Introduction
Gastric cancer is the second leading cause of
cancer-related mortality worldwide, and the overall survival of
patients with this tumor entity remains poor, as diagnosis
frequently occurs at an advanced stage. (1,2). Despite
the implementation of multimodality treatment strategies for
locally advanced tumors, patients with gastric cancer persistently
exhibit a poor prognosis, possibly due to the continuing lack of
effective prognostic clinicopathological factors and predictive
markers for response assessment in the neoadjuvant setting,
preventing individualization of therapy (3,4).
Therefore, predictive and prognostic markers in the multimodality
therapy of gastric cancer are required.
microRNAs (miRNAs) are a class of endogenous, small
non-coding RNAs that regulate gene expression by inhibiting mRNA
translation (5). Recent studies have
indicated that miRNAs are key in the carcinogenesis of solid
tumors, including gastric cancer (6).
In fact, Ueda et al (7)
assessed the association between intratumoral miRNA expression and
the progression and prognosis in the tissues samples of 353 gastric
cancer patients. It was demonstrated that low expression levels of
let-7 g and miR-433, as well as high expression levels of miR-214,
were significantly associated with reduced overall survival
independent of clinical covariates (7). In addition, by analyzing tumor tissue
samples from 100 gastric cancer patients, Li et al (8) developed a seven-miRNA signature that was
strongly associated with relapse-free and overall survival.
Furthermore, Liu et al (9)
investigated 68 patients with locally advanced gastric cancer
undergoing neoadjuvant chemotherapy followed by surgical resection,
and revealed that low levels of let-7i miRNA were significantly
correlated with local invasion, lymphatic metastasis and a poor
histopathological response. However, thus far, only a small number
of studies have assessed whether miRNAs in the serum could serve as
prognostic or predictive factors in the multimodality treatment of
patients with gastric cancer (10–16). For
example, Song et al (12)
analyzed the expression levels of serum miR-21 in 103 gastric
cancer patients and demonstrated that high levels of miR-21
expression were associated with an increased tumor size and an
advanced pathological tumor stage, but not with patient prognosis
(10).
Therefore, the aim of the present study was to
characterize different miRNA profiles in the serum of three gastric
cancer patients undergoing multimodality therapy based on
histopathological response. In addition, the present study aimed to
validate these miRNA profiles in a larger series of patients,
including patients that had undergone primary resection and
neoadjuvant therapy, to identify possible prognostic and predictive
markers.
Patients and methods
Patients
The present retrospective translational study
consisted of 32 patients with gastric cancer who underwent either
primary surgical resection (n=14) or neoadjuvant therapy followed
by surgical resection (n=18) at the Department of General,
Visceral, Pediatric and Vascular Surgery, University of Heidelberg
(Heidelberg, Germany) or the Department of General, Visceral and
Pediatric Surgery, University of Düsseldorf (Düsseldorf, Germany)
between January 2008 and December 2012. Patients with
histologically confirmed gastric cancer, with no evidence of
distant metastases who had or had not received neoadjuvant therapy
were included in the study. Table I
indicates the demographic and histopathological data of the patient
cohort. Serum samples were used in accordance with the local
policies of the Institutional Review Boards of the University of
Heidelberg and the University of Düsseldorf.
 | Table I.Patient characteristicsa. |
Table I.
Patient characteristicsa.
| Characteristic | n | % |
|---|
| Gender |
|
|
| Male | 18 | 56 |
|
Female | 14 | 44 |
| Neoadjuvant
therapy |
|
|
| No | 14 | 44 |
| Yes | 18 | 56 |
| Type of surgical
resection |
|
|
| Subtotal
gastrectomy | 14 | 44 |
| Total
gastrectomy | 16 | 50 |
|
Transhiatal extended
gastrectomy | 2 | 6 |
| T classification |
|
|
| ypT1 | 5 | 16 |
| ypT2 | 7 | 22 |
|
ypT3/4 | 20 | 62 |
| M classification |
|
|
| M0 | 32 | 100 |
| M1 | 0 | 0 |
| R
classificationb |
|
|
| R0 | 22 | 69 |
| R1 | 8 | 25 |
| Histopathological
response |
|
|
|
Minor | 14 | 44 |
|
Major | 4 | 12 |
Staging
Tumor-node-metastasis staging was performed
according to the criteria of the International Union Against Cancer
(17). Clinical staging consisted of
endoscopy, endoscopic ultrasound in the majority of patients, a
computed tomography (CT) scan of the thorax and abdomen, and
positron emission tomography in a small number of selected cases
that presented ambiguous CT results.
Therapeutic strategy
A total of 14 patients underwent primary surgical
treatment, while 18 patients received neoadjuvant chemotherapy
followed by surgical resection. In the cases treated with
neoadjuvant chemotherapy, the following regimens were typically
administered: Epirubicin (50 mg/m2), oxaliplatin (100
mg/m2) and capecitabine (800 mg/m2) or
5-fluorouracil (2,600 mg/m2), leucovorin (200
mg/m2), oxaliplatin (85 mg/m2) and docetaxel
(50 mg/m2). Restaging was performed 2–3 weeks after the
completion of neoadjuvant therapy. With regard to the surgical
treatment, 14 patients underwent a subtotal gastrectomy, 16
patients underwent a total gastrectomy and 2 patients underwent a
transhiatal extended gastrectomy with D2-lymphadenectomy.
Histopathological grading of tumor
response
An objective histopathological examination was
performed to assess the extent of tumor regression in each case.
The resected specimens were fixed in formalin (10%), embedded in
paraffin, cut into 5-µm slices, and stained with hematoxylin and
eosin (Sigma-Aldrich, Munich, Germany). The prepared sections were
analyzed to determine the effect of neoadjuvant therapy on the
histopathological stage and regression. The number of vital
residual tumor cells in the specimens was determined using a
microscope. Specifically, histopathological regression was
classified as a major histopathological response (i.e., responder)
when <10% vital residual tumor cells remained in the excised
specimen. By contrast, a minor histopathological response (i.e.,
non-responder) was defined as a specimen containing >10% vital
residual tumor cells (18).
Serum samples and study design
Serum samples were collected from the 32 patients
with gastric cancer prior to undergoing primary surgery or directly
after completion of neoadjuvant therapy. Microarray-based analysis
was employed to identify miRNAs that were differentially expressed
between patients defined as histopathological responders and
non-responders. This intratumoral miRNA profiling included a subset
of six patient samples, including three patients exhibiting a major
response and three patients exhibiting a minor response.
Subsequently, the predictive values of the differentially-expressed
miRNAs were assessed using reverse transcription-polymerase chain
reaction (RT-PCR)-based analysis. RT-PCR analysis was performed on
the serum samples of an additional 12 patients undergoing
neoadjuvant therapy followed by surgical resection and all 14
patients undergoing primary surgical treatment. All methods are
described below.
Total RNA isolation from serum
samples
To allow subsequent normalization of extracellular
miRNA levels, Simian virus (SV) 40-miRNA (2 pmol/200 µl; Qiagen,
Hilden, Germany) was added to the serum samples prior to RNA
isolation. RNA was isolated from the serum samples using QIAzol
lysis reagent, according to the manufacturer's instructions
(Qiagen). RNA quantity was determined using a ND-1000 NanoDrop
spectrophotometer at an absorbance of 260 nm (Thermo Fisher
Scientific, Wilmington, DE, USA) and the quality of the RNA samples
was assessed by performing microcapillary electrophoresis (2100
BioAnalyzer; Agilent Technologies, Inc., Waldbronn, Germany).
miRNA profiling using RT-PCR
arrays
Serum RNA samples obtained from major and minor
responders were subjected to miRNA profiling using a TaqMan® human
microRNA array (version 2.0; Applied Biosystems Life Technologies,
Carlsbad, CA, USA). RT of 300 ng total RNA from each sample was
performed using Megaplex™ RT primers, human pools A and B, and a
TaqMan miRNA RT kit (Applied Biosystems Life Technologies) to
obtain complementary (c)DNA. The cDNA samples were loaded onto the
microfluidic cards and miRNA profiling was performed by
quantitative PCR (qPCR) array analysis using TaqMan Universal PCR
Master Mix (Applied Biosystems Life Technologies) and the 7900HT
fast real-time PCR system (Applied Biosystems Life Technologies).
PCR conditions were as follows: Initial enzyme activation step at
95°C for 10 min, followed by 45 cycles at 60°C for 1 min and 95°C
for 20 sec. Sequence Detection System software (version 2.2;
Applied Biosystems Life Technologies) was used to read the
expression signals. Subsequently, these signals were normalized and
interpreted by employing the ΔΔ cycle threshold (Ct) method, as
well as cluster analysis. Ward's method and Manhattan distance
interpretation were performed for this cluster analysis.
miRNA quantification by RT-PCR and
subsequent qPCR
miRNA expression levels were analyzed by performing
two-step qPCR using the miScript II RT kit and the miScript SYBR®
Green PCR kit (Qiagen). Primers for miR-21, miR-29b, miR-221 and
SV-40 were used for cDNA synthesis, and the GeneGlobe search center
(Qiagen) was used to identify appropriate primers for qPCR. PCR
using 2 ng cDNA was carried out in a 20 µl assay under the
following conditions: 45 cycles of 95°C for 15 sec, 59°C for 30 sec
and 45°C for 45 sec. All experiments were performed in triplicate
and in agreement with the manufacturer's instructions. Cellular
miRNA levels were normalized using RNA U6 small nuclear 2 as the
reference RNA. By contrast, spike-in SV40-miRNA (Qiagen) was used
for normalization of extracellular miR-21, miR-29a and miR-221
levels.
Statistical analysis
Data were analyzed using non-parametric tests,
including the Wilcoxon rank sum test for comparisons of paired
data, the Kruskal-Wallis test for comparing greater than two groups
and the Mann-Whitney test for comparing unpaired data. The maximal
χ2 method, which was initially developed by Miller and
Siegmund (19), and Halpern (20), was adapted to identify the miRNA
expression value that resulted in optimal segregation of the cohort
into poor and good prognosis groups (in terms of estimated survival
time). In addition, the log-rank test used to determine the
strength of the groupings. P<0.05 was considered to indicate a
statistically significant difference. Statistical analyses were
performed using SPSS software for Windows (version 18.0; SPSS,
Inc., Chicago, IL, USA). For cluster analysis and ΔΔCt, RealTime
StatMiner® software (Integromics, Granada, Spain) was used.
Results
miRNA profiles in the serum samples of
gastric cancer patients depending on therapeutic response
Blood samples were obtained from 32 patients with
gastric cancer. A total of 18 patients underwent neoadjuvant
therapy followed by surgical resection and 14 patients underwent
primary surgical resection. The blood samples were collected after
neoadjuvant therapy prior to surgical resection or directly prior
to primary surgery. Comprehensive miRNA profiling was performed
using PCR-based microarray analyses from a subset of six
pre-operatively treated patients (three patients exhibiting a major
response and three patients exhibiting a minor response). The
initial analysis assessed the differences between the miRNA
profiles of the study patients and the miRNA profiles of healthy
subjects. A cluster analysis was performed between the study
patients and the healthy control subjects depending on the
therapeutic response (no response versus response; Fig. 1). Furthermore, direct comparisons
between the miRNA profiles of the responder and non-responder
patients revealed that 112 miRNAs exhibited a divergent expression
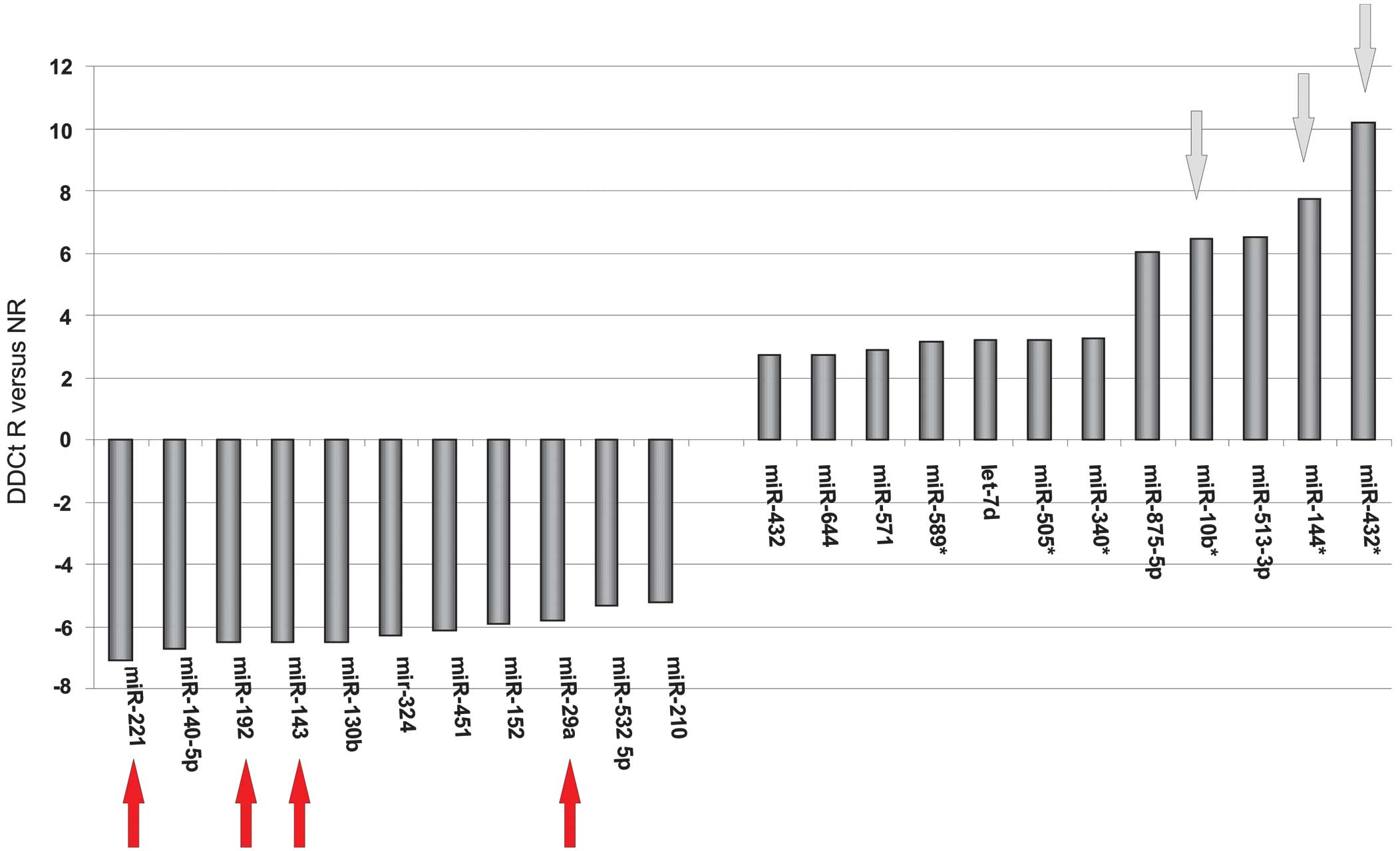
level of >2.5 between the two groups. In particular, miR-144*,
miR-432*, miR-875-5p and miR-10b expression levels were decreased
in the responders compared with the non-responders (Fig. 1).
Based on the microarray results, miR-432*, miR-144*,
miR10b, miR-29a, miR-143, miR-192 and miR-221 exhibited
differential expression between the responders and non-responders
(Fig. 2). Therefore, these miRNAs
were selected for additional validation in all study patients by
single RT-PCR analyses. Quality control demonstrated that miR-432,
miR-144, miR10b, miR-875-5p and miR-192 expression may be measured
in a linear concentration-dependent manner in a limited detection
range, while miR-29a and miR-221 exhibited an adequate detection
range. In addition, analysis of the current literature was used to
select miR-21 for additional validation analyses (13). Therefore, miR-21, miR-29b and miR-221
were selected for subsequent analysis.
miR-21, miR-29b and miR-221 expression
following neoadjuvant therapy, and the association with
histopathological response
miRNA expression levels in serum samples collected
prior to primary surgical resection (n=14) and after neoadjuvant
therapy followed by surgical resection (n=18) were compared. It was
revealed that the neoadjuvant therapy did not significantly affect
the expression of all the investigated miRNA and none of the
selected miRNAs were significantly associated with
histopathological response at any time-point (P>0.05; data not
shown).
miR-21, miR-29b and miR-221 expression
in serum samples of gastric cancer patients depending on
clinicopathological factors
The miRNA expression analyses performed prior to
neoadjuvant treatment as well as prior to direct surgical resection
revealed that none of the three selected miRNAs significantly
correlated with clinicopathological factors or survival (P>0.05;
data not shown). Furthermore, no significant associations were
detected when performing the analyses for all 32 study patients
concurrently.
Discussion
The current translational pilot study aimed to
identify possible prognostic and predictive markers in the
multimodality treatment of gastric cancer. The miRNA profile was
characterized in the serum of patients exhibiting gastric cancer
who had undergone multimodality therapy or primary resection. The
microarray analyses identified that a differential miRNA profile
was expressed depending on the histopathological response of
patients with locally advanced gastric cancer undergoing
multimodality treatment. However, subsequent single RT-PCR analyses
revealed no significant predictive or prognostic impact of the
miRNAs in the two analyzed subgroups.
Methods for assessing prognosis and response are
required to individualize multimodality therapy in gastric cancer,
therefore, various studies have been conducted to characterize
novel effective prognostic/predictive markers in this tumor entity
(21,22). For example, one of the largest
studies, conducted by Ueda et al (7), analyzed tissue samples of 353 gastric
cancer patients and identified that low expression levels of let-7
g and miR-433, as well as a high expression level of miR-214, were
significantly associated with reduced overall survival independent
of clinical covariates. However, the use of these possible markers
is invasive, as an endoscopy is the most minimally invasive way to
obtain an appropriate tissue biopsy for molecular analysis.
Therefore, the identification of a marker in the peripheral blood
(i.e., a non-invasive marker) would be valuable for effective
prognostic and predictive assessment in the multimodality treatment
of patients with gastric cancer.
In fact, recent studies investigating serum samples
of gastric cancer patients revealed that miRNA expression can be
detected in the blood and may have prognostic impact (10–16).
However, to the best of our knowledge, thus far, no data exists
regarding the predictive role of specific miRNAs in the serum of
patients with gastric cancer.
By performing microarray analyses, the present study
identified a differential miRNA expression profile depending on the
histopathological response of patients with locally advanced
gastric cancer undergoing multimodality treatment. However, the
current study failed to identify a significant predictive impact of
these miRNAs in the single RT-PCR analyses. The following miRNAs
were detected as possible predictive factors in the neoadjuvant
treatment of gastric cancer followed by surgical resection:
miR-432*, miR-144*, miR10b, miR-29a, miR-143, miR-192 and miR-221.
Although these miRNAs have not previously been described as
predictive factors in gastric cancer, miR-10b, miR-221, miR-29a and
miR-192 in particular have been detected in other malignant tumors
as possible response predictors (23–28). For
example, Shen et al (23)
demonstrated that the expression status of miR-21 and miR-10b in
patients with non-small lung cancer was associated with disease
progression, survival and response to adjuvant therapy with
gefitinib. In addition, Preis et al (24) indicated that intratumoral miR-10b
expression correlated with the response to neoadjuvant therapy and
survival in patients with pancreatic ductal adenocarcinoma.
Furthermore, evidence indicates that even miR-221 may serve as an
effective factor for response prediction in human cancer. For
example, Zhao et al (25)
detected that plasma miR-221 expression may be a predictive
biomarker for sensitivity to neoadjuvant chemotherapy in patients
with breast cancer. This data is in accordance with a recent review
conducted by Garofalo et al (26) regarding the role of miR-221 and
miR-222 in tumor progression, and the response to antitumor
therapy. The study described the two miRNAs as oncogenes or tumor
suppressors that are dysregulated in different tumor entities and
may serve a prognostic/predictive markers, as well as therapeutic
tools, in cancer. In addition, Nagano et al (27) revealed that miR-29a induces resistance
to gemcitabine in pancreatic cancer cells via the Wnt/β-catenin
signaling pathway. Finally, our previous study recently revealed
that, in patients with locally advanced esophageal cancer
undergoing neoadjuvant chemoradiation followed by esophagectomy,
miR-192 and miR-194 levels in pre-therapeutic biopsies are
considered to be indicators of a major histopathological response
(28).
The present study may have several limitations. For
example, it is a retrospective translational pilot study with a
small number of study patients and therefore has the associated
disadvantages. In addition, the patients in the present study were
a heterogeneous group, as the cohort included patients that had and
had not received neoadjuvant therapy. Other possible limitations of
the present study include the time-point at which the blood samples
were drawn.
In conclusion, the current pilot study did not
identify significant prognostic or predictive value in the selected
miRNAs upon single RT-PCR analysis, however, the microarray results
revealed differential miRNA expression profiles depending on the
extent of histopathological regression. Therefore, prospective
translational studies with a high number of study patients are
required to validate the results of the present study and to
implement miRNAs as predictive factors in the multimodality
treatment of patients with locally advanced gastric cancer.
Acknowledgements
The present study was funded by a grant from the
Deutsche Forschungsgemeinschaft (no. VA506/3-1).
References
|
1
|
Are C, Rajaram S, Are M, et al: A review
of global cancer burden: trends, challenges, strategies and a role
for surgeons. J Surg Oncol. 107:221–6. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cunningham D, Allum WH, Stenning SP, et
al: MAGIC Trial Participants: Perioperative chemotherapy versus
surgery alone for resectable gastroesophageal cancer. N Engl J Med.
355:11–20. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ychou M, Boige V, Pignon JP, et al:
Perioperative chemotherapy compared with surgery alone for
resectable gastroesophageal adenocarcinoma: an FNCLCC and FFCD
multicenter phase III trial. J Clin Oncol. 29:1715–21. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ruan K, Fang X and Ouyang G: MicroRNAs:
novel regulators in the hallmarks of human cancer. Cancer Lett.
285:116–126. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang F, Sun GP, Zou YF, Hao JQ, Zhong F
and Ren WJ: MicroRNAs as promising biomarkers for gastric cancer.
Cancer Biomark. 11:259–267. 2012.PubMed/NCBI
|
|
7
|
Ueda T, Volinia S, Okumura H, et al:
Relation between microRNA expression and progression and prognosis
of gastric cancer: a microRNA expression analysis. Lancet Oncol.
11:136–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li X, Zhang Y, Zhang Y, Ding J, Wu K and
Fan D: Survival prediction of gastric cancer by a seven-microRNA
signature. Gut. 59:579–585. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu K, Qian T, Tang L, Wang J, Yang H and
Ren J: Decreased expression of microRNA let-7i and its association
with chemotherapeutic response in human gastric cancer. World J
Surg Oncol. 10:2252012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu R, Zhang C, Hu Z, et al: A
five-microRNA signature identified from genome-wide serum microRNA
expression profiling serves as a fingerprint for gastric cancer
diagnosis. Eur J Cancer. 47:784–791. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu H, Zhu L, Liu B, et al: Genome-wide
microRNA profiles identify miR-378 as a serum biomarker for early
detection of gastric cancer. Cancer Lett. 316:196–203. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Song MY, Pan KF, Su HJ, et al:
Identification of serum microRNAs as novel non-invasive biomarkers
for early detection of gastric cancer. PLoS One. 7:e336082012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Song J, Bai Z, Zhang J, et al: Serum
microRNA-21 levels are related to tumor size in gastric cancer
patients but cannot predict prognosis. Oncol Lett. 6:1733–1737.
2013.PubMed/NCBI
|
|
14
|
Xu X, Yang X, Xing C, Zhang S and Cao J:
miRNA: The nemesis of gastric cancer (Review). Oncol Lett.
6:631–641. 2013.PubMed/NCBI
|
|
15
|
Tong F, Cao P, Yin Y, Xia S, Lai R and Liu
S: MicroRNAs in gastric cancer: from benchtop to bedside. Dig Dis
Sci. 59:24–30. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu HH, Lin WC and Tsai KW: Advances in
molecular biomarkers for gastric cancer: miRNAs as emerging novel
cancer markers. Expert Rev Mol Med. 16:e12014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sobin LH, Gospodarowicz MK and Wittekind
C: TNM Classification of Malignant Tumours (7th). Wiley-Blackwell.
Oxford: 2009.
|
|
18
|
Becker K, Reim D, Novotny A, et al:
Proposal for a multifactorial prognostic score that accurately
classifies 3 groups of gastric carcinoma patients with different
outcomes after neoadjuvant chemotherapy and surgery. Ann Surg.
256:1002–1007. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Miller R and Siegmund D: Maximally
selected chi-square statistics. Biometrics. 38:1011–1016. 1982.
View Article : Google Scholar
|
|
20
|
Halpern J: Maximally selected chi-square
statistics for small samples. Biometrics. 38:1017–1023. 1982.
View Article : Google Scholar
|
|
21
|
Yasui W, Oue N, Aung PP, Matsumura S,
Shutoh M and Nakayama H: Molecular-pathological prognostic factors
of gastric cancer: a review. Gastric Cancer. 8:86–94. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pietrantonio F, De Braud F, Da Prat V, et
al: A review on biomarkers for prediction of treatment outcome in
gastric cancer. Anticancer Res. 33:1257–1266. 2013.PubMed/NCBI
|
|
23
|
Shen Y, Tang D, Yao R, et al: microRNA
expression profiles associated with survival, disease progression
and response to gefitinib in completely resected non-small-cell
lung cancer with EGFR mutation. Med Oncol. 30:7502013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Preis M, Gardner TB, Gordon SR, et al:
MicroRNA-10b expression correlates with response to neoadjuvant
therapy and survival in pancreatic ductal adenocarcinoma. Clin
Cancer Res. 17:5812–5821. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao R, Wu J, Jia W, et al: Plasma miR-221
as a predictive biomarker for chemoresistance in breast cancer
patients who previously received neoadjuvant chemotherapy.
Onkologie. 34:675–680. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Garofalo M, Quintavalle C, Romano G, Croce
CM and Condorelli G: miR221/222 in cancer: their role in tumor
progression and response to therapy. Curr Mol Med. 12:27–33. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nagano H, Tomimaru Y, Eguchi H, et al:
MicroRNA-29a induces resistance to gemcitabine through the
Wnt/β-catenin signaling pathway in pancreatic cancer cells. Int J
Oncol. 43:1066–1072. 2013.PubMed/NCBI
|
|
28
|
Odenthal M, Bollschweiler E, Grimminger
PP, et al: MicroRNA profiling in locally advanced esophageal cancer
indicates a high potential of miR-192 in prediction of
multimodality therapy response. Int J Cancer. 133:2454–2463. 2013.
View Article : Google Scholar : PubMed/NCBI
|