Introduction
Esophageal carcinoma (EC) is the eighth most
aggressive and malignant type of cancer, with a high incidence that
varies according to geographic location and ethnicity (1). Despite progress in the development of
diagnostic and therapeutic options, the survival rates for EC
patients remain poor. Therefore, the identification of novel genes
involved in the tumorigenesis and development of EC is urgently
required.
Long non-coding RNAs (lncRNAs) are a class of RNAs
that have been reported to be involved in the regulation, invasion,
proliferation and apoptosis of multiple tumors (2,3). The
association between H19 expression and the progression of various
types of cancer has been demonstrated in previous studies. One
study found that the overexpression of lncRNA H19 enhanced the
carcinogenesis and metastasis of gastric cancer (4). MALAT-1, an abundant lncRNA present in
many human cell types, has been suggested to regulate the
alternative splicing of a subset of pre-messenger (m)RNAs by
modulating serine/arginine splicing factor activity. This factor in
turn regulates tissue or cell-type-specific alternative splicing in
a phosphorylation-dependent manner (5). However, the role of H19 in EC is yet to
be elucidated.
The epithelial-to-mesenchymal transition (EMT) has
an important role in the invasion of various types of cancer by
transforming adherent and polarized epithelial cells into invasive
and motile mesenchymal cells (6,7). A number
of transcription factors involved in EMTs, including Twist and
Snail, increase the expression level of mesenchymal markers,
including fibronectin, collagen and Vimentin, and decrease the
expression of epithelial markers, including E-cadherin. The
breakdown of tight junctions results in the loss of epithelial
markers and the acquisition of mesenchymal markers (8–10).
In the present study, the expression levels of H19
in EC were investigated, in order to elucidate the role of H19 in
EC.
Materials and methods
Clinical samples
EC samples with corresponding adjacent esophageal
tissues were obtained from 133 patients who had undergone routine
surgery at The Fourth Affiliated Hospital of Nantong Medical
College (Yancheng, China) between June 2007 and May 2012, and from
The First Affiliated Hospital of Nanjing Medical University
(Nanjing, China) between March 2010 and June 2013. The tissues were
stored at −80°C until the RNA extraction was performed. The
institutional committee approved the experiments. The present study
was approved by the Ethical Committee of Nantong Medical College,
and each patient provided written informed consent.
Cell culture
In total, five EC cell lines (TE-1, TE-10, Eca-1,
Eca-109 and KYSE1170) and one normal control cell line (HEEC) were
purchased from the Shanghai Institute of Biochemistry and Cell
Biology (Shanghai, China) and cultured in RPMI-1640 medium
supplemented with 10% fetal bovine serum (FBS) (Invitrogen Life
Technologies, Carlsbad, CA, USA). All cells were maintained in a
humidified 37°C incubator with 5% CO2.
Isolation of total RNA and reverse
transcription quantitative-polymerase chain reaction (RT-qPCR)
RNA was extracted from the tissue samples using
TRIzol reagent (Invitrogen Life Technologies, Shanghai, China).
Subsequently, complemetary DNA was synthesized using a reverse
transcriptase kit (Takara Bio, Inc., Otsu, Japan) according to the
manufacturers' instructions. The relative expression levels of H19
mRNA were determined using a SYBR Green real-time PCR kit (Takara
Bio, Inc.) and normalized to GAPDH. RT-PCR was performed using the
ABI 7500 Fast Real-Time PCR system (Applied Biosystems Life
Technologies, Foster City, CA, USA) and the following gene-specific
primers: Forward, 5′-ATCGGTGCCTCAGCGTTCGG-3′ and reverse,
5′-CTGTCCTCGCCGTCACACCG-3′ for H19; forward,
5′-CTGTCCTCGCCGTCACACCG-3′ and reverse, 5′-GGCATGGACTGTGGTCATGAG-3′
for GAPDH. All primers were designed using the National Center for
Biotechnology Information Primer-BLAST tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome).
PCR was performed under the following conditions: Denaturation at
at 50°C for 2 min, followed by 40 cycles of 95°C for 15 sec and
60°C for 1 min. Protein expression was quantified using the
2−∆CT method, as previously described (11).
Transwell assay
The invasive ability of the cell lines was
determined using a polycarbonate membrane, Boyden chamber insert
with an 8-µm-pore size in a Transwell apparatus (EMD Millipore,
Billerica, MA, USA). The transfected cells were first treated with
trypsin/EDTA solution (Invitrogen Life Technologies) and then
washed once with a serum-containing RPMI-1640 medium. In total,
1×105 cells in 0.2 ml serum-free RPMI-1640 medium were
seeded into the Transwell apparatus. Next, RPMI-1640 supplemented
with 600 µl 10% FBS was added to the lower chamber. In addition, an
invasion assay was performed following an identical procedure, with
the exception that the Transwell chamber filters were coated with
45 µg Matrigel (BD Biosciences, San Jose, CA, USA). Subsequent to a
24-h incubation at 37°C in a 5% CO2 incubator, the cells
on the upper surface of the insert were removed using a cotton
swab. The cells that had invaded to the lower surface of the insert
were fixed in 100% precooling methanol (Lindi, Shanghai, China) for
10 min, stained in 0.5% crystal violet (Beyotime Institute of
Biotechnology, Shanghai, China) for 30 min, rinsed in
phosphate-buffered saline (Sigma-Aldrich, St. Louis, MO, USA) and
analyzed using a microscope (XSP-4C; Changfang, Shanghai, China).
Invasive ability values were obtained by counting three fields per
membrane and then presented as the average of three independent
experiments.
Cell proliferation assay
The various cell lines were seeded into 96-well
plates at a density of 2000 cells/well. In total, 20 µl MTT (0.5
mg/ml) was added into each well and incubated at 37°C for 4 h.
Next, 200 µl DMSO was added to each well in order to dissolve the
precipitate. The optical density was then measured at 490 nm using
a microplate reader (Model 550; Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). Three independent experiments were performed in
quintuplicate.
Western blot analysis
The total proteins were extracted from the cultured
cells and then quantified using a bicinchoninic acid assay
(Beyotime Institute of Biotechnology). Next, the proteins were
fractionated by 5% SDS-PAGE (Beyotime Institute of Biotechnology),
transferred to a polyvinylidene fluoride membrane (Beyotime
Institute of Biotechnology), blocked in 4% dry milk at room
temperature for 1 h and then immunostained using primary polyclonal
rabbit anti-human E-cadherin (dilution, 1:500; cat. no. ab15148;
Abcam, Cambridge, MA, USA), anti-human fibronectin (dilution,
1:1,000; cat. no. ab61214; Abcam), anti-human vimentin (dilution,
1:5,000; cat. no. ab71144; Abcam) and anti-human GAPDH (dilution,
1:5,000; cat. no. ab9385; Abcam) antibodies at 4°C overnight. The
membranes were washed four times with PBS/0.1% Tween 20 solution
(Sigma-Aldrich) then incubated with horseradish
peroxidase-conjugated polyclonal goat anti-rabbit IgG (dilution,
1:2,000; cat. no. ab6721; Abcam) secondary antibodies for 1 h at
37°C. The results were then visualized using a chemiluminescent
detection system (Pierce ECL western blotting substrate detection
system; Thermo Fisher Scientific, Pittsburgh, PA, USA) and exposed
by the Molecular Imager ChemiDoc XRS System (Bio-Rad Laboratories,
Inc.). The integrated density of the bands was quantified using
Image Lab 4.1 software (Bio-Rad Laboratories, Inc.).
Transfection of small interfering RNAs
(siRNAs)
The cells were seeded into six-well plates and
transfected with 50 nM H19-targeting siRNA (siRNA/H19; GenePharma,
Shanghai, China) using Lipofectamine® 2000 (Invitrogen Life
Technologies) according to the manufacturer's instructions.
Non-targeting siRNA (siRNA/control) was used as the control. The
transfection efficiency was monitored by RT-qPCR. RT-PCR was
performed using the ABI 7500 Fast Real-Time PCR system (Applied
Biosystems Life Technologies, Foster City, CA, USA) and the
following gene-specific primers: Forward,
5′-ATCGGTGCCTCAGCGTTCGG-3′ and reverse, 5′-CTGTCCTCGCCGTCACACCG-3′
for H19; forward, 5′-CTGTCCTCGCCGTCACACCG-3′ and reverse,
5′-GGCATGGACTGTGGTCATGAG-3′ for GAPDH. PCR was performed under the
following conditions: Denaturation at 50°C for 2 min, followed by
40 cycles at 95°C for 15 sec and 60°C for 1 min. Protein expression
was quantified using the 2−∆CT method, as previously
described (11).
Plasmid construction and cell
transduction
The H19 sequence was synthesized and subcloned into
pCDNA3.1 (Invitrogen Life Technologies) to generate pCDNAH19.
Aberrant expression of H19 was achieved by transfection with
pCDNAH19. An empty pCDNA vector was used as the control. The
Eca-109 cells were cultured on a six-well plate, and transfected
with the pCDNA-H19 or empty vector using Lipofectamine 2000
(Invitrogen Life Technologies) according to the manufacturer's
instructions. The expression level of H19 was detected by qPCR
using the aforementioned primers, and was performed under the
following conditions: Denaturation at 50°C for 2 min, followed by
40 cycles at 95°C for 15 sec and 60°C for 1 min. Protein expression
was quantified using the 2−∆CT method, as previously
described (11).
Statistical analysis
The expression levels of H19 in the tissues were
evaluated using χ2 tests. All P-values are two-sided.
P<0.05 was considered to indicate a statistically significant
difference. Statistical analysis was performed using Stata 11
(StataCorp LP, College Station, TX, USA), and presented with Graph
PAD prism version 4.0 software (GraphPad Software, Inc., La Jolla,
CA, USA).
Results
H19 expression is increased in human
EC tissues and cell lines
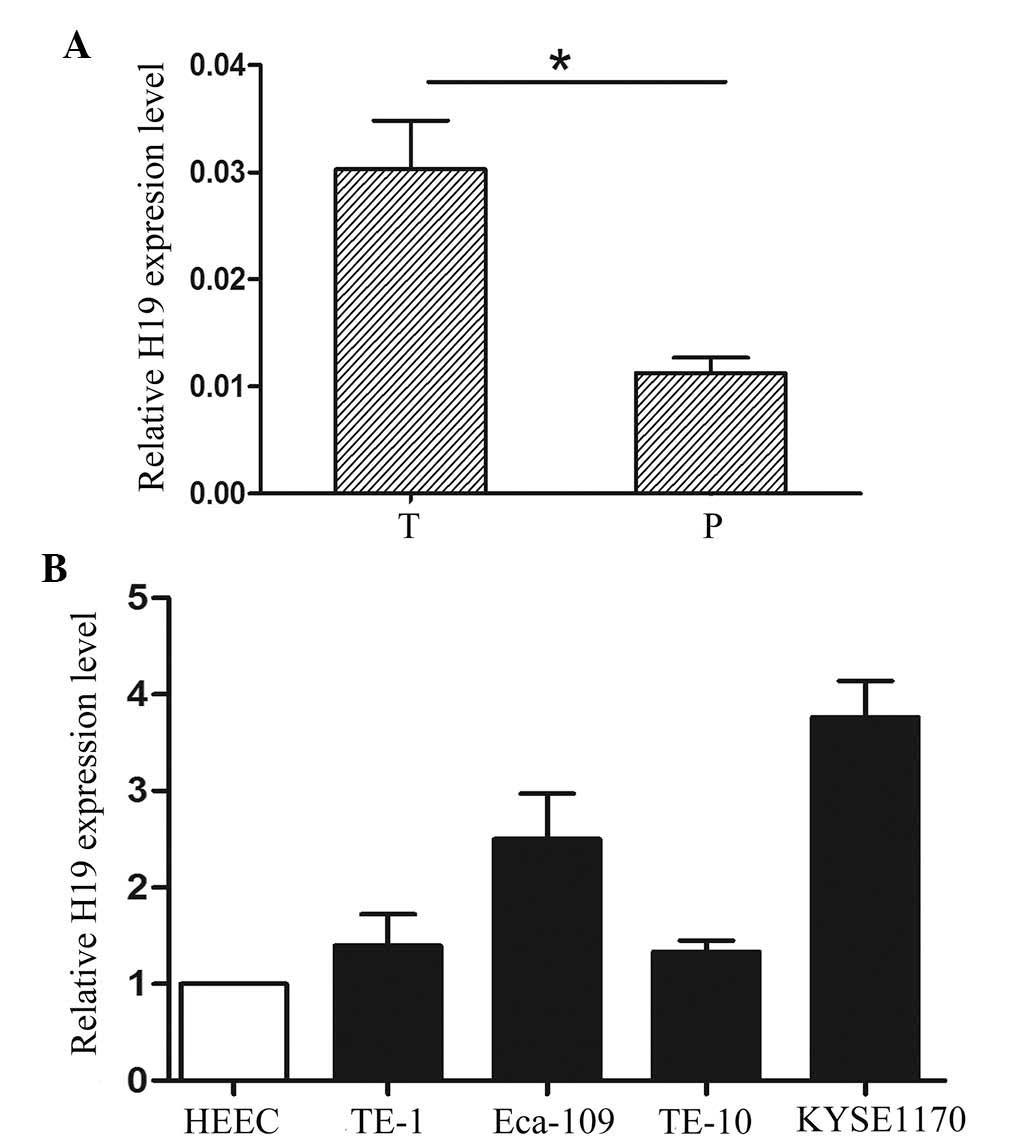
The results of the RT-qPCR analysis revealed that
the expression of H19 was higher in the 133 EC tissues compared
with that of the corresponding adjacent tissues (Fig. 1A). The cases were divided into H19
low- and high-expression groups. The median was used as the cut-off
value. The correlation between the expression of H19 and the
clinicopathological characteristics of the patients with EC are
shown in Table I. A marked
correlation was evident between H19 and tumor depth (P=0.007),
tumor stage (P=0.001) and metastasis (P=0.000). By contrast, no
positive associations with gender, age or histological
differentiation were noted. In addition, the expression of H19 was
analyzed in the EC cell lines (TE-1, TE-10, Eca-1, Eca-109 and
KYSE1170) and in the normal control cell line (HEEC). Compared with
the HEEC cells, H19 expression was significantly increased in the
EC cell lines (Fig. 1B). These
findings suggested that the aberrant expression of H19 may be
involved in the development and progression of EC.
 | Table I.Expression levels of H19 in esophageal
cancer and corresponding adjacent tissues. |
Table I.
Expression levels of H19 in esophageal
cancer and corresponding adjacent tissues.
| Factors | Patients, n | H19 low expression
(≤median), n | H19 high expression
(>median), n | P-value |
|---|
| Total | 133 | 67 | 66 |
|
| Age, years |
|
|
| 0.536 |
|
<64 | 60 | 32 | 28 |
|
| ≥64 | 73 | 35 | 38 |
|
| Gender |
|
|
| 0.663 |
| Male | 65 | 34 | 31 |
|
|
Female | 68 | 33 | 35 |
|
| Histology |
|
|
| 0.931 |
| AC | 66 | 33 | 33 |
|
| SCC | 67 | 34 | 33 |
|
| Tumor depth |
|
|
| 0.007 |
| Tis,
T1 | 65 | 25 | 40 |
|
| T2, T3,
T4 | 68 | 42 | 26 |
|
| Stage |
|
|
| 0.001 |
| 0, I | 63 | 22 | 41 |
|
| II, III,
IV | 70 | 45 | 25 |
|
| Metastasis |
|
|
| 0.000 |
| Yes | 40 | 30 | 10 |
|
| No | 93 | 37 | 56 |
|
H19 regulates EC cell invasion in
Eca-109 cells
Northern blot analysis has previously revealed that
H19 is increased in EC (12).
However, the potential mechanisms of H19 in the development of EC
are yet to be elucidated. The present study used a Transwell assay
in order to determine whether H19 had an effect on the invasion of
EC cells. Based on the expression of H19 in the EC cell lines,
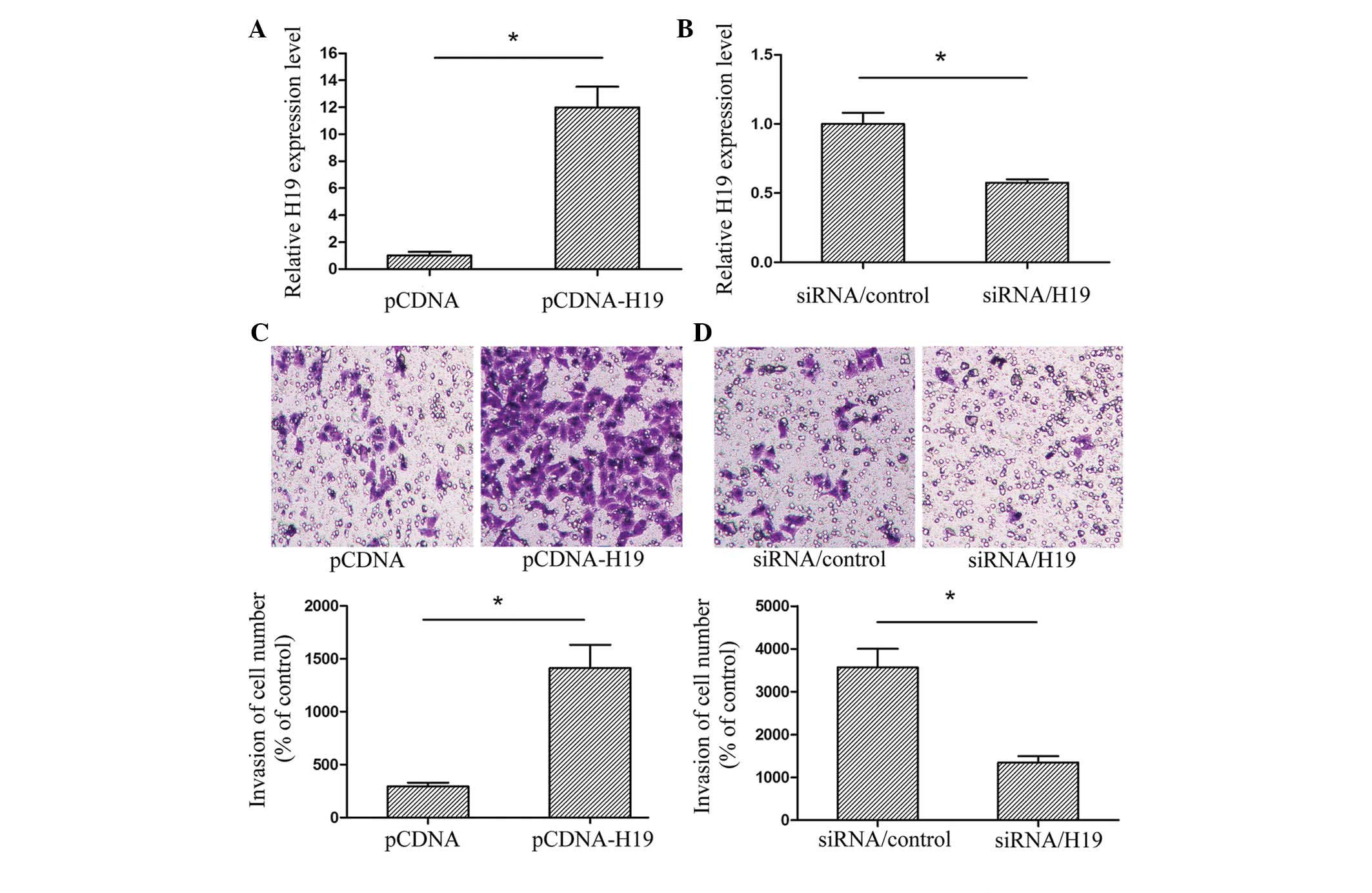
Eca-109 cells were selected for analysis. The Eca-109 cells were
transfected with pCDNA, pCDNA-H19, siRNA/control or siRNA/H19, and
the transfection efficiency was then validated using RT-qPCR
(Fig. 2A and B). The assay revealed
that upregulated H19 expression promoted Eca-109 cell invasion,
whereas a downregulation of H19 inhibited the invasion ability of
EC cell lines (Fig. 2C and D). The
results indicated that H19 may have an important role in regulating
the metastasis of EC.
Aberrant expression of H19 regulates
cell proliferation in vitro
An MTT assay was performed in order to investigate
whether H19 had an effect upon the proliferation of the EC cell
lines. The survival rate of the cells transfected with pCDNA-H19
was markedly higher than that of the controls, whereas the survival
rate of the cells transfected with siRNA/H19 was lower than that of
the controls (Fig. 3A and B). The
data indicated that aberrant expression of H19 was able to regulate
cell proliferation in vitro.
H19 regulates EMT
The Eca-109 cells were transfected with pCDNA,
pCDNA-H19, siRNA/control or siRNA/H19 in order to determine whether
H19 was involved in the EMT. The expression of the epithelial
marker, E-cadherin, and the mesenchymal markers, fibronectin and
Vimentin, was investigated using western blot analysis. At the
protein level, the upregulation of H19 expression by pCDNA-H19
resulted in decreased E-cadherin expression and increased Vimentin
and fibronectin expression. By contrast, the suppression of
expression by siRNA/H19 resulted in increased E-cadherin expression
and decreased Vimentin and fibronectin expression (Fig. 3C). Taken together, these findings
indicated that H19 may be involved in the regulation of EMT marker
expression in EC cell lines.
Discussion
EC is one of the most common causes of
cancer-associated mortalities worldwide (13). The standard treatment for patients
with early-stage disease, who have been diagnosed in accordance
with the tumor, node and metastasis classification, is surgical
resection. However, the majority of these patients subsequently
develop metastasis, even following successful surgery (14). Identifying the mechanisms that
underlie metastasis is therefore required, in order to improve
treatment outcomes.
Previous data has identified that lncRNAs have
regulatory roles in cancer proliferation, invasion and prognosis.
In a previous study of EC, HNF1A-AS1 knockdown significantly
inhibited cell proliferation and anchorage-independent growth,
suppressed S-phase entry, and inhibited cell migration and invasion
in multiple in vitro models of esophageal adenocarcinoma
(15). In a further study, HOTAIR
directly decreased the expression of WIF-1 by inducing promoter
region histone H3K27 methylation and activation of the
Wnt/β-catenin signaling pathway (16). The results of the present study
indicated that the expression of H19 was higher in EC tissues
(n=133) compared with that of the corresponding adjacent tissues.
Furthermore, it was revealed that the aberrant expression of H19
affected the invasion potential of EC cell lines in
vitro.
Decreased E-cadherin and increased Vimentin and
Snail expression are characteristic of EMT; a process known to be
significant in cancer invasion (17).
Previous studies have established that EMT is associated with tumor
invasiveness, metastasis and prognosis (18,19).
Furthermore, a number of studies have identified functional
associations between lncRNAs and key effectors of EMT during
carcinogenesis and embryonic development, including LincRNA-ROR
(20), MALAT-1 (21) and BANCR (22). In addition to its role in cancer
progression, EMT contributes to chronic epithelial injury (23), which leads to tissue fibrosis and
organ failure (24,25). The present study also revealed that an
overexpression of H19 led to a decreased expression of the
epithelial marker, E-cadherin, and increased expression of
mesenchymal markers, Vimentin and Snail. The downregulation of H19
had the opposite effect. These results suggested that H19 may
promote EC invasion by inducing EMT.
In conclusion, higher H19 expression levels were
detected in EC tumor tissues than in corresponding adjacent
tissues. The H19 expression levels were associated with tumor
depth, stage and metastasis. Furthermore, H19 was able to regulate
the invasion and proliferation of EC cells, and induce EMT in
vitro.
Acknowledgements
The authors would like to thank Dr Linjie Si (The
First Affiliated Hospital of Nanjing Medical University, Nanjing,
China) for providing the EC samples.
References
|
1
|
Zhang F, Yang Z, Cao M, et al: MiR-203
suppresses tumor growth and invasion and down-regulates MiR-21
expression through repressing Ran in esophageal cancer. Cancer
Lett. 342:121–129. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang F, Li X, Xie X, Zhao L and Chen W:
UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma
and embryo, influencing cell growth and promoting invasion. FEBS
Lett. 582:1919–1927. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wu ZH, Wang XL, Tang HM, et al: Long
non-coding RNA HOTAIR is a powerful predictor of metastasis and
poor prognosis and is associated with epithelial-mesenchymal
transition in colon cancer. Oncol Rep. 32:395–402. 2014.PubMed/NCBI
|
|
4
|
Li H, Yu B, Li J, et al: Overexpression of
lncRNA H19 enhances carcinogenesis and metastasis of gastric
cancer. Oncotarget. 5:2318–2329. 2014.PubMed/NCBI
|
|
5
|
Gutschner T, Hämmerle M, Eissmann M, et
al: The noncoding RNA MALAT1 is a critical regulator of the
metastasis phenotype of lung cancer cells. Cancer Res.
73:1180–1189. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Y, Wen M, Kwon Y, et al: CUL4A
induces epithelial-mesenchymal transition and promotes cancer
metastasis by regulating ZEB1 expression. Cancer Res. 74:520–531.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu J, Ruan B, You N, et al:
Downregulation of miR-200a induces EMT phenotypes and CSC-like
signatures through targeting the β-catenin pathway in hepatic oval
cells. PLoS One. 8:e794092013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dong H, Xie L, Tang C, et al: Snail1
correlates with patient outcomes in E-cadherin-preserved
gastroesophageal junction adenocarcinoma. Clin Transl Oncol.
16:783–791. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu Y, Li H, Feng J, et al: Lin28 induces
epithelial-to-mesenchymal transition and stemness via
downregulation of let-7a in breast cancer cells. PLoS One.
8:e830832013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bao YX, Cao Q, Yang Y, et al: Expression
and prognostic significance of golgiglycoprotein73 (GP73) with
epithelial-mesenchymal transition (EMT) related molecules in
Hepatocellular Carcinoma (HCC). Diagn Pathol. 8:1972013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li W, Jiang G, Zhou J, et al:
Down-regulation of miR-140 induces EMT and promotes invasion by
targeting Slug in esophageal cancer. Cell Physiol Biochem.
34:1466–1476. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hibi K, Nakamura H, Hirai A, et al: Loss
of H19 imprinting in esophageal cancer. Cancer Res. 56:480–482.
1996.PubMed/NCBI
|
|
13
|
Shigaki H, Baba Y, Watanabe M, Murata A,
Ishimoto T, Iwatsuki M, Iwagami S, Nosho K and Baba H: PIK3CA
mutation is associated with a favorable prognosis among patients
with curatively resected esophageal squamous cell carcinoma. Clin
Cancer Res. 19:2451–2459. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Koshy M, Esiashvilli N, Landry JC, Thomas
CR Jr and Matthews RH: Multiple management modalities in esophageal
cancer: combined modality management approaches. Oncologist.
9:147–159. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang X, Song JH, Cheng Y, et al: Long
non-coding RNA HNF1A-AS1 regulates proliferation and migration in
oesophageal adenocarcinoma cells. Gut. 63:881–890. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ge XS, Ma HJ, Zheng XH, et al: HOTAIR, a
prognostic factor in esophageal squamous cell carcinoma, inhibits
WIF-1 expression and activates Wnt pathway. Cancer Sci.
104:1675–1682. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kitamura K, Seike M, Okano T, et al:
MiR-134/487b/655 cluster regulates TGF-β-induced
epithelial-mesenchymal transition and drug resistance to gefitinib
by targeting MAGI2 in lung adenocarcinoma cells. Mol Cancer Ther.
13:444–453. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo S, Xu X, Tang Y, et al: miR-15a
inhibits cell proliferation and epithelial to mesenchymal
transition in pancreatic ductal adenocarcinoma by down-regulating
Bmi-1 expression. Cancer Lett. 344:40–46. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yamada S, Fuchs BC, Fujii T, et al:
Epithelial-to-mesenchymal transition predicts prognosis of
pancreatic cancer. Surgery. 154:946–954. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hou P, Zhao Y, Li Z, et al: LincRNA-ROR
induces epithelial-to-mesenchymal transition and contributes to
breast cancer tumorigenesis and metastasis. Cell Death Dis.
5:e12872014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ying L, Chen Q, Wang Y, Zhou Z, Huang Y
and Qiu F: Upregulated MALAT-1 contributes to bladder cancer cell
migration by inducing epithelial-to-mesenchymal transition. Mol
Biosyst. 8:2289–2294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun M, Liu XH, Wang KM, et al:
Downregulation of BRAF activated non-coding RNA is associated with
poor prognosis for non-small cell lung cancer and promotes
metastasis by affecting epithelial-mesenchymal transition. Mol
Cancer. 13:682014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vitalone MJ, Naesens M, Sigdel T, Li L,
Hseih S and Sarwal MM: The dual role of epithelial-to-mesenchymal
transition in chronic allograft injury in pediatric renal
transplantation. Transplantation. 92:787–795. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
López-Novoa JM and Nieto MA: Inflammation
and EMT: An alliance towards organ fibrosis and cancer progression.
EMBO Mol Med. 1:303–314. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mucsi I and Rosivall L:
Epithelial-mesenchymal transition in renal tubular cells in the
pathogenesis of progressive tubulo-interstitial fibrosis. Acta
Physiol Hung. 94:117–131. 2007. View Article : Google Scholar : PubMed/NCBI
|