Introduction
Breast cancer is the most common malignancy in
females worldwide and its incidence has increased rapidly over
recent decades (1). It is
well-established that polyamines are low molecular weight,
positively-charged compounds that are highly-expressed in numerous
malignancies, including breast cancer (2). Previous studies have indicated that
polyamines may affect various processes in transcription, RNA
stabilization, ion channel gating and carcinogenesis, while
inhibition of polyamine synthesis appears to be important in
inhibiting proliferation, decreasing apoptosis and suppressing
angiogenesis (3–5). Ornithine decarboxylase (ODC) is a
rate-limiting enzyme that contributes to polyamine synthesis
(6). Increased polyamine levels and
ODC activity have been identified in various types of human cancer,
particularly in breast cancer. Therefore, inhibiting ODC activity
was able to suppress cancer development in animal models (7–10). ODC
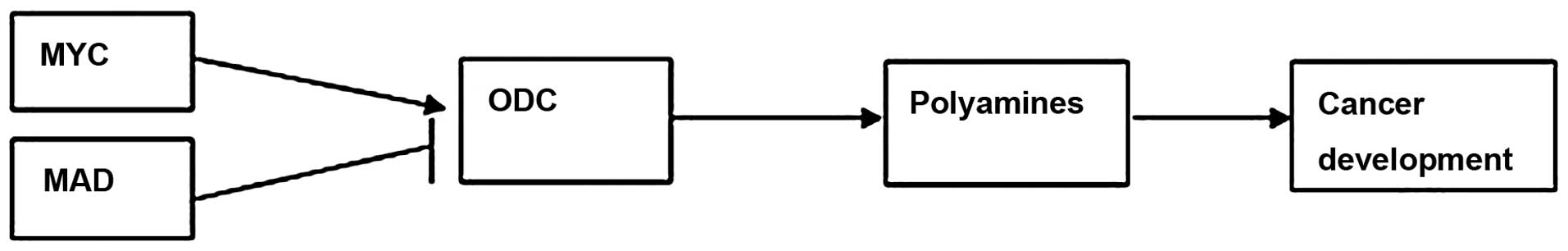
expression is regulated by the MYC and MAD transcription factors,
which include the MYC-associated factor X (MAX) network
transcriptional repressor, MAX interactor 1 (MXI1), MAD1 and MAD4
(11). When MYC forms a heterodimer
with MAX, it can bind to DNA at the E-box sequence (CACGTG) and
activate ODC gene expression. By contrast, MAD combines with MAX,
which can suppress the transcription of the ODC gene (Fig. 1) (12,13).
ODC G316A is a functional single nucleotide
polymorphism (SNP) that is located between two promoter
region/transcription factor binding sites. Furthermore, the
transcription of the ODC gene can be altered by this SNP (14). Previous studies have demonstrated that
the ODC G316A polymorphism exhibits prognostic efficacy in
colorectal adenoma recurrence and is associated with the survival
of patients with colorectal cancer (15,16).
However, the effects of this SNP on the clinical outcomes in breast
cancer remain unclear. Therefore, the aim of the present
population-based study was to investigate the association between
the ODC G316A SNP and breast cancer-specific mortality by
genotyping 300 samples from breast cancer patients that were
enrolled at the Affiliated Cancer Hospital of Zhengzhou University
(Zhengzhou, China) between 2002 and 2003. Subsequently, the
functional significance of the ODC G316A SNP in the MCF-7 breast
cancer cell line was investigated.
Materials and methods
Epidemiological studies
Study population
The present study investigated 300 patients with
stage I–III breast cancer that were enrolled at the Affiliated
Cancer Hospital of Zhengzhou University between 2002 and 2003, with
follow-up visits conducted until May 2013. For all breast cancer
patients recruited to the present study, written informed consent
was obtained from the patients or the patient's families, allowing
the collection of paraffin-embedded tissue samples and the release
of the patients' medical data. Furthermore, the present study was
approved by the Ethics Committee of the Affiliated Cancer Hospital
of Zhengzhou University. Clinical and demographic data, including
vital status and follow-up details, were obtained by associating
follow-up and regional cancer registry databases. Information
regarding lifestyle factors and a detailed family history were
collected from all the study participants during face-to-face
interviews. Tumor grade was determined according to the 2013 breast
cancer guidelines of the National Comprehensive Cancer Network
(17). The morphological features
(tubule formation, nuclear pleomorphism and mitotic count) of the
tumors were assessed and each assigned a score of 1–3. The scores
for each category were combined to determine the histological
grades of the tumors as follows: Grade 1 (low), score, 3–5; grade 2
(intermediate), score, 6–7; and grade 3 (high), score, 8–9. The
‘tumor node metastasis’ staging was derived from existing American
Joint Committee on Cancer (AJCC) codes, where available, and
conversion of the extent of disease codes was performed as
previously reported (18,19).
DNA extraction
Subsequently, DNA was extracted from 300
paraffin-embedded breast cancer tissue samples obtained during
surgery between May and June 2013, which were previously stored at
the Affiliated Cancer Hospital of Zhengzhou University, using a
QIAamp DNA FFPE Tissue kit (Qiagen, Hilden, Germany) (20), according to the manufacturers
instructions.
Primer design
The ODC sequence was obtained from GenBank
(accession no. NC_009142.1) and polymerase chain reaction (PCR)
primers were designed using the Premier Primer software (version
5.0; Premier Biosoft, Palo Alto, CA, USA). The forward (F) 1 and
reverse (R) 1 primers were used to amplify a 547-bp region, while
the F2 and R2 primers were used amplify a 178-bp region within the
first 547-bp amplification. The two PCR primer pairs are indicated
in Table I.
 | Table I.Oligonucleotide primers used to
amplify the ornithine decarboxylase gene. |
Table I.
Oligonucleotide primers used to
amplify the ornithine decarboxylase gene.
| Primer | Sequence | Size of DNA product,
bp |
|---|
| F1 |
5-GGTGCTATAAGTAGGGAGCGGC-3 | 547 |
| R1 |
5-CGAAGGGTTGGGAAAGAGGC-3 | 547 |
| F2 |
5-CCTGGGGGCTCTGAGGT-3 | 178 |
| R2 |
5-AGGAAGCGGCGCCTCAA-3 | 178 |
Nested PCR-restriction fragment length
polymorphism (RFLP)
ODC G316A genotypes were generated using nested
PCR-RFLP technology. DNA amplification was performed in a 25-µl
reaction volume consisting of 12.5 µl 2X PCR buffer for KOD FX
(Toyobo Co., Ltd., Osaka, Japan), 5 µl 2 mM deoxynucleotide
triphosphate, 2 µl of each primer, 0.1 µl KOD, 2.4 µl water and 1
µl DNA. The standard conditions for PCR were as follows: 95°C for 2
min; followed by 40 cycles each at 95°C for 30 sec and 62°C for 1
min; and a final extension at 72°C for 5 min. All the reactions
were performed in a Perkin Elmer 2400 thermocycler (Perkin Elmer,
Foster City, CA, USA). The nested PCR products were analyzed by
detecting the polymorphic PstI site using capillary
electrophoresis (QIAxcel Advanced System; Qiagen).
Experimental studies
Cell culture
MDA-435 cells and the MCF-7 human breast cancer cell
line, which exhibited an ODC AG genotype, were maintained in
Dulbecco's modified Eagle's medium (Invitrogen Life Technologies,
Carlsbad, CA, USA) and all media used were supplemented with 10%
fetal bovine serum. Furthermore, cells were maintained at 37°C in a
humidified atmosphere of 5% CO2.
Western blot analysis
Cells were harvested and lysed, and proteins were
separated on a 12.5% SDS-PAGE gel (15). Next, the proteins were transferred by
electrophoresis onto a Hybond-C membrane (GE Healthcare Life
Sciences, Little Chalfont, UK), which was blocked with Blotto A (5%
blocking grade dry milk in Tris-buffered saline/Tween-20; Santa
Cruz Biotechnology, Inc., Dallas, TX, USA), and probed using
monoclonal mouse anti-human c-MYC (1:300; cat. no. sc-41; Santa
Cruz Biotechnology, Inc.) and monoclonal mouse anti-human anti-MXI1
(1:300; cat. no. sc-130627; Santa Cruz Biotechnology, Inc.) primary
antibodies in Blotto A. The primary antibodies were incubated at
4°C overnight, followed by incubation with a monoclonal goat
anti-mouse Ig horseradish peroxidase-tagged secondary antibody
(1:1,000; cat. no. sc-2005; Santa Cruz Biotechnology, Inc.) for 1 h
at room temperature. Chemiluminescent detection was conducted using
an electrochemiluminescence western blotting detection reagent (GE
Healthcare Life Sciences) and exposing on a Biomax XAR film (Kodak,
Rochester, NY, USA).
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed using a ChIP Assay Kit,
according to the manufacturer's instructions (Cell Signaling
Technology, Inc., Danvers, MA, USA). Briefly, the cells were
treated with 1% formaldehyde to cross-link the DNA and proteins
prior to disruption of the DNA-protein complexes by sonication [25%
ultrasound; 14 cycles (4.5 sec each)] to produce fragment lengths
of 200–1,000 bp. The lysates were then diluted 10 times with
immunoprecipitation dilution buffer containing 1 mmol/l
phenylmethanesulfonyl fluoride. Antibodies for c-MYC and MXI1
(Santa Cruz Biotechnology, Inc.) were used to treat the samples, in
order to induce chromatin precipitation. However, one sample was
left untreated as a minus-antibody control. Immunoprecipitation was
performed overnight at 4°C with centrifugation at 234.8 × g for 15
min. Subsequently, immune complexes were obtained by adding 60 µl
salmon sperm DNA/protein A agarose slurry (Merck Millipore,
Darmstadt, Germany) and incubating for 1 h at 4°C with rotation,
followed by gentle centrifugation (335.4 × g for 1 min). Protein A
agarose pellets were washed with low-salt, high-salt, LiCl and
Tris-EDTA buffer. The complexes were then eluted twice by adding
250 µl elution buffer (0.1 mol/l NaHCO3 and 1% SDS), and
DNA-protein cross-links were reversed using 0.2 mol/l NaCl and
heating at 65°C for 4 h for all the samples, including the input
DNA and minus-antibody DNA controls. Finally, DNA was re-suspended
in 30 µl double-distilled H2O.
Statistical analysis
All the statistical data were analyzed using SPSS
statistical software (version 17.0; SPSS Inc., Chicago, IL, USA). A
χ2 test was used to describe the breast cancer cases
overall and in regard to their ODC genotype. Survival curves were
plotted using the Kaplan-Meier method and analyzed using the
log-rank test. Furthermore, Coxs proportional hazards model was
used to identify prognostic factors for survival. For all data,
P<0.05 was considered to indicate a statistically significant
difference.
Results
Epidemiological study
Clinicopathological characteristics of breast
cancer based on ODC G316A SNP genotypes
In order to enhance sensitivity, nested PCR was used
to amplify the products. The first set of primers were designed to
amplify a 547-bp region within the ODC gene, while the second
primer pair was designed to amplify a 178-bp region within the
547-bp region, and these were used in the nested PCR assay.
Capillary electrophoresis technology was subsequently employed to
determine the sequence of the PCR products. For the 178-bp region
of the ODC gene, PstI enzyme digestion and PCR-RFLP were
used to genotype the ODC G316A SNP. The results of capillary
electrophoresis were termed C1-C300. For the ODC AG genotype, the
178, 50 and 120-bp regions were obtained. However, only a single
region (178-bp) was identified for ODC GG, whereas the 50 and 20-bp
regions were detected for ODC AA. A total of 300 stage I–III breast
cancer cases admitted at the Affiliated Cancer Hospital of
Zhengzhou University were used in the present case-only analysis.
Nested PCR-RFLP was used to genotype the patients, and the median
follow-up duration was 10 years and 3 months. The cohort included
156 (52%) ODC GG and 144 (48%) ODC GA/AA cases. Clinicopathological
data for the breast cancer cases based on ODC genotype are
indicated in Table II.
 | Table II.Descriptive analysis for breast cancer
cases overall and based on ODC genotype. |
Table II.
Descriptive analysis for breast cancer
cases overall and based on ODC genotype.
|
|
| ODC G316A
genotype |
|
|---|
|
|
|
|
|
|---|
| Category | Total breast cancer
cases (n=300), n (%) | GG (n=156), n
(%) | GA/AA (n=144), n
(%) | P-value |
|---|
| Age,
yearsa |
|
|
| 0.280 |
| ≤45 | 125 (41.67) | 68 (43.59) | 57 (39.58) |
|
|
>45 | 175 (58.33) | 88 (56.41) | 87 (60.42) |
|
| Stage at
diagnosis |
|
|
| 0.482 |
| I | 90 (30.00) | 48 (30.77) | 42 (29.17) |
|
| II | 108 (36.00) | 56 (35.90) | 52 (36.11) |
|
|
III | 102 (34.00) | 52 (33.33) | 50 (34.72) |
|
| Family history of
cancer |
|
|
| 0.948 |
|
Yes | 106 (35.33) | 54 (34.62) | 52 (36.11) |
|
| No | 194 (64.67) | 102 (65.38) | 92 (63.89) |
|
| Tumor grade |
|
|
| 0.787 |
| 1 | 64 (21.33) | 35 (22.44) | 29 (20.14) |
|
| 2 | 191 (63.67) | 97 (62.18) | 94 (65.28) |
|
| 3 | 45 (15.00) | 24 (15.38) | 21 (14.58) |
|
| ER and PR
status |
|
|
| 0.848 |
|
Positive | 187 (62.33) | 99 (63.46) | 88 (61.11) |
|
|
Negative | 113 (37.67) | 57 (36.54) | 56 (38.89) |
|
| Her-2 status |
|
|
| 0.675 |
|
Positive | 128 (42.67) | 65 (41.67) | 63 (43.75) |
|
|
Negative | 172 (57.33) | 91 (58.33) | 81 (56.25) |
|
| Ki-67 status |
|
|
| 0.715 |
|
Positive | 167 (55.67) | 85 (54.49) | 82 (56.94) |
|
|
Negative | 133 (44.33) | 71 (45.51) | 62 (43.05) |
|
| Surgical
treatment |
|
|
| 0.669 |
|
Yes | 300 (100.00) | 156 (100.00) | 144 (100.00) |
|
| No | 0 (0.00) | 0 (0.00) | 0 (0.00) |
|
| Radiation
therapy |
|
|
| 0.963 |
|
Yes | 165 (55.00) | 86 (55.12) | 79 (54.86) |
|
| No | 135 (45.00) | 70 (44.87) | 65 (45.14) |
|
| Chemotherapy |
|
|
| 0.981 |
|
Yes | 246 (82.00) | 128 (82.05) | 18 (81.94) |
|
| No | 54 (18.00) | 28 (17.95) | 26 (18.06) |
|
Cancer-specific survival time based on ODC G316A
genotypes
Of the 300 stage I–III breast cancer cases, 168
patients (56%) had succumbed to the disease prior to the analysis.
In total, 72 mortalities (42.86%) occurred in patients carrying the
ODC GG genotype, compared with 96 mortalities (57.14%) in patients
with the GA/AA genotypes. A statistically significant improvement
in breast cancer-specific survival was observed among breast cancer
cases homozygous for the ODC G-allele (10-year survival, 53.85%)
compared with cases exhibiting at least one A-allele (ODC GA/AA;
10-year survival, 33.33%; P<0.001). Furthermore, breast
cancer-specific survival analysis by stage revealed no
statistically significant differences in the survival of patients
with AJCC stages I (P=0.537) or II (P=0.482; data not shown).
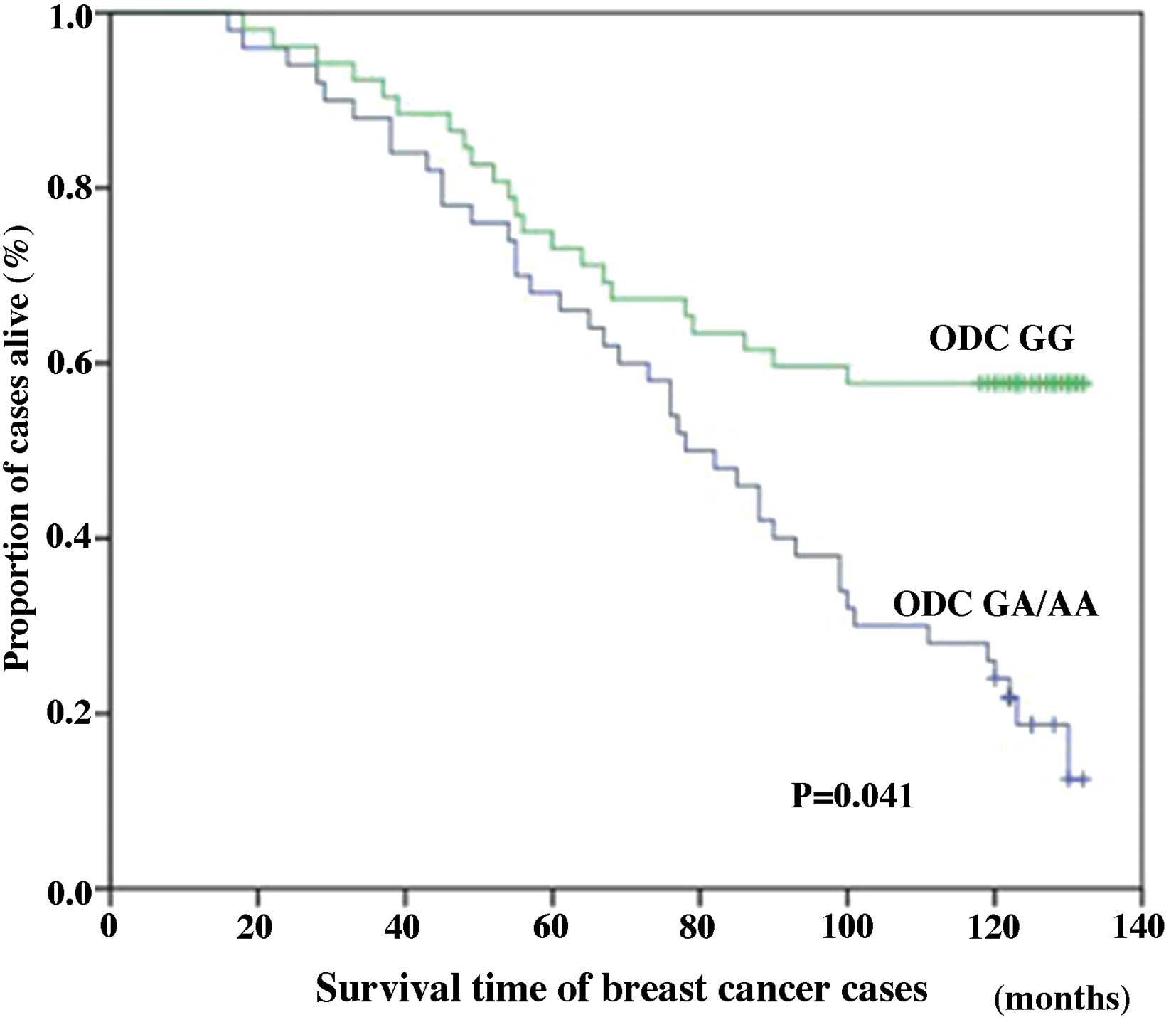
However, among cases of stage III breast cancer, the ODC GG
genotype was significantly associated with improved 10-year breast
cancer-specific survival (38.46%) compared with the ODC GA/AA
genotype cases (20.00%; P=0.041; Fig.
2). Among all the cases (stages I–III), differences in the
genotype-specific breast cancer survival were statistically
significant, with the ODC G316A SNP acting as an independent
predictor of breast cancer-specific survival. Compared with the ODC
GG genotype cases [hazard ratio (HR), 1; 95% confidence interval
(CI), 1, reference], the breast cancer-specific risk of mortality
was significantly greater for the ODC GA/AA genotype (HR, 1.57; 95%
CI, 1.16–4.23; P=0.037; Table
III).
 | Table III.Breast cancer-specific survival
analysis of patients with breast cancer based on the ODC G316A
genotype. |
Table III.
Breast cancer-specific survival
analysis of patients with breast cancer based on the ODC G316A
genotype.
|
| Breast
cancer-specific mortality |
|---|
|
|
|
|---|
| ODC G316A
genotype | Events, n | At risk, n | HR (95%
CI)a |
|---|
| GG | 32 | 156 | 1 (reference) |
| GA/AA | 40 | 144 | 1.57
(1.16–3.34) |
Experimental studies
Western blot analysis
The expression of specific E-box binding proteins,
including the transcriptional activator c-MYC and the
transcriptional repressor MXI1, was established by western blotting
of MCF-7 cell proteins (Fig. 3).
c-MYC and MXI1 were expressed in the MDA-435 and MCF-7 cells
(Fig. 3).
ChIP
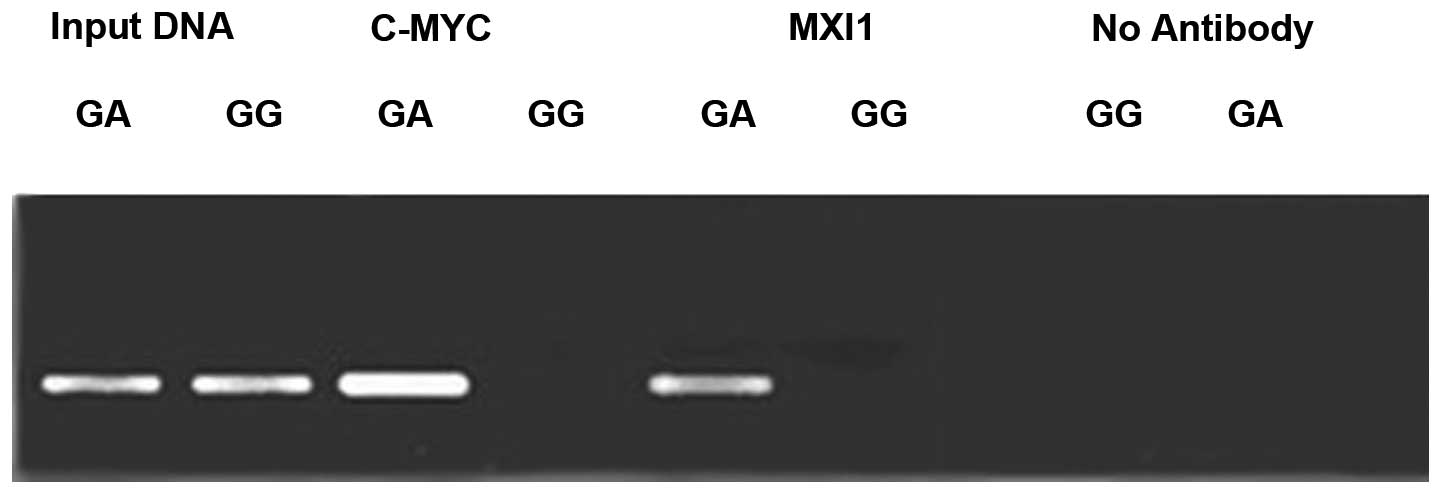
To investigate the affinity of the c-MYC and MXI1
proteins for the ODC G316A allele, ChIP analysis of the +316 bp
region of the ODC promoter was conducted using antibodies against
the aforementioned E-box binding proteins (Fig. 4). The results revealed that protein
bands for the c-MYC and MXI1 proteins were present in the MCF-7
cell lines, but not in the MDA-435 cell lines. These results
indicate that c-MYC and MXI1 proteins may selectively bind to ODC
G316A allele A, rather than the G.
Discussion
Accumulating evidence has indicated that polyamine
catabolism is involved in the response to therapeutic agents,
apoptosis and the stress response, and is important in the
development of psoriasis, parasitic infection and cancer (21–23). In
recent years, a number of anticancer compounds relevant to
polyamine biosynthetic single enzyme inhibitors have been
developed, including the α-difluoromethylornithine and
methylglyoxal bis-guanylhydrazone. However, the application of
these compounds in a clinical setting is limited due to their
poorly-tolerated adverse reactions (24,25).
Ornithine decarboxylase (ODC) is the most significant and
rate-limiting enzyme in the pathway of polyamine synthesis, which
is critical in cell proliferation and highly expressed in a variety
of cancer types, including breast cancer (6,26–28). Previous studies have indicated that
the overexpression of ODC is common in carcinogenesis and cancer
progression (6,26–28).
Therefore, certain studies have proposed that it may be an
important biological marker in the evaluation of biological
behavior and the prognosis of various types of cancer (13,26).
Recently, an increasing number of studies have focused on ODC as a
potential target for cancer therapy (24,25). In
particular, ODC has been proposed to be a promising candidate
target for natural products in cancer chemoprevention (29). Thus, future investigation of ODC
inhibitors present in nature may facilitate the identification of
novel cancer chemopreventive agents. In 2000, Guo et al
(14) identified a single nucleotide
polymorphism (SNP) in the gene regulatory region of ODC, known as
ODC G316A. This SNP is located between two significant gene
regulatory regions, namely the MYC/MAX/MAD protein binding region,
CACGTG E-box, which is known to regulate ODC transcription.
However, only a limited number of studies have focused on the
association between the ODC G316A genotype and breast cancer
survival, as well as the underlying mechanism. In 2003, Martinez
et al (30) reported that the
ODC G316A polymorphism was able to independently reduce the risk of
adenoma recurrence by inhibiting synthesis and activating
catabolism of the tumor cells. Furthermore, in colorectal cancer,
the ODC polymorphism appeared to act as a genetic marker for
predicting colon cancer recurrence. Furthermore, the ODC G316A
genotype was found to be a prognostic factor in colorectal adenoma
recurrence and survival (15,16).
However, thus far, no evidence exists to indicate
that this SNP exhibits the same function in breast cancer as in
colorectal cancer. In 2009, Brown et al (31) reported that the ODC G316A polymorphism
(SNP no. rs2302615) may be less important in individuals with an
inherited predisposition for breast cancer than in individuals who
develop sporadic breast cancer (32).
However, our study involved the ODC G316A polymorphism (SNP no.
rs1045900). Therefore, the present study used nested-PCR-RFLP to
genotype the ODC G316A SNP in breast cancer. Of the 300 cases of
breast cancer investigated, 156 were ODC GG cases and 144 were ODC
GA/AA cases. Additional analysis demonstrated that patients with
the GG phenotype exhibited significantly higher 10-year survival
rates compared with those with the GA/AA phenotype. Furthermore,
according to the results of the present study, patients with an A
allele (ODC GA/AA patients) exhibited significantly lower 10-year
survival rates compared with patients presenting the ODC GG
phenotype, indicating that the ODC G316A SNP may be a useful marker
for predicting the survival of patients with breast cancer. The
results of the present study were in accordance with the report by
Hubner et al (15); however,
the present study was conducted in a Chinese population and used
the simpler and more economical technology of nested-PCR-RFLP to
genotype breast cancer. Overall, the current results indicated that
the ODC G316A genotype is associated with breast cancer
survival.
The ODC A allele has been reported to be associated
with poor survival in colorectal cancer (16). Martinez et al (30) reported that MAD1 (as well as MXI1)
selectively suppressed the activity of the ODC promoter containing
the A allele, as opposed to the G allele, in the HT29 human colon
cancer-derived cell line. Furthermore, Hubner et al
(15) achieved the same results using
ChIP in different human colon cancer-derived cell lines (HCT116 and
HT29). However, due to insufficient evidence, it was unclear
whether the same outcome would occur in human breast cancer-derived
cell lines. A previous study of the current group identified that
c-MYC and MXI1 protein expression levels are associated with the
prognosis of breast cancer patients (32). Furthermore, the present study employed
western blotting and ChIP to examine the association between the
two E-box proteins and the ODC SNP. In cultured cells, the E-box
activator c-MYC and repressor MXI1 were found to preferentially
bind to ODC minor A alleles, rather than major G alleles.
Consistent with the results of the present study, Zell et al
(16) reported that c-MYC, MAD1 and
MAD4 preferred binding to ODC minor A-alleles, rather than major
G-alleles, in vitro.
In conclusion, the present study genotyped ODC G316A
in patients with breast cancer and investigated the clinical
outcome of the ODC G316A SNP on breast cancer-specific mortality.
The current results indicated that MXI1 preferentially binds to the
ODC A-allele, thus, contributing to breast cancer progression.
Furthermore, the findings of the present study indicated that the
G316A SNP may be used to assess the risk of progression in patients
with breast cancer.
Acknowledgements
The present study was supported by the Henan Medical
Science Foundation (grant no. 201203152) and the National Nature
Science Foundation of China (grant no. 81372269). The authors would
like to thank The Affiliated Cancer Hospital of Zhengzhou
University (Zhengzhou, China) and its staff, and Dr. Xibin Sun
(Department of Cancer Prevention, Henan Cancer Hospital, Zhengzhou,
China) for the statistical analysis.
References
|
1
|
DeSantis C, Siegel R, Bandi P and Jemal A:
Breast cancer statistics, 2011. CA Cancer J Clin. 61:409–418. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gerner EW and Meyskens FL Jr: Polyamines
and cancer: old molecules, new understanding. Nat Rev Cancer.
4:781–792. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wallace HM: The physiological role of the
polyamines. Eur J Clin Invest. 30:1–3. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Babbar N, Ignatenko NA, Casero RA Jr and
Gerner EW: Cyclooxygenase-independent induction of apoptosis by
sulindac sulfone is mediated by polyamines in colon cancer. J Biol
Chem. 278:47762–47775. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Takigawa M, Enomoto M, Nishida Y, Pan HO,
Kinoshita A and Suzuki F: Tumor angiogenesis and polyamines:
alpha-difluoromethylornithine, an irreversible inhibitor of
ornithine decarboxylase, inhibits B16 melanoma-induced angiogenesis
in ovo and the proliferation of vascular endothelial cells in
vitro. Cancer Res. 50:4131–4138. 1990.PubMed/NCBI
|
|
6
|
Gödderz D, Schäfer E, Palanimurugan R and
Dohmen RJ: The N-terminal unstructured domain of yeast ODC
functions as a transplantable and replaceable ubiquitin-independent
degron. J Mol Biol. 407:354–367. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen Y, Megosh LC, Gilmour SK, Sawicki JA
and OBrien TG: K6/ODC transgenic mice as a sensitive model for
carcinogen identification. Toxicol Lett. 116:27–35. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Young L, Salomon R, Au W, Allan C, Russell
P and Dong Q: Ornithine decarboxylase (ODC) expression pattern in
human prostate tissues and ODC transgenic mice. J Histochem
Cytochem. 54:223–229. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lan L, Trempus C and Gilmour SK:
Inhibition of ornithine decarboxylase (ODC) decreases tumor
vascularization and reverses spontaneous tumors in ODC/Ras
transgenic mice. Cancer Res. 60:5696–5703. 2000.PubMed/NCBI
|
|
10
|
Guo Y, Cleveland JL and OBrien TG:
Haploinsufficiency for odc modifies mouse skin tumor
susceptibility. Cancer Res. 65:1146–1149. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Grinberg AV, Hu CD and Kerppola TK:
Visualization of Myc/Max/Mad family dimers and the competition for
dimerization in living cells. Mol Cell Biol. 24:4294–4308. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Walhout AJ, Gubbels JM, Bernards R, van
der Vliet PC and Timmers HT: c-Myc/Max heterodimers bind
cooperatively to the E-box sequences located in the first intron of
the rat ornithine decarboxylase (ODC) gene. Nucleic Acids Res.
25:1493–1501. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Funakoshi-Tago M, Sumi K, Kasahara T and
Tago K: Critical roles of Myc-ODC axis in the cellular
transformation induced by myeloproliferative neoplasm-associated
JAK2 V617F mutant. PLoS One. 8:e528442013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guo Y, Harris RB, Rosson D, Boorman D and
OBrien TG: Functional analysis of human ornithine decarboxylase
alleles. Cancer Res. 60:6314–6317. 2000.PubMed/NCBI
|
|
15
|
Hubner RA, Muir KR, Liu JF, Logan RF,
Grainge MJ and Houlston RS: Members of the UKCAP Consortium:
Ornithine decarboxylase G316A genotype is prognostic for colorectal
adenoma recurrence and predicts efficacy of aspirin
chemoprevention. Clin Cancer Res. 14:2303–2309. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zell JA, Ziogas A, Ignatenko N, et al:
Associations of a polymorphism in the ornithine decarboxylase gene
with colorectal cancer survival. Clin Cancer Res. 15:6208–6216.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Theriault RL, Carlson RW, Allred C, et al:
National Comprehensive Cancer Network: Breast cancer, version
3.2013: featured updates to the NCCN guidelines. J Natl Compr Canc
Netw. 11:753–760. 2013.PubMed/NCBI
|
|
18
|
Harris L, Fritsche H, Mennel R, et al:
American Society of Clinical Oncology: American Society of Clinical
Oncology 2007 update of recommendations for the use of tumor
markers in breast cancer. J Clin Oncol. 25:5287–5312. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Singletary SE, Allred C, Ashley P, et al:
Revision of the American Joint Committee on Cancer staging system
for breast cancer. J Clin Oncol. 20:3628–3636. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Leen S, Steven VL, Marie DA, et al: Study
assessing the quality of quantification of estrogen receptor
protein expression by immunohistochemistry and gene expression in
breast cancer. Patholog Res Int. 2014:3726532014.PubMed/NCBI
|
|
21
|
Casero RA and Pegg AE: Polyamine
catabolism and disease. Biochem J. 421:323–338. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Thomas T and Thomas TJ: Polyamine
metabolism and cancer. J Cell Mol Med. 7:113–126. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wallace HM and Fraser AV: Inhibitors of
polyamine metabolism: review article. Amino Acids. 26:353–365.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wallace HM and Fraser AV: Inhibitors of
polyamine metabolism: review article. Amino Acids. 26:353–365.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Levin VA, Uhm JH, Jaeckle KA, et al: Phase
III randomized study of postradiotherapy chemotherapy with
alpha-difluoromethylornithine-procarbazine,
N-(2-chloroethyl)-N-cyclohexyl-N-nitrosurea, vincristine (DFMO-PCV)
versus PCV for glioblastoma multiforme. Clin Cancer Res.
6:3878–3884. 2000.PubMed/NCBI
|
|
26
|
Deng X and Pei D: Ornithine decarboxylase
and glutamate decarboxylase 65 as prognostic markers of gallbladder
malignancy: a clinicopathological study in benign and malignant
lesions of the gallbladder. Mol Med Rep. 7:413–418. 2013.PubMed/NCBI
|
|
27
|
Wilson SM, Hawel L III, Pastorian KE and
Byus CV: A stable, inducible, dose-responsive ODC overexpression
system in human cell lines. Biochim Biophys Acta. 1732:103–110.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Love RR, Astrow SH, Cheeks AM and
Havighurst TC: Ornithine decarboxylase (ODC) as a prognostic factor
in operable breast cancer. Breast Cancer Res Treat. 79:329–334.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Luqman S: Ornithine decarboxylase: a
promising and exploratory candidate target for natural products in
cancer chemoprevention. Asian Pac J Cancer Prev. 13:2425–2427.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Martinez ME, OBrien TG, Fultz KE, et al:
Pronounced reduction in adenoma recurrence associated with aspirin
use and a polymorphism in the ornithine decarboxylase gene. Proc
Natl Acad Sci USA. 100:7859–7864. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Brown I, Halliday S, Greig H, et al:
Genetic polymorphism in ornithine decarboxylase and risk of breast
cancer. Fam Cancer. 8:307–311. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu LP, Sun Y, Li W, Mai L, Guo YJ and Fan
QX: MYC and MXI1 protein expression: potential prognostic
significance in women with breast cancer in China. Oncol Res Treat.
37:118–123. 2014. View Article : Google Scholar : PubMed/NCBI
|