Introduction
Melanoma is the most aggressive type of cutaneous
malignancy and is responsible for 80% of mortalities from skin
cancer, despite accounting for only 4% of all cases of
dermatological cancer (1). Typically,
melanoma is histologically diagnosed with the aid of specific
protein markers, including S100, melanoma-associated antigen
recognized by T cells-1 (MART-1) and human melanoma black-45
(HMB-45). Additionally, the diagnosis of malignant melanoma (MM) is
commonly supported by the detection of BRAF mutations
(2).
By contrast, clear cell sarcoma (CCS), previously
known as melanoma of the soft parts, is an uncommon type of
malignant tumor. CCS typically presents as a deep soft-tissue mass,
and microscopically and immunohistochemically demonstrates evidence
of melanocytic differentiation (3).
While the majority of CCS occurs in the deep soft
tissue, superficial variants have recently been reported in the
dermis and/or superficial subcutis. Such CCS may be confused with
primary melanoma or MM, as CCS and MM can stain positively for S100
and HMB-45 (4,5). Determination of the differential
diagnosis of these tumors is facilitated by the result of a
previously conducted cytogenetic study that identified the presence
of a characteristic t(12;22) chromosomal translocation involving
the EWSR1 gene in 70–90% of CCS cases (6).
The current study reports the case of a 71-year-old
male who presented with a suspicious lymph node mass. The final
diagnosis of melanoma was difficult despite the support of
immunohistochemical analyses, thus, cytogenetic analysis was
performed to detect chromosomal rearrangements of the 22q12 region
and exclude a diagnosis of CCS. Subsequently, a previously
undescribed amplification of the chromosomal area adjacent to the
EWSR1 gene in the centromeric direction was revealed. The
study describes this chromosomal aberration and supports its
presence by conducting molecular investigations into potential
expression alterations of all genes included in the amplified area.
Written informed consent was obtained from the patient.
Case report
A 71-year-old male was admitted to the Department of
Melanoma and Soft Tissue Tumor Surgery of the National Cancer
Institute IRCCS ‘Fondazione G. Pascale’ (Naples, Italy) due to the
presence of a suspicious lymph node mass. An examination of the
medical history revealed that the patient had underdone a surgical
procedure six years previously to remove a skin tumor with a
diagnosis of MM. Following surgical excision of the lymph node
mass, macroscopic examination of the sample was used to
characterize a whitish-colored mass measuring 8.0×7.5×6.5 cm, with
interspersed areas grossly consistent with necrosis. Focally, the
lesion reached the margins of the exeresis (along the minor axis)
and was surrounded by adipose tissue. Microscopically, the lesion
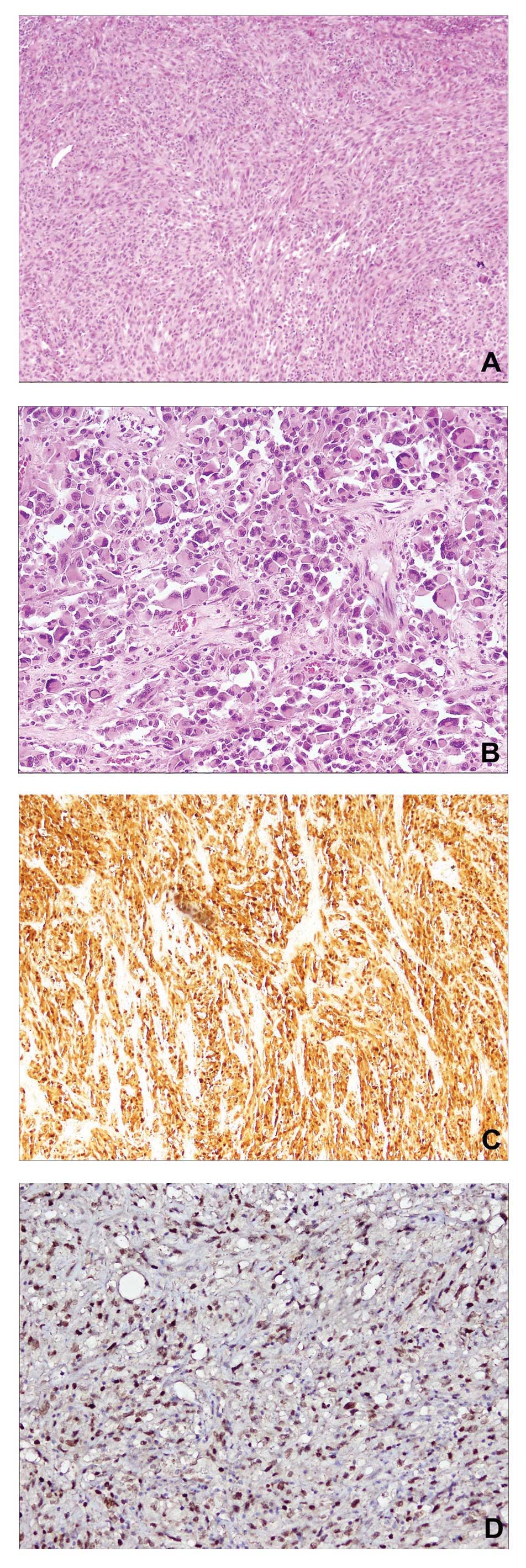
was characterized by a hypercellular neoplasia with epithelioid and
pleomorphic spindle cells, occasionally accompanied by the presence
of abundant collagen (Fig. 1A and B).
In addition to focal necrosis, evaluation of the mitotic activity
index revealed 15 mitotic figures in 10 high-power fields (15/10
hpf), and residual lymph node parenchyma was identified on the mass
boundary. Furthermore, immunohistochemical analysis identified
strong positivity for S100 (polyclonal rabbit anti-human clone
S100; cat. no. 760–2523; dilution, 1:100; Ventana Medical Systems,
Inc., Tuscon, AZ, USA; Fig. 1C) and
MITF (monoclonal mouse anti-human clone 34CA5; cat. no. NCL-MITF;
dilution, 1:20; Leica Microsystems Ltd., Wetzlar, Germany; Fig. 1D) protein expression, and negativity
for HMB-45 (monoclonal mouse anti-human clone gp100; cat. no.
790–4366; dilution, 1:100; Ventana Medical Systems, Inc.), MART-1
(monoclonal mouse anti-human A103; cat. no. ORG-8953; dilution,
1:50; Leica Microsystems Ltd.), muscle-specific actin (monoclonal
mouse anti-human clone HHF35; cat. no. NCL-L-MSA; dilution, 1:500;
Leica Microsystems Ltd.) and desmin (monoclonal mouse anti-human
clone D33; cat. no. 760–2513; dilution, 1:50; Ventana Medical
Systems, Inc.) markers.
To clarify the diagnosis, a molecular analysis was
performed to assess the mutational status of the BRAF gene.
In brief, a Food and Drug Administration-approved and CE-in
vitro diagnostics-marked quantitative polymerase chain reaction
(PCR)-based assay (cobas® 4800 BRAF V600 mutation test; Roche
Molecular Systems, Inc., Branchburg, NJ, USA) was employed to
identify potential mutations in codon 600, however, no mutations
were identified in the BRAF gene. Finally, to resolve the
problem of a differential diagnosis with CCS, cytogenetic analysis
was performed to investigate potential chromosomal rearrangements
of the 22q12 region associated with the EWSR1 gene
translocation. In brief, the fluorescence in situ
hybridization (FISH) method in conjunction with a Ewing sarcoma
breakpoint region 1 (EWSR1) probe (Vysis LSI EWSR1 Dual Color Break
Apart Probe; Abbott Molecular) was used to reveal a clear orange
amplification signal relative to an ~500-kb region adjacent to the
5′ EWSR1 gene in the centromeric direction (Fig. 2).
Analysis of the amplified 22q12 chromosomal region
(http://omim.org/geneMap/22/135?start=-3&limit=10&highlight=135)
was performed, and the expression of only three genes adjacent to
EWSR1, termed KREMEM1, ZNRF3 and XBP1,
were identified (OMIM nos. 609898, 612062 and 194355, respectively;
Fig. 3).
To verify possible gene overexpression associated
with chromosomal amplification, specific primers for each of the
three genes were designed to enable the performance of gene
expression analysis (Table I).
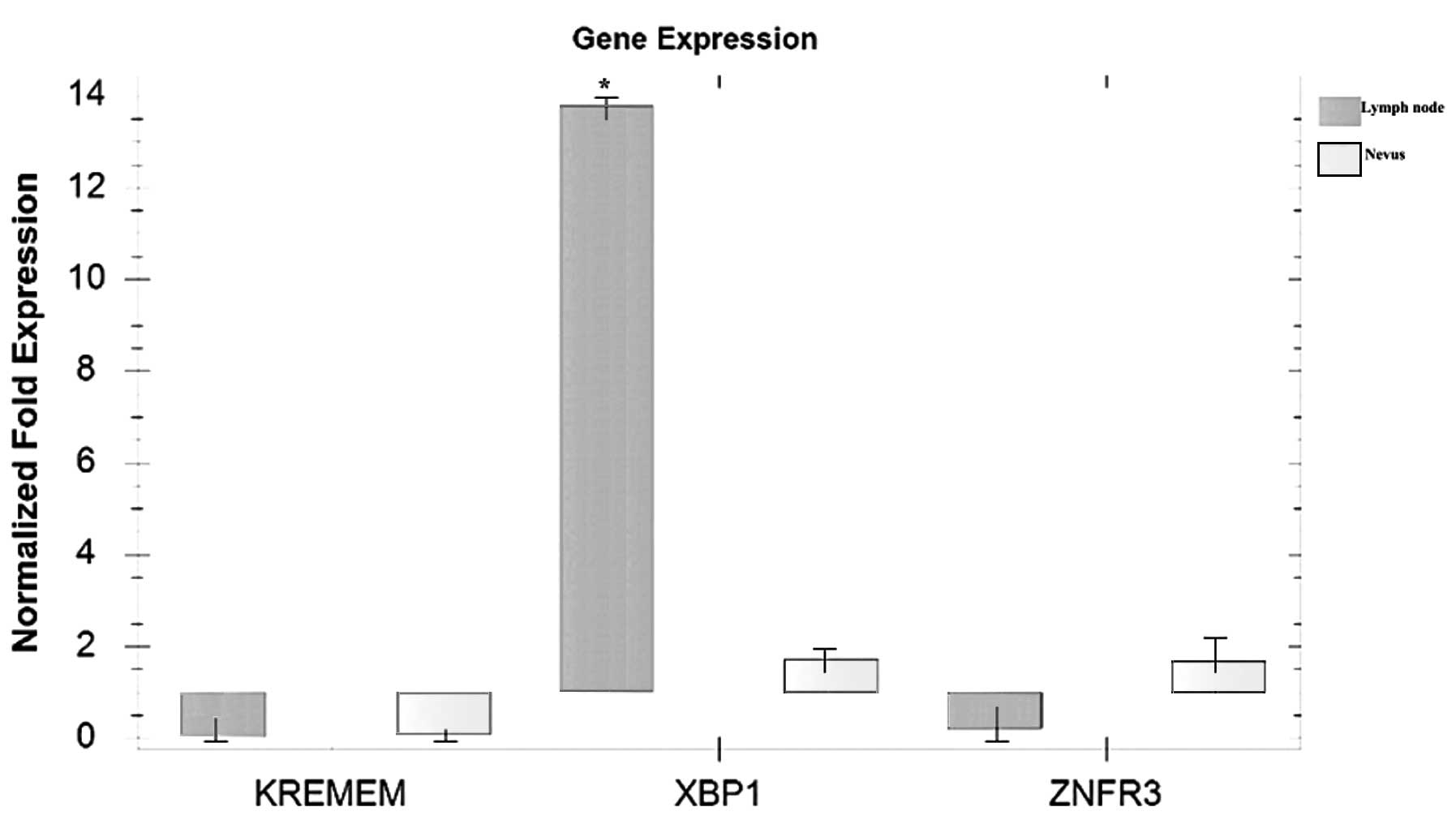
Qualitative PCR analysis revealed marked elevation only in
XBP1 gene expression in the lymph node specimen compared
with in the nevus sample, which was used as the non-neoplastic
control (Fig. 4). However, variation
in the gene or protein expression levels of X box-binding protein 1
(XBP1) may be observed in a larger series of melanoma samples,
thus, a quantitative, large-scale study is required to clarify the
results of the present study.
 | Table I.Primers used for polymerase chain
reaction analysis of the KREMEM1, ZNRF3 and
XBP1 genes. |
Table I.
Primers used for polymerase chain
reaction analysis of the KREMEM1, ZNRF3 and
XBP1 genes.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) | Product length,
bp | GenBank reference
number |
|---|
| ZNRF3 |
GGAGACCAGCAACCTCTCAC |
CCATGCTTGTCCTCGTAGGG | 116 | NM_001206998.1 |
| KREMEN1 |
GGGATGGAGTCAGGCTATGC |
TGTTGCATTCGGTACTGGCT | 85 | NM_001039570.2 |
| XBP1 |
AGCCAAGGGGAATGAAGTGAG |
CTGCAGAGGTGCACGTAGTC | 76 | NM_005080.3 |
The clinical history of the patient, as well as the
support of the in situ and molecular investigations, allowed
a conclusive diagnosis of lymph node metastasis from melanoma with
the presence of unconventional cytogenetic abnormalities to be
determined in the present patient.
Discussion
The current study describes the case of a male
patient with a previous history of melanoma, who presented with a
suspicious lymph node mass. The clinical history, supported by
morphological and immunohistochemical analysis, allowed a final
diagnosis of lymph node metastasis from melanoma to be established,
despite the analysis only demonstrating positivity for the S100
marker. For diagnostic and therapeutic purposes, the sample was
additionally evaluated to determine the mutational state of the
BRAF gene, revealing an absence of nucleotide
substitution.
Finally, considering the importance of the
differential diagnosis between MM and CCS, particularly when the
lesion occurs in the superficial side of the dermis, cytogenetic
analysis was performed. Cytogenetic analysis was determined as
essential for providing evidence of the potential presence of an
EWSR1 translocation with the ATF1 gene, which
characterizes the majority of CCS cases.
FISH analysis identified the presence of gene
translocations, however, an amplification of the 5′ region of the
EWSR1 gene in the centromeric direction was also identified.
The presence of this chromosomal aberration has been sporadically
described in Ewing's sarcoma (7),
however, it has not previously been described in cutaneous
melanoma, and a recent study reported aberrations of chromosome 22
in only a small percentage of acral melanomas (8). Thus, to the best of our knowledge, the
current case represents the first description of the amplification
of the 22q12 region in an MM patient. The chromosome region
involved in the amplification and detected by the probe was ~500 kb
in length, and, in addition to EWSR1, includes three other
genes: KREMEM1, ZNRF3 and XBP1.
KREMEM1, also termed KRM1, is a transmembrane
receptor that functionally cooperates with Dickkopf-related protein
1 to block wingless-type MMTV integration site family
(Wnt)/β-catenin signaling (9,10). The highest expression levels were
previously described in heart, lung, kidney, skeletal muscle and
neuroblastoma cell lines (10). Zinc
and ring finger 3 (ZNRF3) is a zinc finger protein that exerts its
activity as a cell-surface transmembrane E3 ubiquitin ligase. The
protein acts as a negative regulator of the Wnt signaling pathway
by mediating the ubiquitination and subsequent degradation of the
Wnt receptor complex components Frizzled and low-density
lipoprotein receptor-related protein 6. In particular, ZNRF3
appears to act as a tumor suppressor in colorectal tumor cell lines
(11). XBP1, previously termed XBP2,
is a transcription factor that regulates major histocompatibility
complex (MHC) class II genes by binding to a promoter element
referred to as X box. Furthermore, XBP1 is essential for hepatocyte
growth, the differentiation of plasma cells and immunoglobulin
secretion. Abnormal expression of MHC molecules in melanoma has
been widely described in the literature (12), however, none of the three genes
located in the 22q12 chromosomal area have previously been directly
associated with the pathogenesis of melanoma. Thus, possible
alterations in the gene expression levels were evaluated in the
present sample. The amplification of various chromosomal regions in
tumors is often associated with the amplification/overexpression of
specific genes (13,14). In the present case, only one of the
three genes, XBP1, demonstrated abnormal expression levels
compared with the non-neoplastic sample. Although the gene and
protein expression values require quantitative confirmation in a
larger series of melanoma cases, the data indicates specific
functional characteristics of this marker. Notably, XBP1 is
activated during endoplasmic reticulum (ER) stress by unfolded
protein response (UPR) and in particular, is activated by binding
with inositol-requiring enzyme 1, the most highly conserved
signaling node of the UPR (15,16).
Cancer cells commonly undergo chronic ER stress, to which the cells
have to adapt in order to survive and proliferate. In melanoma
cells, intrinsic activation of the ER stress response is driven by
oncogenic activation of mitogen-activated protein kinase
kinase/extracellular signal-regulated kinase (17). Furthermore, a previous study observed
that inhibition of XBP1 expression decelerated melanoma cell
proliferation and enhanced apoptosis induced by pharmacological ER
stress inducers (17). Melanoma is
the most frequent type of skin malignancy and is typically
characterized by a poor prognosis associated with high metastatic
capacity. Although numerous molecular pathways have been described
for melanoma progression, the molecular mechanisms that result in
metastatic development are not fully understood (1).
In conclusion, in the present study cytogenetic
analysis revealed chromosomal rearrangement of chromosome 22q12,
which was associated with the amplification of the XBP1
gene. To the best of our knowledge, this is the first study to
demonstrate an association between 22q12 chromosomal amplification
and melanoma. Therefore, the identification of novel molecular
markers may be useful for the improved diagnosis of melanoma, as
well as for potentially indicating the prognostic and predictive
value of various types of therapy.
Abbreviations:
|
CCS
|
clear cell sarcoma
|
|
EWSR1
|
Ewing sarcoma breakpoint region 1
|
|
MHC
|
major histocompatibility complex
|
|
XBP
|
X box-binding protein
|
|
ER
|
endoplasmic reticulum
|
|
UPR
|
unfolded protein response
|
References
|
1
|
Palmieri G, Capone M, Ascierto ML, et al:
Main roads to melanoma. J Transl Med. 7:862009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bhandaru M, Ardekani GS, Zhang G, et al: A
combination of p300 and Braf expression in the diagnosis and
prognosis of melanoma. BMC Cancer. 14:3982014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang L, Chen Y, Cui T, Knösel T, Zhang Q,
Geier C, Katenkamp D and Petersen I: Identification of biomarkers
to distinguish clear cell sarcoma from malignant melanoma. Hum
Pathol. 43:1463–1470. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kiuru M, Hameed M and Busam KJ: Compound
clear cell sarcoma misdiagnosed as a Spitz nevus. J Cutan Pathol.
40:950–954. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Falconieri G, Bacchi CE and Luzar B:
Cutaneous clear cell sarcoma: report of three cases of a
potentially underestimated mimicker of spindle cell melanoma. Am J
Dermatopathol. 34:619–625. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Song JS, Choi J, Kim JH, Jang SJ and Cho
KJ: Diagnostic utility of EWS break-apart fluorescence in situ
hybridization in distinguishing between non-cutaneous melanoma and
clear cell sarcoma. Pathol Int. 60:608–613. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Szuhai K, IJszenga M, Tanke HJ, Taminiau
AH, de Schepper A, van Duinen SG, Rosenberg C and Hogendoorn PC:
Detection and molecular cytogenetic characterization of a novel
ring chromosome in a histological variant of Ewing sarcoma. Cancer
Genet Cytogenet. 172:12–22. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Buckley PG, Mantripragada KK, Benetkiewicz
M, et al: A full-coverage, high-resolution human chromosome 22
genomic microarray for clinical and research applications. Hum Mol
Genet. 11:3221–3229. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mao B, Wu W, Davidson G, et al: Kremen
proteins are Dickkopf receptors that regulate Wnt/beta-catenin
signalling. Nature. 417:664–667. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nakamura T, Aoki S, Kitajima K, Takahashi
T, Matsumoto K and Nakamura T: Molecular cloning and
characterization of Kremen, a novel kringle-containing
transmembrane protein. Biochim Biophys Acta. 1518:63–72. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Koo BK, Spit M, Jordens I, et al: Tumour
suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis
of Wnt receptors. Nature. 488:665–669. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Degenhardt Y, Huang J, Greshock J,
Horiates G, Nathanson K, Yang X, Herlyn M and Weber B: Distinct MHC
gene expression patterns during progression of melanoma. Genes
Chromosomes Cancer. 49:144–154. 2010.PubMed/NCBI
|
|
13
|
Dei Tos AP, Doglioni C, Piccinin S, Sciot
R, Furlanetto A, Boiocchi M, Dal Cin P, Maestro R, Fletcher CD and
Tallini G: Coordinated expression and amplification of the MDM2,
CDK4, and HMGI-C genes in atypical lipomatous tumours. J Pathol.
190:531–536. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cantile M, Galletta F, Franco R, et al:
Hyperexpression of HOXC13, located in the 12q13 chromosomal region,
in well-differentiated and dedifferentiated human liposarcomas.
Oncol Rep. 30:2579–2586. 2013.PubMed/NCBI
|
|
15
|
Hetz C, Martinon F, Rodriguez D and
Glimcher LH: The unfolded protein response: integrating stress
signals through the stress sensor IRE1α. Physiol Rev. 91:1219–1243.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
He Y, Sun S, Sha H, et al: Emerging roles
for XBP1, a sUPeR transcription factor. Gene Expr. 15:13–25. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Croft A, Tay KH, Boyd SC, et al: Oncogenic
activation of MEK/ERK primes melanoma cells for adaptation to
endoplasmic reticulum stress. J Invest Dermatol. 134:488–497. 2014.
View Article : Google Scholar : PubMed/NCBI
|