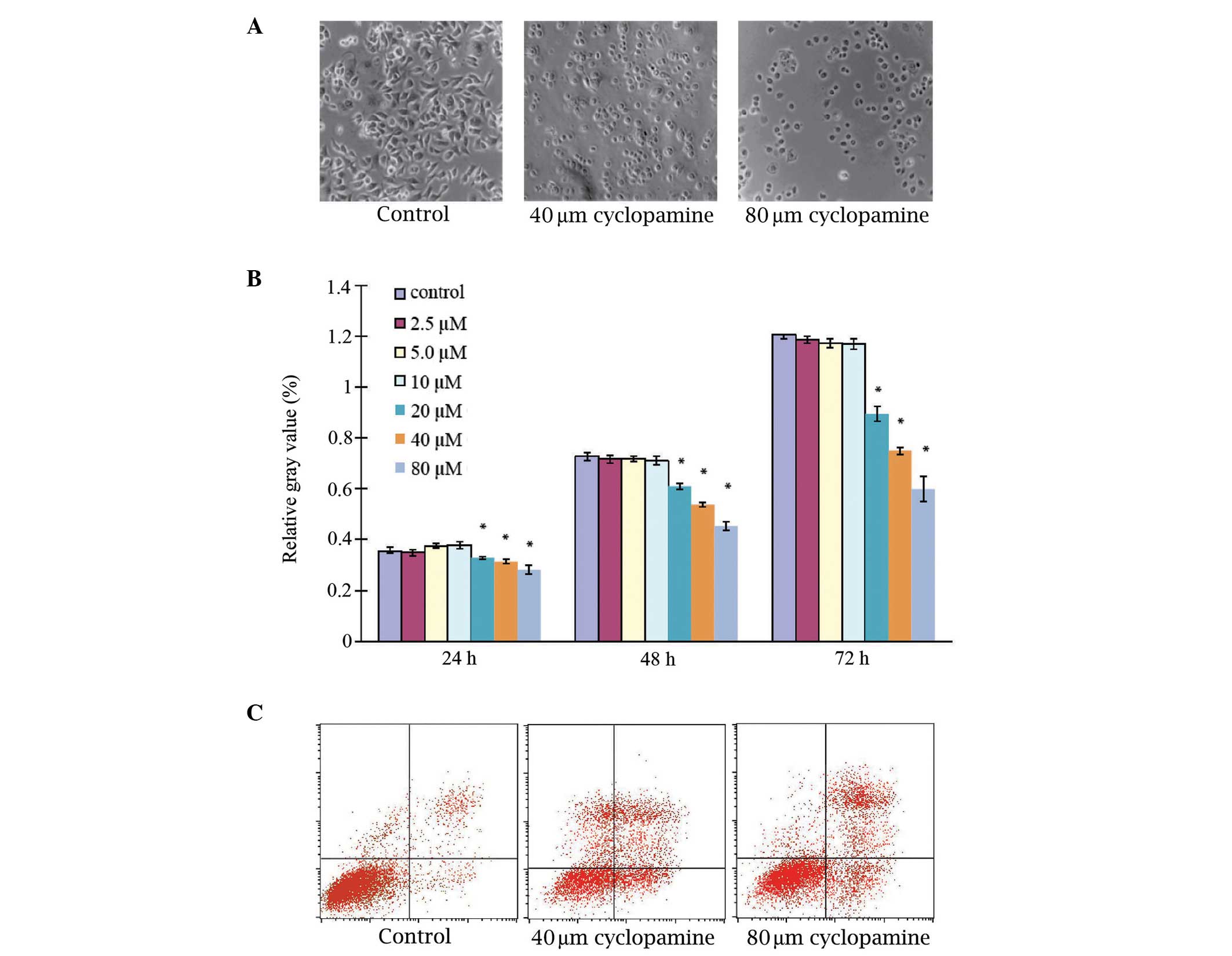
Examination of the effects of cyclopamine
administration on AGS cell proliferation identified that the
untreated AGS cells grew as an adherent monolayer, established
cylindrical shapes and exhibited nuclei that were located at the
proximal pole of the cell bodies (Fig.
1A). Upon AGS cell treatment with 40 and 80 µM cyclopamine for
48 h, cell growth was markedly inhibited, exhibiting a diminished
three-dimensional appearance and increased inter-cellular gaps.
Quantitative measurement identified that cyclopamine inhibited the
growth of the AGS cells in a dose-dependent manner; however, when
the AGS cells were treated with 2.5, 5 and 10 µM cyclopamine for
24, 48 or 72 h, the proliferation rates were not significantly
different to those under control conditions (P>0.05), indicating
that cyclopamine at a concentration range of 2.5–10 µM may not
affect cell proliferation. However, cyclopamine significantly
inhibited cell proliferation at higher concentrations (20, 40 and
80 µM; P<0.05; Fig. 1B).
Cyclopamine induces apoptosis in AGS
cells
Examination of the effects of cyclopamine on AGS
cell apoptosis was conducted by flow cytometric analysis. Annexin
staining was used to determine the effect on apoptosis 24 h after
the treatment of the cancer cells with 40 or 80 µM cyclopamine
(Fig. 1C; Table I). Under controlled conditions
(untreated), no increase in AGS cell apoptosis was observed,
however, the administration of cyclopamine appeared to induce
significant apoptosis in the AGS cells. The early and late
apoptotic rates were 2.34±0.90 and 4.05±0.87%, respectively, for
the control group. After 24 h of cyclopamine treatment, the
proportion of early and late apoptotic cells was increased in a
dose-dependent manner and was significantly higher than that of the
control group (P<0.05).
 | Table I.Percentage of cell apoptosis induced
by 40 and 80 µM cyclopamine. |
Table I.
Percentage of cell apoptosis induced
by 40 and 80 µM cyclopamine.
| Apoptosis | Control | 40 µmol/l | 80 µmol/l |
|---|
| Early | 2.34±0.90 |
13.53±1.27a |
20.89±7.72a,b |
| Late | 4.05±0.87 |
16.12±1.63a |
22.06±0.98a,b |
Cyclopamine reduces motility and
invasiveness of AGS cells
The ability to invade a reconstituted basement
membrane is an important phenomenon that distinguishes cancer cells
from other cell types (20). Thus,
the effect of cyclopamine on cellular motility and invasion of the
AGS cells was evaluated by treatment with doses of cyclopamine low
enough to not affect AGS cell proliferation and apoptosis. The
cancer cells were untreated or treated with cyclopamine at
concentrations of 2.5, 5 and 10 µM, and maintained for 24 h. As
hypothesized, the AGS cells demonstrated a moderate rate of
invasion under the control conditions, however, upon cyclopamine
treatment, baseline invasion was diminished. A dose-response effect
was observed such that 10 µM resulted in the least degree of
invasion (Fig. 2).
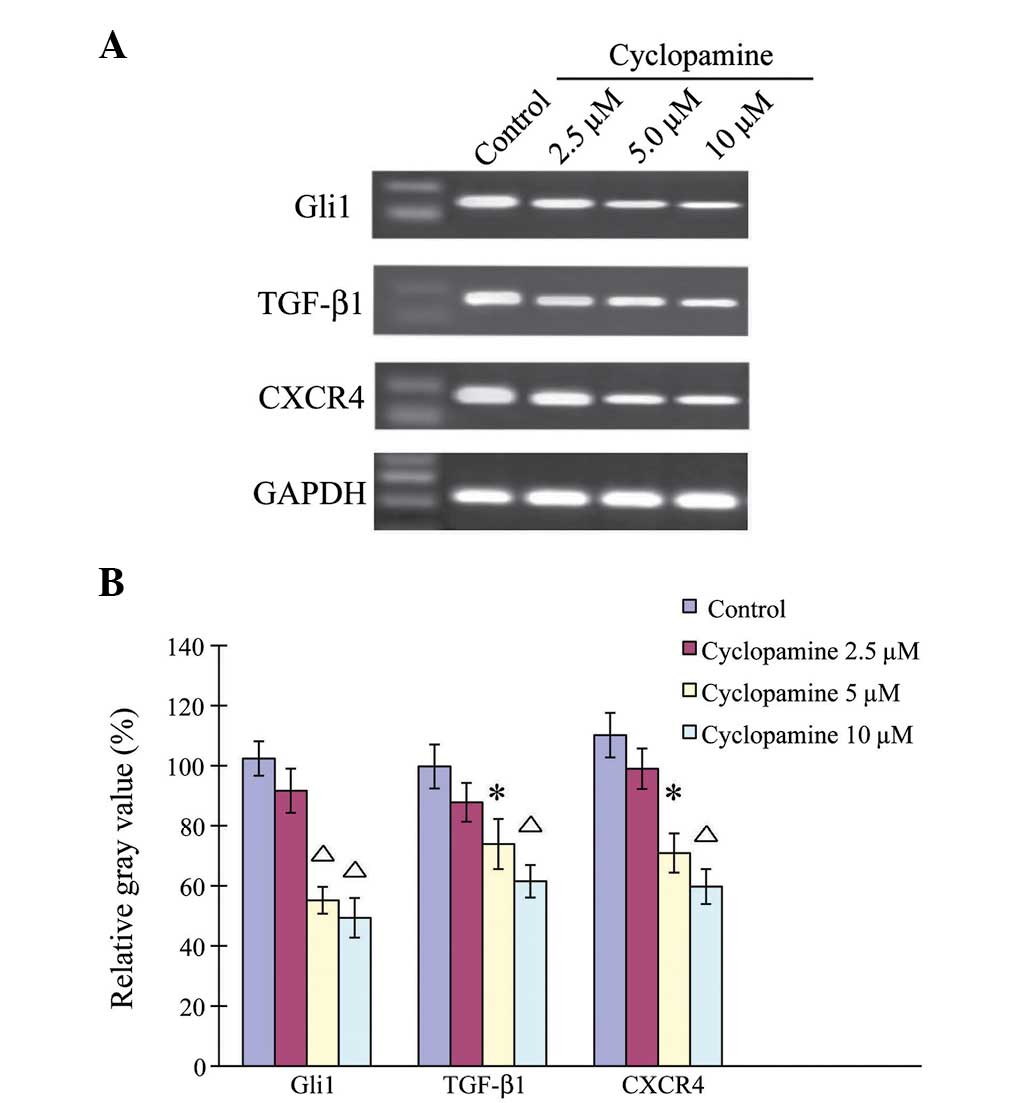
Cyclopamine downregulates
Hh-associated genes in AGS cells
The effects of cyclopamine on gene regulation were
then examined in the AGS cells (Fig.
3). The AGS cells were treated with 2.5, 5 and 10 µM
cyclopamine for 24 h. Quantitative measurement showed that
cyclopamine downregulated the genes in the AGS cells in a
dose-dependent manner. When the AGS cells were treated with 2.5 µM
cyclopamine for 24 h, the gene expression levels of Gli1, TGF-β1
and CXCR4 were similar to those under control conditions
(P>0.05). When the AGS cells were treated with 5 or 10 µM
cyclopamine, the Gli1, TGF-β1 and CXCR4 gene expression levels were
significantly downregulated (P< 0.05).
Cyclopamine downregulates
Hh-associated proteins in AGS cells
Consistent with its effect on mRNA expression level,
cyclopamine additionally reduced Hh-associated protein expression
levels in a dose-dependent manner (Fig.
4). When the AGS cells were treated with 2.5 µM cyclopamine for
24 h, the protein expression levels of Gli1, TGF-β1 and CXCR4 were
similar to those under control conditions (P>0.05). However, the
AGS cells that were treated with 5 or 10 µM cyclopamine exhibited
significantly downregulated Gli1, TGF-β1 and CXCR4 protein
expression levels (P<0.05).
Cyclopamine inhibits TGF-β1 secretion
in AGS cells
Following the observations that cyclopamine appears
to inhibit cancer cell invasion and downregulate the mRNA and
protein expression levels of Shh-associated genes, the effect of
cyclopamine on the TGF-β signaling pathway in the AGS cells was
examined in attempt to elucidate the mechanism of these
observations. As indicated in Table
II, when the AGS cells were treated with 2.5 µM cyclopamine for
24 h, the quantity of TGF-β1 identified in the collected
supernatant was similar to that observed under control conditions
(P>0.05). However, in the AGS cells treated with 5 and 10 µM
cyclopamine, TGF-β1 secretion was significantly reduced
(P<0.05).
 | Table II.Effect of cyclopamine administration
on TGF-β1 secretion in AGS cells. |
Table II.
Effect of cyclopamine administration
on TGF-β1 secretion in AGS cells.
| Cyclopamine,
µmol/l | TGF-β1, µg/l |
|---|
| 0.0 (control) | 5.935±0.825 |
| 2.5 | 5.268±0.638 |
| 5.0 |
3.527±0.539a |
| 10.0 |
1.947±0.635b |
Discussion
The Hh signaling pathway was initially recognized
for its role in modulating embryonic cell proliferation and
differentiation (1–4); however, more recently, it has been
demonstrated that Hh is important in the proliferation of various
types of cancer cells, including lung, pancreatic and gastric
cancer cells (12,14,16,24–26).
While the mechanisms of the Hh signaling pathway in
promoting gastric tumorigenesis and regulating downstream target
genes are largely unknown, various lines of evidence indicate that
a number of key factors, such as TGF-β1 and CXCR4, are actively
involved. Previous studies have demonstrated that TGF-β mRNA is
overexpressed in gastric carcinoma (27,28), and
that the Hh pathway may promote cancer cell mobility via activation
of the TGF-β/activin receptor-like kinase-Smad3 pathway in gastric
cancer cell lines, such as MKN-28 (29). In addition, it has previously been
demonstrated that TGF-β may induce cancer migration via the c-Jun
N-terminal kinase or extracellular signal-regulated kinase pathways
(30). The chemokine receptor, CXCR4,
and its cognate ligand, C-X-C ligand type 12, are expressed in
various types of tissue and have been proposed as regulators of the
directional trafficking and invasion of tumor cells, such as
breast, endometrial and prostate cancer cells (31–34).
Furthermore, CXCR4 is expressed in gastric carcinoma, as well as
gastric cancer cell lines, and appears to be highly associated with
lymph node metastasis and a high tumor stage (35).
The present study demonstrated that by blocking the
Hh signaling pathway with cyclopamine, the proliferation and
migration of gastric cancer AGS cells could be significantly
reduced. Furthermore, it was identified that the mRNA and protein
levels of Gli1, TGF-β1 and CXCR4 were coordinately downregulated in
the cyclopamine-treated AGS cells, and that the quantity of TGF-β1
secreted into the culture supernatant was significantly reduced
following a 24-h treatment with 5 and 10 µM cyclopamine. These
findings are in agreement with a number of previously conducted
studies (25,36,37). To
further elucidate the role of Hh as an important regulator in AGS
cells, the present study demonstrated that the Hh signaling pathway
appears to regulate tumor invasion and metastasis via TGF-β1 and
CXCR4. Furthermore, the current study demonstrated that blocking
the Hh signaling pathway downregulated TGF-β1 and CXCR4 expression,
thus, inhibiting human gastric cancer cell invasion and metastasis.
In conclusion, the present study identified that blocking the Hh
signaling pathway by cyclopamine administration may serve as a
potential therapeutic strategy for the prevention and treatment of
gastric cancer invasion in human cancer patients.
References
|
1
|
Hooper JE and Scott MP: Communicating with
Hedgehogs. Nat Rev Mol Cell Biol. 6:306–317. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ingham PW and McMahon AP: Hedgehog
signaling in animal development: paradigms and principles. Genes
Dev. 15:3059–3087. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
McMahon AP, Ingham PW and Tabin CJ:
Developmental roles and clinical significance of hedgehog
signaling. Curr Top Dev Biol. 53:1–114. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pasca di Magliano M and Hebrok M: Hedgehog
signalling in cancerformation and maintenance. Nat Rev Cancer.
3:903–911. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ruiz i Altaba A, Mas C and Stecca B: The
Gli code: an information nexus regulating cellfate, stemness and
cancer. Trends Cell Biol. 17:438–447. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bale AE and Yu KP: The hedgehogpathway and
basal cell carcinomas. Hum Mol Genet. 10:757–762. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Daya-Grosjean L and Couvé-Privat S: Sonic
hedgehog signaling in basal cell carcinomas. Cancer Lett.
225:181–192. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Berman DM, Karhadkar SS, Hallahan AR, et
al: Medulloblastoma growth inhibition by hedgehog pathway blockade.
Science. 297:1559–1561. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Watkins DN, Berman DM, Burkholder SG, Wang
B, Beachy PA and Baylin SB: Hedgehog signalling within airway
epithelialprogenitors and in small-cell lung cancer. Nature.
422:313–317. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gialmanidis IP, Bravou V, Amanetopoulou
SG, Varakis J, Kourea H and Papadaki H: Overexpression of hedgehog
pathwaymolecules and FOXM1 in non-small cell lung carcinomas. Lung
Cancer. 66:64–74. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Berman DM, Karhadkar SS, Maitra A, et al:
Widespread requirement for Hedgehog ligand stimulation in growth of
digestive tract tumours. Nature. 425:846–851. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mori Y, Okumura T, Tsunoda S, Sakai Y and
Shimada Y: Gli-1 expression is associated with lymph nodemetastasis
and tumor progression in esophageal squamous cell carcinoma.
Oncology. 70:378–389. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qualtrough D, Buda A, Gaffield W, Williams
AC and Paraskeva C: Hedgehog signalling in colorectal tumour cells:
induction of apoptosis with cyclopamine treatment. Int J Cancer.
110:831–837. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
ten Haaf A, Bektas N, von Serenyi S, et
al: Expression of the glioma-associated oncogene homolog (GLI) 1 in
human breast cancer is associated with unfavourable overall
survival. BMC Cancer. 9:2982009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Karhadkar SS, Bova GS, Abdallah N, et al:
Hedgehog signalling in prostateregeneration, neoplasia and
metastasis. Nature. 431:707–712. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liao X, Siu MK, Au CW, et al: Aberrant
activation of hedgehog signaling pathway in ovarian cancers: effect
onprognosis, cellinvasion and differentiation. Carcinogenesis.
30:131–140. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Feng YZ, Shiozawa T, Miyamoto T, et al:
Overexpression of hedgehog signalingmolecules and its involvement
in the proliferation of endometrial carcinoma cells. Clin Cancer
Res. 13:1389–1398. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen JK, Taipale J, Cooper MK and Beachy
PA: Inhibition of Hedgehog signaling by direct binding of
cyclopamine to Smoothened. Genes Dev. 16:2743–2748. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen JK, Taipale J, Young KE, Maiti T and
Beachy PA: Small molecule modulation of Smoothened activity. In:
Proc Natl Acad Sci USA. 99. pp. 14071–14076. 2002; View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ma X, Chen K, Huang S, et al: Frequent
activation of the hedgehog pathway in advanced gastric
adenocarcinomas. Carcinogenesis. 26:1698–1705. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee SY, Han HS, Lee KY, et al: Sonic
hedgehog expression in gastriccancer and gastric adenoma. Oncol
Rep. 17:1051–1055. 2007.PubMed/NCBI
|
|
22
|
Ohta M, Tateishi K, Kanai F, et al:
p53-Independent negative regulation of p21/cyclin-dependent
kinase-interacting protein 1 by the sonic
hedgehog-glioma-associated oncogene 1 pathway in gastric carcinoma
cells. Cancer Res. 65:10822–10829. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fukaya M, Isohata N, Ohta H, et al:
Hedgehog signal activation in gastric pitcell and in diffuse-type
gastric cancer. Gastroenterology. 131:14–29. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yoo YA, Kang MH, Kim JS and Oh SC: Sonic
hedgehog signaling promotesmotility and invasiveness of gastric
cancer cells through TGF-beta-mediated activation of the ALK5-Smad
3 pathway. Carcinogenesis. 29:480–490. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nagai S, Nakamura M, Yanai K, et al: Gli1
contributes to the invasiveness of pancreatic cancer through matrix
metalloproteinase-9 activation. Cancer Sci. 99:1377–1384. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Feldmann G, Dhara S, Fendrich V, et al:
Blockade of hedgehog signaling inhibits pancreatic cancerinvasion
and metastases: a new paradigm for combination therapy in solid
cancers. Cancer Res. 67:2187–2196. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Naef M, Ishiwata T, Friess H, Büchler MW,
Gold LI and Korc M: Differential localization of transforming
growth factor-beta isoforms in human gastricmucosa and
overexpression in gastric carcinoma. Int J Cancer. 71:131–137.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ebert MP, Yu J, Miehlke S, et al:
Expression of transforming growth factor beta-1 in gastriccancer
and in the gastric mucosa of first-degree relatives of patients
with gastric cancer. Br J Cancer. 82:1795–1800. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nag S, Qin J, Srivenugopal KS, et al: The
MDM2-p53 pathway revisited. J Biomed Res. 27:254–271. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fu H, Hu Z, Wen J, Wang K and Liu Y:
TGF-beta promotesinvasion and metastasis of gastric cancer cells by
increasing fascin1 expression viaERK and JNK signal pathways. Acta
Biochim Biophys Sin (Shanghai). 41:648–656. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Raman D, Baugher PJ, Thu YM and Richmond
A: Role of chemokines in tumor growth. Cancer Lett. 256:137–165.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Salvucci O, Bouchard A, Baccarelli A, et
al: The role of CXCR4 receptor expression in breast cancer: a large
tissue microarray study. Breast Cancer Res Treat. 97:275–283. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kodama J, Hasengaowa, Seki N, Kusumoto T
and Hiramatsu Y: Expression of the CXCR4 and CCR7 chemokine
receptors in human endometrial cancer. Eur J Gynaecol Oncol.
28:370–375. 2007.PubMed/NCBI
|
|
34
|
Engl T, Relja B, Marian D, et al: CXCR4
chemokine receptor mediates prostate tumor cell adhesion through
alpha5 and beta3 integrins. Neoplasia. 8:290–301. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee HJ, Kim SW, Kim HY, et al: Chemokine
receptor CXCR4 expression, function, and clinical implications in
gastric cancer. Int J Oncol. 34:473–480. 2009.PubMed/NCBI
|
|
36
|
Yoon JW, Gilbertson R, Iannaccone S,
Iannaccone P and Walterhouse D: Defining a role for Sonic hedgehog
pathway activation in desmoplastic medulloblastoma by identifying
GLI1 target genes. Int J Cancer. 124:109–119. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Katoh M: Integrative genomic analyses of
CXCR4: transcriptional regulation of CXCR4 based onTGFbeta, Nodal,
Activinsignaling and POU5F1, FOXA2, FOXC2, FOXH1, SOX17, and GFI1
transcription factors. Int J Oncol. 36:415–420. 2010.PubMed/NCBI
|